Abstract
Cultures across the globe, especially Western societies, are burdened by chronic diseases such as obesity, metabolic syndrome, cardiovascular disease, and cancer. Several factors, including diet, genetics, and sedentary lifestyle, are suspected culprits to the development and progression of these health maladies. Fatty acids are primary constituents of cellular physiology. Humans can acquire fatty acids by de novo synthesis from carbohydrate or protein sources or by dietary consumption. Importantly, regulation of their metabolism is critical to sustain balanced homeostasis, and perturbations of such can lead to the development of disease. Here, we review de novo and dietary fatty acid metabolism and highlight recent advances in our understanding of the relationship between dietary influences and genetic variation in fatty acid metabolism and their role in chronic diseases.
Keywords: De novo lipogenesis, Dietary fatty acids, Genetic variation, Fatty acid metabolism
Introduction
The epidemic spread of obesity and metabolic syndrome is raising public health concerns across the globe. The factors that distinguish these diseases are also known contributors to cardiovascular disease and cancer, the two most common causes of death in the United States (Hoyert and Xu, 2012). Epidemiologic studies have shown an increased prevalence of these diseases in immigrating populations to Western societies (Nasseri and Moulton, 2011; Seeff and McKenna, 2003). One of the major hypotheses to explain this observation is the abrupt and dramatic change in diet upon emigration, particularly changes in dietary fatty acid consumption. A prevailing body of literature suggests not only the quantity, but more importantly, the quality, of dietary fat consumption modulates disease (Berquin et al., 2011; Berquin et al., 2007; Suburu and Chen, 2012). Specifically, dietary omega-6 polyunsaturated fatty acid (PUFA) can be pro-inflammatory mediators, while omega-3 PUFA acts as anti-inflammatory mediators (Serhan and Petasis, 2011). As such, tipping the balance of an immigrant's dietary consumption can lead to a chronic state of inflammation and promote the development and progression of cardiovascular disease and cancer.
New evidence suggests various populations may differentially metabolize dietary fatty acids (Teslovich et al., 2010), implying that the risk for chronic inflammation, metabolic syndrome, and cardiovascular disease may be inheritable. Even the progression of cancer, which is well known to be a disease of genetic alterations, can be exacerbated by pro-inflammatory lipid metabolism (Wang and Dubois, 2010). Discrepancies observed in the incidence of cardiovascular disease and cancer in various ethnic populations have been attributed to socioeconomic variance (Marmot et al., 2012). However, more recent data suggests genetic diversity in the form of single nucleotide polymorphisms (SNPs) found in several lipid metabolism genes may be a major contributing factor to these epidemiological health disparities (Illig et al., 2010; Kathiresan et al., 2009; Teslovich et al., 2010). Despite the potential inheritance of genetic risk factors for cardiovascular disease and cancer progression, advancements in the new field of metabolomics is yielding promising work to identify metabolic biomarkers of disease that may fuel diagnostic testing and/or predict health outcome. The purpose of this article is to review de novo and dietary fatty acid metabolism and highlight the most recent findings in genetic variation found in fatty acid metabolism genes as they relate to dietary fat consumption and various diseases.
Fatty Acids: a biological necessity
Fatty acids are fundamental molecules of cellular biology. Composed of hydrogenated carbons with a carboxyl moiety at the alpha carbon, mammalian fatty acids are divided into three major groups based on the quantity of double bonds found within the carbon chain: saturated fatty acids (SAFA), monounsaturated fatty acids (MUFA), and polyunsaturated fatty acids (PUFA). SAFA are fully hydrogenated, containing zero double bonds. They are a primary constituent of glycerolipids as well as phospholipids and sphingolipids found in cellular membrane structures. SAFA may also act as post-translational modifiers, thereby dictating the activity and location of cellular signaling proteins. MUFA contain a single double bond, most commonly found between the 9th and 10th carbons from the alpha carbon, and occasionally between the 7th and 8th carbons. MUFA are also primary constituents of cellular membrane structures and glycerolipids. PUFA have more than one, and as many as six, double bonds in their carbon chain. PUFA are found in phospholipids of membrane structures, and may also act as precursors to a variety of lipid signaling molecules. The most unique characteristic of PUFA metabolism is the inability for mammals to synthesize them de novo. While mammals express the necessary enzymes to convert carbohydrate and protein-derived carbons into SAFA and MUFA, they lack the desaturase enzymes required for producing the limiting substrate for PUFA synthesis. Therefore, PUFA are considered essential fatty acids that must be acquired from the diet. It is important to note that not only the degree of saturation, but also the carbon chain length can differentiate fatty acids and their biological roles. Within each class of fatty acids the carbon chain length may vary greatly, with as few as 12 and as many as 30 carbons. The chain length of fatty acids in cellular membrane can modify the membrane properties, such as fluidity and formation of microdomains and signaling platforms, ultimately altering susceptibility to cell death or survival (Iwabuchi et al., 2010; Sassa et al., 2012).
