Abstract
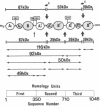
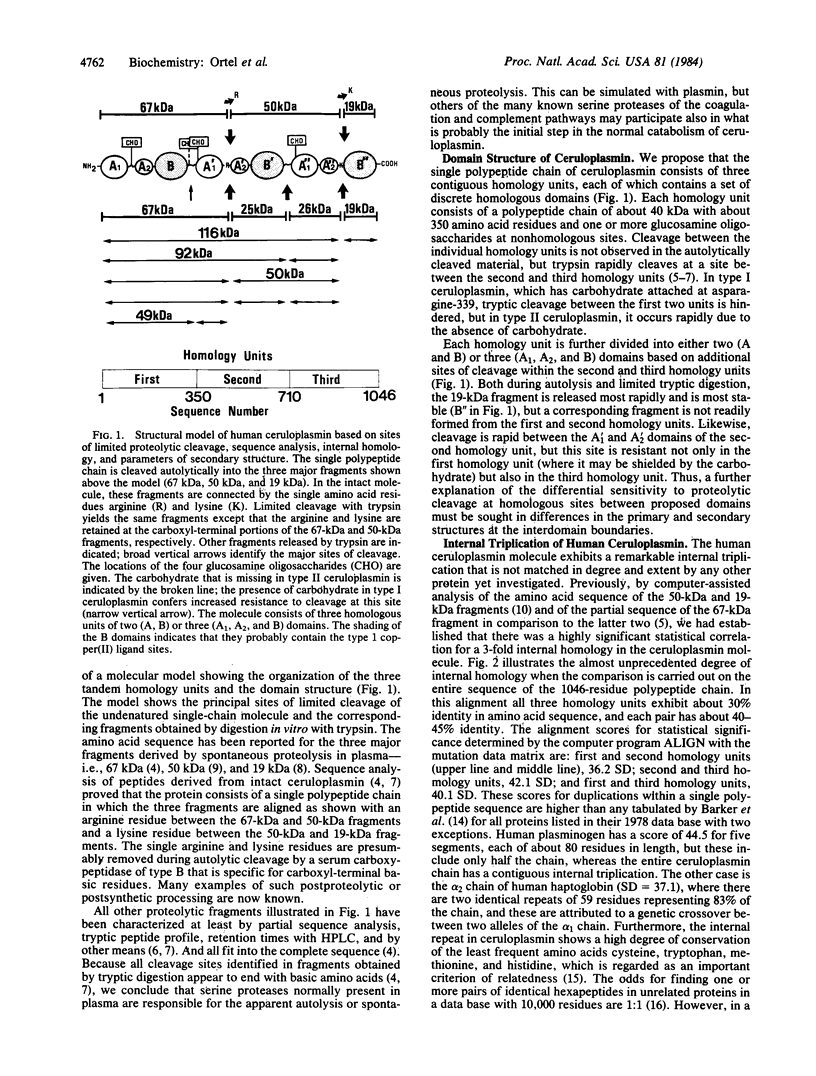
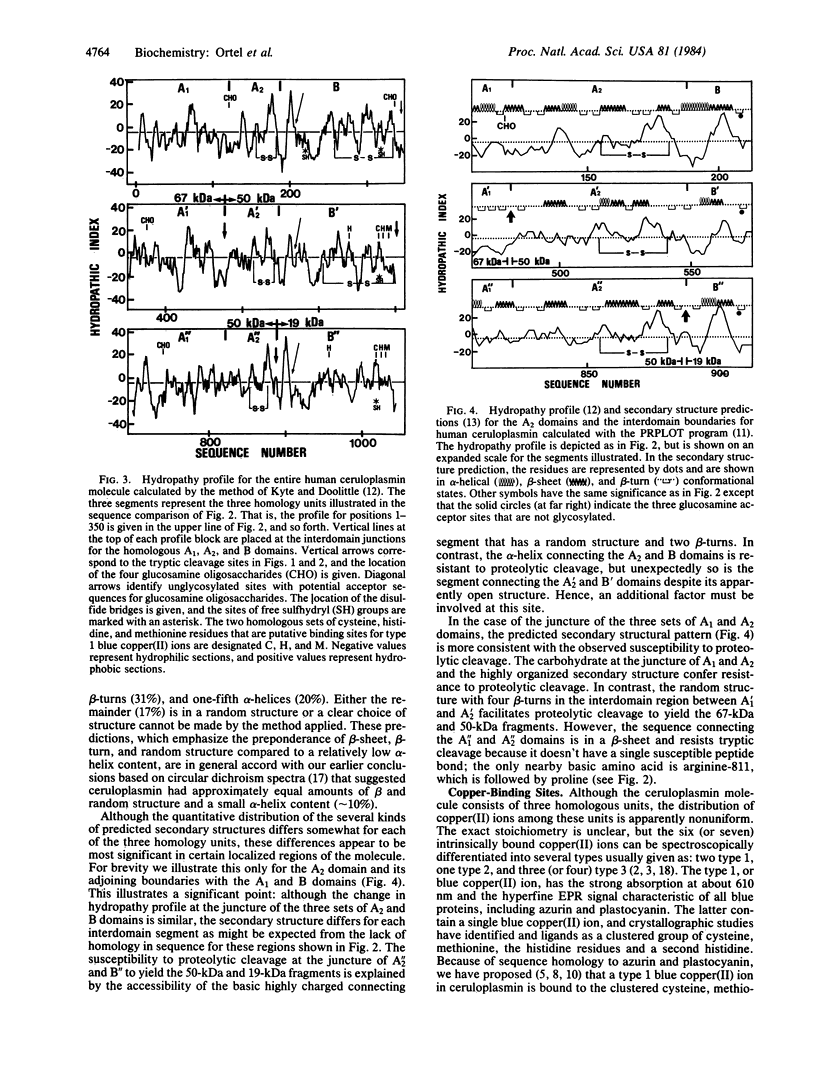
A molecular model for the structure of human ceruloplasmin is proposed that is based on the determination of the complete amino acid sequence, studies of the products of limited proteolytic cleavage, calculations of the hydrophilic/hydrophobic character (hydropathy profile), and predictions of the local secondary structure. This multicopper oxidase (Mr approximately 132,000) consists of a single polypeptide chain (1046 amino acid residues) with four attached glucosamine oligosaccharides. Computer-assisted statistical analysis of the internal repetition in the amino acid sequence confirms that the entire polypeptide chain is divided into three contiguous homology units, each containing about 350 amino acid residues. Each homology unit is subdivided into three domains, designated A1, A2, and B, that differ in structure and probably in function. Calculations of the hydropathy profile and predictions of the secondary structure support a molecular model based on internal repetition of three homology units and help to identify characteristic features of the interdomain junctions. The alignment scores for internal duplication of pairings of the three homology units of ceruloplasmin exceed the scores yet reported for contiguous internal duplication of any other protein. This highly significant evidence for intragenic repetition suggests that the ceruloplasmin molecule evolved by tandem triplication of ancestral genes coding for a primordial copper oxidase.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F. Similar amino acid sequences: chance or common ancestry? Science. 1981 Oct 9;214(4517):149–159. doi: 10.1126/science.7280687. [DOI] [PubMed] [Google Scholar]
- Dwulet F. E., Putnam F. W. Complete amino acid sequence of a 50,000-dalton fragment of human ceruloplasmin. Proc Natl Acad Sci U S A. 1981 Feb;78(2):790–794. doi: 10.1073/pnas.78.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwulet F. E., Putnam F. W. Internal duplication and evolution of human ceruloplasmin. Proc Natl Acad Sci U S A. 1981 May;78(5):2805–2809. doi: 10.1073/pnas.78.5.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieden E. Caeruloplasmin: a multi-functional metalloprotein of vertebrate plasma. Ciba Found Symp. 1980;79:93–124. doi: 10.1002/9780470720622.ch6. [DOI] [PubMed] [Google Scholar]
- Hervé M., Garnier A., Tosi L., Steinbuch M. Spectroscopic and photoreduction studies of copper chromophores in ceruloplasmin. Eur J Biochem. 1981 May;116(1):177–183. doi: 10.1111/j.1432-1033.1981.tb05316.x. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. On the use of sequence homologies to predict protein structure: identical pentapeptides can have completely different conformations. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1075–1078. doi: 10.1073/pnas.81.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston I. B., Kingston B. L., Putnam F. W. Primary structure of a histidine-rich proteolytic fragment of human ceruloplasmin. II. Amino acid sequence of the tryptic peptides. J Biol Chem. 1980 Apr 10;255(7):2886–2896. [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- MacGillivray R. T., Mendez E., Shewale J. G., Sinha S. K., Lineback-Zins J., Brew K. The primary structure of human serum transferrin. The structures of seven cyanogen bromide fragments and the assembly of the complete structure. J Biol Chem. 1983 Mar 25;258(6):3543–3553. [PubMed] [Google Scholar]
- Noyer M., Putnam F. W. A circular dichroism study of undegraded human ceruloplasmin. Biochemistry. 1981 Jun 9;20(12):3536–3542. doi: 10.1021/bi00515a036. [DOI] [PubMed] [Google Scholar]
- Ortel T. L., Takahashi N., Putnam F. W. Separation of limited tryptic fragments of human ceruloplasmin by gel-permeation high-performance liquid chromatography. J Chromatogr. 1983 Aug 26;266:257–263. doi: 10.1016/s0021-9673(01)90899-4. [DOI] [PubMed] [Google Scholar]
- Takahashi N., Bauman R. A., Ortel T. L., Dwulet F. E., Wang C. C., Putnam F. W. Internal triplication in the structure of human ceruloplasmin. Proc Natl Acad Sci U S A. 1983 Jan;80(1):115–119. doi: 10.1073/pnas.80.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Ortel T. L., Putnam F. W. Single-chain structure of human ceruloplasmin: the complete amino acid sequence of the whole molecule. Proc Natl Acad Sci U S A. 1984 Jan;81(2):390–394. doi: 10.1073/pnas.81.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitkamp L. R. Evidence for linkage between the loci for transferrin and ceruloplasmin in man. Ann Hum Genet. 1983 Oct;47(Pt 4):293–297. doi: 10.1111/j.1469-1809.1983.tb00999.x. [DOI] [PubMed] [Google Scholar]