Unlike PUFA, SAFA and MUFA may be acquired from the diet or produced de novo. Despite identical biochemical structures, it remains unclear whether dietary and de novo fatty acids are equivalent or two separate pools of fatty acids used for distinct biological functions in the body. A number of features distinguishing de novo and dietary fatty acid metabolism directed questions of why dietary SAFA and MUFA are likely to fall short in compensating for impaired de novo fatty acid synthesis. First, the major biological function of fatty acid synthesis is to store energy from carbohydrate-derived carbon precursors as compact fatty acids. This process occurs in the cytosol of cells and is performed by a series of enzymes beginning with the production of acetyl-CoA by ATP citrate lyase (ACLY). Acetyl-CoA is then metabolized by the rate limiting enzyme of the fatty acid synthesis pathway, acetyl-CoA carboxylase 1 (ACACA) to produce the limiting reagent, malonyl-CoA. The multifunctional enzyme, fatty acid synthase (FASN) then produces saturated, short (14:0) to medium (18:0) chain fatty acids by sequentially adding malonyl-CoA to the growing acyl chain through a series of biochemical reactions, with palmitate (16:0) representing about 80-90% of its total product (Jayakumar et al., 1995; Kuhajda et al., 1994). FASN-derived SAFA can be further modified by various elongation and desaturase enzymes. These de novo synthesized fatty acids can then be esterified and converted into triglyceride molecules for storage.
Expression of FASN is observed throughout the body, with the most prominent expression in the liver, brain, and abdominal adipose, where energy storages are important for cell survival during periods of physiological or pathological stress. In contrast, other tissues maintain a relatively low rate of fatty acid synthesis, and circulating fatty acids from the diet were believed to suffice as a sustainable source of fat for these tissue types (Jayakumar 1995 and Semenkovich 1995). However, more recent evidence suggests fatty acid synthesis plays a critical role in the function and survival of non-energy-storing tissues as well. Acylation of proteins regulates their activation, location, and propensity for receptor binding (Miura and Treisman, 2006); in particular, palmitoylation of Wnt was shown to be dependent on de novo fatty acid synthesis. Wnt palmitoylation facilitates its receptor binding (Janda et al., 2012) and thereby regulates its ability to activate downstream signaling proteins (Fiorentino et al., 2008). Studies of conditional, tissue-specific knockout of FASN in mice have shown that de novo fatty acids are constituents of endogenous lipid ligands (Chakravarthy et al., 2009a) and are required for PPARα activation in the liver (Chakravarthy et al., 2005), macrophage (Schneider et al., 2010), and hypothalamus (Chakravarthy et al., 2007). Additionally, conditional knockout of FASN in the heart of mice demonstrated that fatty acid synthesis regulates calcium signaling and adaptation to stress in the myocardium (Razani et al). Whole body knockout of FASN is embryonic lethal, as fertilized blastocysts fail to implant in the uterus (Chirala et al., 2003), suggesting fatty acid synthesis is critical for development and survival of rapidly proliferating tissues. Indeed, FASN is also critical for the proliferation and survival of most cancers (Menendez and Lupu, 2007), which utilize de novo fatty acids to facilitate membrane biogenesis (Swinnen et al., 2003) and prevent reactive oxygen species-induced apoptotic cell death by lipid peroxidation (Rysman et al., 2010). Whether FASN is most critical for its fatty acid products or possibly acts as a signaling protein itself is still unclear. Most in vitro studies can rescue the phenotype of FASN knockdown with addition of exogenous palmitate (Kridel et al., 2004; Migita et al., 2009; Rysman et al., 2010), while some cannot (Razani et al., 2011). Additionally, while dietary fat consumption can regulate de novo lipogenesis (Ringseis and Eder, 2011), no animal study has shown a phenotypic rescue of FASN knockout by dietary fat. This may possibly be attributed to the physiological differences between de novo and dietary fat metabolism.
Unlike de novo fatty acid metabolism where fatty acids can be immediately used inside the cell, dietary fatty acids require the presence of enzymes, transporters, and chaperone proteins to facilitate their absorption, transport, and uptake by cells in the body. Fatty acids absorbed from the gut are loaded onto lipoprotein particles in the form of triglycerides and enter circulation through the lymphatic system. Lipoprotein particles then bind to their respective receptors for cellular uptake. Release of the fatty acids from the lipoprotein particles requires the activity of lipoprotein lipase (LPL) for fatty acid storage or energetic use in peripheral tissues, chiefly liver, muscle, adipose, and cardiac tissue; therefore, the use of dietary fatty acids by peripheral tissues is largely dependent upon the expression and activity of LPL (Mead et al., 2002; Merkel et al., 2002).
Following their release by lipase enzymes, free fatty acids can be modified by elongation or desaturation. The metabolism of de novo and dietary fatty acids can vary greatly; however, they may also compete as substrates for the same elongation and desaturation enzymes. Mammals possess seven known elongase enzymes (ELOVL1-7) with various substrate specificities that mediate the elongation of fatty acids through the addition of malonyl-CoA (Guillou et al., 2010). Recall that malonyl-CoA is produced by ACACA, the rate limiting enzyme of the de novo fatty acid synthesis pathway; hence, the activity of ACACA may contribute to both de novo and dietary fatty acid metabolism. Introduction of double bonds into fatty acids is mediated by the activity of the desaturase enzymes, SCD-1 (stearoly-CoA desaturase 1), FADS1 (fatty acid desaturase 1), and FADS2 (fatty acid desaturase 2), each of which insert a double bond at specific locations in the fatty acid carbon chain. Humans also possess a third FADS3 (fatty acid desaturase 3) enzyme; however, its function remains elusive. SCD-1 (also known as, delta 9 desaturase) is specific for the conversion of SAFA to MUFA, while FADS1 (delta 6 desaturase) and FADS2 (delta 5 desaturase) are specific to PUFA (Guillou et al., 2010); hence, SCD1 can metabolize both de novo and dietary fatty acids, while FADS1 and FADS2 are specific to dietary fatty acids. Enzymes known to be strictly responsible for de novo fatty acid metabolism include FASN, ELOVL1, ELOVL3, and ELOVL6, while enzymes that metabolize only dietary fatty acids include ELOVL2, ELOVL5, FADS1, and FADS2. Several enzymes participate in the metabolism of both de novo and dietary fatty acids, including ACLY, ACACA, SCD1, ELOVL4, and ELOVL7. Variation in any of these enzymes may lead to altered fatty acid metabolism and can modulate the propensity for the development and progression of various diseases (Guillou et al., 2010; Kihara, 2012).
Dietary Fat Metabolism Studies – The critical need for standardized methods
An exceptionally large and continuously growing body of literature has investigated the role of dietary fatty acids in disease progression, but conflicting results have lead to disparate conclusions, and for many, an elusive uncertainty of how dietary fatty acids modulate disease. Although the enzymatic pathways of dietary fatty acid metabolism are fairly well understood, new knowledge of the role of dietary fat in disease progression is hindered by several complexities in the design and methodology of research studies, consequently making it extremely difficult to identify causal roles for specific dietary fatty acids in disease. A major confounding factor throughout epidemiological research is the study design. There are two main approaches to investigating dietary and genetic influence on disease. The first approach distinguishes a population with common ailments and identifies common genetic variants and dietary habits that possibly modulate their phenotype. This approach is very common, but may unfortunately lead to biased results based on the concept of searching for significant data. An alternative to this method is the second approach, whereby the study distinguishes a population with specific genetic variants and identifies how specific dietary interventions modulate their phenotype (Perez-Martinez et al., 2012). This intervention-based approach is more controlled and can successfully address the direct influence of uniquely formulated diets (Shaw et al., 2009). It also focuses on the interaction between genetic signatures and metabolism, a topic we will return to later in this review.
Another concern for studies seeking to investigate dietary interactions with disease is that dietary fatty acids are never consumed in isolation. Whether researching humans or animal models, the complexity of fatty acid composition in foods makes it very difficult to attribute a phenotype to any given fatty acid. Certain oils and foods may be enriched with particular fatty acids. For example, corn oil is enriched in linoleic omega-6 PUFA and olive oil is enriched in oleic MUFA (Beare-Rogers et al., 2001), but the biochemical make up of diets used in most studies is largely undisclosed, and as discussed previously, not all fatty acids are created equal. Some studies focus on a particular group or class of fatty acids and measure total consumption for correlating its role in disease. Unfortunately, such studies often ignore potential confounding factors, such as fatty acid ratios and the counterbalance of other fatty acids and/or dietary components used to make diets isocaloric, which may affect results. For example, the common use of corn oil as a common constituent of a high fat diet complicates conclusions due to the extensive research that omega-6 PUFA are pro-inflammatory and cancer promoting compared to other types of fatty acid (Greene et al., 2011; Wang and Dubois, 2010). A recent study (Baxheinrich et al., 2012) eloquently designed a dietary intervention study that accounted for both quantity and quality of dietary fatty acids consumed by patients with metabolic syndrome to determine the effects of alpha-linolenic acid (ALA) metabolic syndrome and cardiovascular disease risk factors. A health log was first recorded to identify the patients' habits. Patients were then educated and provided with detailed instructions on how to adhere to the prescribed diet, including kitchen scales, recipes, and the intervention oils. The diets of each study arm were carefully calculated to be isocaloric by measuring the same percentage of fat (38%), carbohydrate (42%), and protein (20%). Most importantly, the biochemical make up of the oils was carefully considered, such that the increased quantity of omega-6 PUFA in the high ALA rapeseed arm of the study was compensated for by the inclusion of sunflower oil in the olive oil study arm. Overall, the study concluded that energy restriction, as compared to previous dietary habits, was the largest contributor to changes in diagnostic factors for metabolic syndrome. However, because the study was adequately controlled for both quantitative and qualitative measures, ALA compared to MUFA was more effective in decreasing plasma triacylglyceride concentrations (Baxheinrich et al., 2012).
The time frame in which studies are conducted is another major contributor to controversial results of dietary fat metabolism studies. While some studies may follow subjects for just a few weeks, other studies collect data for several months or even years. A study of longer duration may allow for systemic adaptations to dietary influence and be more representative of dietary impact on disease. However, one must also consider the stringency of subject compliance and accuracy of dietary consumption from questionnaire-derived data for long term studies in humans. Acute dietary intervention may lead to transient changes in liver metabolism, circulating fatty acids, and the composition of red blood cells, but may have little effect on other organs, such as adipose, heart, and other peripheral tissues, all of which may be more relevant to the development and progression of certain chronic diseases. In contrast, chronic exposure to a diet may be more representative of lifestyle and systemic adaptation to diet and more accurately depict how these changes affect disease development and progression. Chronic exposure may even influence epigenetic changes affecting the development and progression of disease (Armitage et al., 2005).
Finally, variance in sample size and, especially, population can lead seemingly similar studies to completely different results, or conversely, repeated investigations of the same population may mislead common knowledge. Increased incidence of cancer and cardiovascular events in certain racial and ethnic populations is a common trend (Casper et al., 2003; Siegel et al., 2011) . Although many studies statistically adjust results for various criteria, it is nearly impossible to adjust for all variables. The population may, in fact, be the most significant variable among dietary research studies since populations are subject to infinite differences of socioeconomic, lifestyle, environmental, cultural, and genetic diversity around the globe. While socioeconomic factors are also becoming more appreciated as critical factors of disease disparity (Devi, 2012), the influence of population genetics and epigenetics among epidemiologic studies has gained particular interest. It is becoming increasingly clear that genetic regulation of lipid metabolism can largely influence various diseases including cardiovascular disease, obesity, metabolic syndrome, and cancer (Illig et al., 2010; Suhre et al., 2011; Teslovich et al., 2010). Therefore, the use of genetic profiling for population studies could help target a population for dietary fat intervention studies by providing a predictive value to the research.
Genetic Variation Modifies Fatty Acid Metabolism
In an era of genomic profiling, whereby genes are well known contributors to disease, our increasing knowledge of the enzymatic pathways in de novo and dietary fatty acid metabolism has fostered research identifying interactions between genetic and dietary modulators of disease. The recent coming of genome-wide association studies (GWAS) has lead to the identification of several new lipid metabolism genes (Teslovich et al., 2010), although it does require extremely laborious and low-throughput functional studies to validate GWAS findings (Burkhardt et al., 2010; Musunuru et al., 2010). Even so, the development of transgenic knockout mouse models for ACLY (Beigneux et al., 2004), ACACA (Abu-Elheiga et al., 2005), FASN (Chirala et al., 2003), SCD-1 (Miyazaki et al., 2001), SCD-2 (Miyazaki et al., 2005), FADS2 (Stoffel et al., 2008; Stroud et al., 2009), ELOVL3 (Westerberg et al., 2004), ELOVL4 (Cameron et al., 2007; Harkewicz et al., 2012; Li et al., 2007), ELOVL5 (Moon et al., 2009), and ELOVL6 (Matsuzaka et al., 2007) have been instrumental in identifying the role of these enzymes in both systemic and localized lipid metabolism and homeostasis. With the continued development of spatially restricted knockout models, we can expect new studies to elucidate the tissue-specific roles of these enzymes. Nevertheless, still enthralling is the continuous discovery of genetic variants in several of these genes found in human populations, many of which have been suggested to modulate disease phenotypes. Specifically, genetic variation has been identified in FASN, SCD-1, ELOVL2, ELOVL5, ELOVL6, LPL, and most notably FADS1 and FADS2 (Garcia-Rios et al., 2011; Illig et al., 2010; Merino et al., 2010; Morcillo et al., 2011; Nguyen et al., 2010). A number of GWAS and other studies have shown that single nucleotide polymorphisms (SNPs) found within lipid metabolism genes can regulate enzyme expression and activity (Corella and Ordovas, 2012; Kihara, 2012; Merino et al., 2010), thereby affecting the efficiency of lipid metabolism. Similar studies have even been conducted in cattle as a means to predict and improve the quality, scored in part by fatty marbling, of meat consumed by humans (Li et al., 2012; Mannen, 2011; Oh et al., 2012). Here, we describe the most recent findings of genetic polymorphisms found within lipid metabolism genes in humans influencing the development and progression of lipid associated diseases, including cardiovascular disease, metabolic syndrome, and cancer.
Genetic variation in de novo fatty acid metabolism enzymes
While enzymes involved in de novo fatty acid metabolism have been implicated in metabolic disease (Chakravarthy et al., 2005), cardiovascular disease (Schneider et al., 2010), and cancer (Menendez and Lupu, 2007) through the use of transgenic mice, very few studies have investigated the putative role of human genetic variation in these genes and susceptibility to disease development and progression. A recent study used data from the European Prospective Investigation into Cancer (EPIC) cohort to investigate genetic variation in lipid metabolism genes and prostate cancer risk. They screened a total of 921 SNPs across 22 different genes involved in fatty acid synthesis, including ACACA, FASN, and SREBF1, the transcriptional regulator of fatty acid synthesis, and found no significant associations with prostate cancer risk (Campa et al., 2011). Despite this singular study, other groups have found significant associations between SNPs within FASN, ELOVL6, and SCD-1 and characteristics of cardiovascular diseases, metabolic syndrome, and cancer (Merino et al., 2010; Morcillo et al., 2011; Moreno-Navarrete et al., 2009; Nguyen et al., 2010).
FASN is a critical mediator of atherosclerosis (Schneider et al., 2010), obesity and inflammation (Chakravarthy et al., 2009b), and cancer (Kuhajda et al., 1994; Menendez and Lupu, 2007); however, disease susceptibility due to genetic variation in FASN is still a nascent topic. Only 3 recent studies pertain to genetic variation in the FASN gene and disease risk. A study published by Campa et. al. (Campa et al., 2009) investigated 20 SNPs in the ChREBP, SREBP-1, and FASN genes of breast cancer and control patients from a Western European population. ChREBP and SREBP-1 are transcription factors known to directly regulate enzymes of the fatty acid synthesis pathway, including FASN (Eberle et al., 2004). They found no significant associations between any of the 20 SNPs and overall breast cancer risk by age or menopausal status. However, 2 SNPs in the FASN gene, rs1140616 and FAS_A0866, were found to be significantly associated with a moderately higher BMI in younger/pre-menopausal women and a marginally lower BMI in older/post-menopausal women, respectively (Campa et al., 2009). Another group (Schleinitz et al., 2010) identified 35 SNPs from sequencing the FASN gene in a German population of adults and children. A total of 4 SNPs were found to be significantly associated with obesity, but the strongest effect was observed for rs2229422, which showed significant association with BMI, waist-to-hip ratio, plasma insulin, and glucose infusion. Most striking from this study was the reaffirmation from 3 previous studies (Korner et al., 2007; Kovacs et al., 2004; Moreno-Navarrete et al., 2009) that a valine to isoleucene amino acid substitution at residue 1483 in FASN, a consequence of the minor allele SNP rs2228305, provides gender-specific protection from obesity in males (Schleinitz et al., 2010). Finally, a study from the Dana-Farber Cancer Institute (Nguyen et al., 2010) identified 4 SNPs in the FASN gene that were significantly associated with prostate cancer in white men of the Unites States. The variant alleles of SNPs rs8066956 and rs6502051 were directly associated with a decreased risk of advanced prostate cancer, and rs8066956 was associated with decreased expression of FASN in prostate tumors (Nguyen et al., 2010), which is a well described predictor of prostate cancer prognosis (Menendez and Lupu, 2007; Shurbaji et al., 1996). Additionally, the variant allele of SNP rs1127678 was associated with increased risk of advanced prostate cancer when stratified for overweight patients bearing a body mass index equal to or greater than 25, and SNP rs4246444 was associated with prostate cancer mortality (Nguyen et al., 2010).
Studies of genetic variation among the fatty acid elongase enzymes are quite sparse. In fact, with the exception of ELOVL2, which metabolizes PUFA, only ELOVL6 has been described to associate with disease on account of its genetic variation. A prospective study showed that minor allele carriers of the rs9997926 and rs6824447 SNPs in ELOVL6 from a Spanish population were at a significantly (p≤0.01) lower risk of being insulin resistant, while minor allele carriers of the rs17041272 SNP displayed an increased risk of insulin resistance. Interestingly, when adjusted for consumption of dietary oils, insulin resistance measurements significantly dropped in carriers of the minor allele for SNP rs6824447 who consumed sunflower oil as opposed to the same minor allele carriers who consumed olive oil. These results suggest the effects of ELOVL6 genetic variation on insulin resistance can be modified by dietary fat consumption (Morcillo et al., 2011). The only other study investigating genetic polymorphisms of non-PUFA metabolizing elongase enzymes described no association between ELOVL4 SNPs and adipose fatty acids, LDL cholesterol, or myocardial infarction (Aslibekyan et al., 2012). More studies are required to determine the potential role of genetic variation in the SAFA and MUFA metabolizing elongase enzymes.
The role of SCD-1 an important fatty acid desaturase enzyme in the regulation of lipid metabolism as it relates to obesity, diabetes, metabolic syndrome, and cancer, has been well characterized (Cohen and Friedman, 2004; Cohen et al., 2003; Guillou et al., 2010; MacDonald et al., 2008). Unlike other de novo lipogenesis enzymes, recent reviews have described numerous studies of SCD-1 variants and dietary interactions (Bjermo and Riserus, 2010; Merino et al., 2010), as the enzyme is known to be regulated by dietary consumption (Cohen and Friedman, 2004) . However, the latest studies (Gong et al., 2011; Stryjecki et al., 2012) have confirmed previous reports of gender specific variation in SCD-1 as a link to metabolic syndrome and inflammation in women. Two common haplotypes across 7 tagSNPs showed a significant (P<0.01) association with metabolic syndrome in Costa Rican women. The study observed a mildly significant association between rs1502593 and systolic blood pressure (p=0.05).Other metabolic characteristics, including fasting blood glucose, waist circumference, and the desaturation index of palmitate showed a similar trend (Gong et al., 2011). Additionally, the rs2060792 SNP of SCD-1 was significantly associated (P<0.05) with plasma palmitate, stearate, and C reactive protein levels in European women (Stryjecki et al., 2012). Overall these studies suggest SCD-1 polymorphisms may play a role in the development of inflammation, insulin resistance, and obesity.
Finally, a few studies have identified associations between characteristics of metabolic syndrome and genetic variants in the ACACB gene (Ma et al., 2011; Phillips et al., 2010; Riancho et al., 2011), the mitochondrial counterpart to ACACA involved in lipid oxidation; however, no study to date has shown similar findings for the ACACA gene in humans. One group identified an interaction between ACACA and the breast cancer susceptibility gene 1, BRCA1 (Magnard et al., 2002), and therefore, examined the human ACACA gene for sequence variation among breast cancer patients. Unfortunately, no single SNP was found to be associated with breast cancer risk.; Nonetheless, low frequency genotypes of haplotype tagging SNPs (htSNPs) across four different SNPs in breast cancer patients suggested a protective effect against breast cancer, and vice versa for the high frequency haplotype. No SNPs in this study were identified in the coding region of ACACA, suggesting functional conservation of the gene (Sinilnikova et al., 2004).A later study by the same group showed that BRCA1 directly regulates lipogenesis by binding to and preventing dephosphorylation of phospho-ACACASer79, an inhibitory phosphorylation, which proposes a tumor suppressor role for BRCA1 (Moreau et al., 2006). Hence, genetic variation in the ACACA gene itself has yet to be identified, but other known genetic breast cancer risk factors, such as mutated BRCA1, may affect lipogenesis by regulating ACACA functionality and activity.
Genetic variation in dietary fatty acid metabolism enzymes
In stark contrast to enzymes responsible for de novo lipogenesis, the fatty acid desaturase enzymes, involved in dietary PUFA metabolism, have been extensively researched and reviewed for genetic variations and their association with disease, including dyslipidemia, coronary artery disease, metabolic syndrome, obesity, and other risk factors for cardiovascular disease (Corella and Ordovas, 2012; Cormier et al., 2012; Glaser et al., 2011; Kathiresan et al., 2009; Kim et al., 2011; Kwak et al., 2011; Lattka et al., 2010; Merino et al., 2010; Sergeant et al., 2012; Simopoulos, 2010). SNPs found within the FADS1 and FADS2 genes of European, Asian, African American, and North American populations have been shown to influence plasma fatty acid concentrations, which can represent desaturase activity when measured as a ratio of fatty acid product to substrate (Bokor et al., 2010; Merino et al., 2011; Sergeant et al., 2012). Even genetic variation in lipoprotein lipase, which is required to release dietary fatty acids from lipoprotein particles, has been shown to regulate plasma lipid concentrations, thereby potentiating risk for metabolic syndrome and cancer (Crous-Bou et al., 2012; Garcia-Rios et al., 2011). Instead of reiterating these well reviewed findings, we note the extensive patterns of global variation in these genes and turn to the interesting prospect of their genetic drift among populations worldwide and the interaction between their genetic variation and regionally exclusive dietary habits.
In 2008, John Speakman proposed the ‘drifty gene hypothesis’ (Speakman, 2008) as a counter argument to James Neel's original ‘thrifty gene hypothesis’ (Neel, 1962). Whereas Neel attributes the current obesity epidemic to the now absent rise and fall of famine that genetically selected for persons of energy, or fat-storing, metabolism (Neel, 1962), Speakman describes the epidemic as a result of genetic drift by random mutation in populations following the release of the human species from predation (Speakman, 2008). More specifically, he argues that the lower intervention limit for survival, set by starvation has not changed in modern society, i.e. humans today have maintained the same survival tactics to last through a famine. On the other hand, the upper intervention limit, set by the risk of predation has dissipated, such that the loss of the body's requirement to evade predators for survival has allowed positive selection for genetic mutations that promote obesity (Speakman, 2008). Speakman also provides an alternative hypothesis to explain modern obesity, which accuses the changes in modern dietary fat consumption. He explains that individual variability in fat oxidation may currently withstand as it was in the past, but due to historically nominal fat consumption, genetic variation never lead to health disparities until humans changed their diet. As such, an increase in dietary fat consumption would lead to negative consequences in individuals of weak or low fat oxidation, driving the development of obesity and/or metabolic and cardiovascular diseases (Speakman, 2008). While Speakman's theory remains simply that, a theory, a few very recent studies published in 2012 have provided evidence to support his hypotheses by studying the genetic evolution of the FADS gene cluster.
The recently published story of the genetic evolution of the Fads gene cluster champions the concept of ecologically induced genetic diversification (Castro et al., 2012). The authors delineate an argument based on the genetic conservation and diversification of the Fads1 and Fads2 genes in various species from teleosts to amphibeans, reptiles, birds and mammals. Overall, the data suggests that the functionally diverse fatty acid desaturase enzymes found in humans today have been shaped over the course of evolution by environmental, dietary, and nutritional selective pressures in combination with genetic events, such as duplications, losses, and mutations (Castro et al., 2012).
A recent study (Ameur et al., 2012) highlighted two haplotypes found in Europeans defined by 28 SNPs that associate with very distinct measurements of long chain PUFA (LC-PUFA) product-to-substrate ratios, identified as haplotype A and D. Haplotype D was characterized as an enzymatically efficient gene signature, showing significantly increased desaturase activity, as depicted by eicosapentaenoic acid to eicosatetraenoic acid (EPA/ETA) and arachidonic acid to dihomo-γ-linolenic acid (AA/DGLA) ratios, whereas haplotype A was considered a less efficient genetic variant. The prevalence of each haplotype was then estimated for geographical ancestry based on the human genome diversity panel (Li et al., 2008). Results showed a nearly fixed population of haplotype D in Africa, while Europe and the Middle East were approximately 75% haplotype D. Eastern Asia and Oceania regions were approximately 50% D, and the Americas were nearly 100% A. To determine when the apparent genetic variation may have arisen, the authors used comparative genomic analysis to identify the haplotypes of rhesus macaques, chimpanzees, gorillas, Denisovans, Neanderthals, a Palaeo-Eskimo, and an Australian Aboriginal. Upon computational clustering they found that all appeared to be largely DD genotype, except the Eskimo and the Aboriginal, which were AA genotype. The authors ultimately concluded that the D haplotype likely appeared in modern humans following the split from Neanderthals, but before their emigration from Africa. Their speculation as to why the D haplotype has reached near fixation in Africa focuses on the requirement for LC-PUFA throughout the sophistication of the brain in hominids. Moreover, positive selection for haplotype D may have been driven in African populations where environmental and dietary supply of AA and DHA (docosahexaenoic acid) were restrictive (Ameur et al., 2012).
Another recent publication (Mathias et al., 2012) found very similar results to the aforementioned report, yet focused on somewhat different conclusions. The group's previous findings associated genetic variants in the FADS gene cluster with a distinct discrepancy in LC-PUFA synthesis between African Americans and European Americans with diabetes or metabolic syndrome (Sergeant et al., 2012). Therefore, they sought to investigate the evolutionary patterns of these genetic variants by analyzing 1092 individuals from 14 different populations. They found that African versus non-African populations displayed decreased nucleotide diversity and large differences in allelic frequencies in the FADS1 region. They went on to identify 9 SNPs around the FADS gene cluster for which the allele associated with increased PUFA metabolism was more frequently observed in African populations compared to non-African. In particular, SNP rs174537 found in FADS1, which is currently reported to have the strongest p-value with LC-PUFAs in literature to date, showed near fixation for the G allele in populations of African ancestry. Computational clustering for the haplotypes revealed that only a relatively small percentage of the African populations identified with the ancestral chimpanzee haplotype. The authors estimated that the positive haplotype selection in African populations occurred approximately 85,000 years ago, prior to the emigration of humans from Africa. Similar to the report by Gyllensten's group (Ameur et al., 2012), the authors speculated that positive selection in Africa likely supported the expansion and movement of populations to more diverse ecological locations (Mathias et al., 2012). Additionally, perhaps the selective pressures following such expansion outside Africa were lost upon the emergence of hunting techniques, including fishing, and the increased technological and social capacity to acquire LC-PUFA from the environment (Mathias et al., 2012). However, in modern Western society, where food is plentiful and consumption of proinflammatory omega-6 PUFA largely outweighs that of omega-3 PUFA, these originally advantageous genetic adaptations in PUFA metabolism have become potential risk factors for metabolic and cardiovascular diseases.
Epigenetic remodeling: Diet, exercise, and metabolomic biomarkers
It is now well established that not only genes and the variation found among them, but also the epigenetic modifications, such as methylation, acetylation, and phosphorylation, on the DNA and/or histones can act as modulators of disease. It is known that even RNA, acting as micro-RNA or long, non-coding RNA, can regulate gene expression. Moreover, it is becoming ever clearer that epigenetic modifications can be readily inherited, particularly during the extremely vulnerable in utero stage of life, and may be predictive of health outcomes in adulthood. Maternal obesity, insulin sensitivity, adiposity, diet, and nutrition can all influence the metabolism of offspring during pregnancy and even beyond parturition. Mounting evidence suggests that epigenetic changes as a result of maternal conditioning are inherently designed to prepare the offspring for the coming environment. However, when malnutrition in utero is a shortcoming to the actual environment where food is plentiful, these in utero induced epigenetic alterations may be misleading, and rather, become susceptibility biomarkers of disease, including obesity, metabolic syndrome, inflammation, cardiovascular disease, and cancer. Now recognized as the Developmental Origins of Health and Disease (DOHaD) Hypothesis, originally postulated by Barker and colleagues (Barker, 2003), this topic has now been extensively reviewed (Armitage et al., 2005; Burdge and Lillycrop, 2010; Ellison, 2005; Gluckman and Hanson, 2004; Martinez et al., 2012; Milagro et al., 2012; Walker and Ho, 2012; Waterland and Michels, 2007).
Fortunately, unlike the genome, the epigenome appears to be quite malleable. That is, even if a mother seems to hold the reins for her child's future health risks in utero, healthy lifestyle choices can still minimize incurred risks. Because humans were evolutionarily adapted to obligatory physical activity, it seems only logical that the now largely sedentary cultural lifestyles are inapt for our current genome (Booth et al., 2002). Aside from diet, studies of exercise intervention suggest regular physical activity helps refine the balance of metabolic homeostasis in patients by reducing risk factors for obesity and metabolic syndrome (Davis et al., 2012; Gremeaux et al., 2012). Unfortunately, the various parameters of combined diet and exercise induced weight loss can make exercise intervention studies difficult to track the cause of the observed effects (Bianchini et al., 2012). More studies with greater power and efficiently controlled parameters are necessary to fully delineate the physiological effects of exercise on health (Metcalf et al., 2012). Nonetheless, literature suggests regular physical activity can positively impact health through a number of mechanisms, including reduced adipocyte size and number, mobilization of fatty acid stores, and modulation of lipolysis and fatty acid uptake (Thompson et al., 2012).
Finally, our understanding of genetic variation in human metabolism has largely increased in recent years due to the introduction of metabolomics. Like other “–omics” research, the ability to peruse the human metabolome has brought insightful knowledge of the functional capacity of the variation hitherto identified in the genome and epigenome. Technology now allows us to reference metabolites and their concentration ratios in GWAS, which provides a systemic perspective on genetic variation in human metabolism (Illig et al., 2010; Suhre et al., 2011). Quite up and coming is the identification of specific metabolites, such as plasma fatty acids or intermediates of the tricarboxylic acid cycle, as markers of disease or predictors of outcome (Kunesova et al., 2012; Shah et al., 2012). By cross-referencing various aspects of “–omics” research, integration of multiple high throughput methods can drive personalized medicine fitting to patient needs and predictive of therapeutic response (Chen et al., 2012).
Conclusions and future perspectives
Fatty acids are essential to life. Whether produced de novo or acquired by diet fatty acids play a critical role as constituents of cellular membrane, posttranslational modifications, signaling ligands, and other cellular processes. There many different kinds of fatty acids that each vary in their carbon chain length and degree of saturation, which are important characteristics that help define the biochemical properties and functions of fatty acids. The metabolism of fatty acids has become a primary focus of biomedical research. Both lipogenesis and dietary fatty acids are proven contributors to diseases such as obesity, metabolic syndrome, inflammation, cardiovascular disease, and cancer. Although the biochemical structure of de novo synthesized and dietary fatty acids shows no discrimination, the two sources may fulfill different biological roles. De novo lipogenesis can be regulated by dietary fatty acid consumption; however, no study to date has identified a compensatory role for de novo fatty acid synthesis by dietary fat. This may be related to the varying aspects of de novo versus dietary lipid metabolism.
As obesity, metabolic syndrome, cardiovascular disease, and cancer trend throughout society, intervention studies, as a means to identify clear recommendations for dietary and lifestyle changes, should take precedence. Such investigations should be well controlled, prospective, and carefully consider both dietary quality and quantity. As genetic variants affecting lipid metabolism are identified, perhaps a more personalized regimen for healthy living can be made to improve the quality of life and decrease the risk for disease. However, irrespective of genetic predisposition, evidence of epigenetic remodeling suggests we can control many of the risk factors contributing to these diseases. With rising technologies, perhaps a high throughput screening for genetic and metabolic biomarkers will aide diagnosis and prognosis, as well as identification of a personalized treatment regimen for these diseases.
Figure 1. Standardizing Methods for Dietary Fat Studies in Humans.
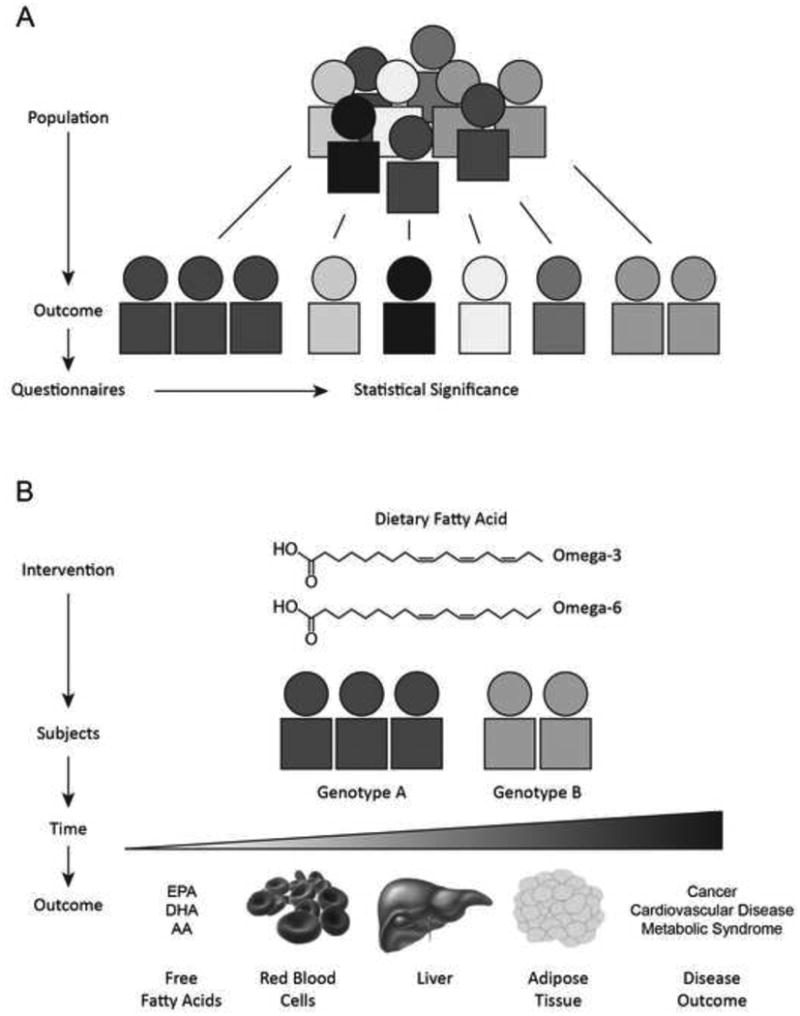
A. Current methodology often stratifies an isolated population by common ailments. Dietary habits are then determined by questionnaires and statistical association with genetic variation is assessed. B. Dietary intervention studies can directly assess the role of diet on health outcome. Here, the intervention with dietary PUFA supplement is tested on a subjects with a specific genetic profile to determine the health outcome, which is dependent on the time duration of the intervention.
Abbreviations
- SNP
single nucleotide polymorphism
- SAFA
saturated fatty acid
- MUFA
monounsaturated fatty acid
- PUFA
polyunsaturated fatty acid
- ACLY
ATP citrate lyase
- ACACA
acetyl-CoA carboxylase 1
- ACACB
acetyl-CoA carboxylase 2
- FASN
fatty acid synthase
- ELOVL
elongation of fatty acid
- LPL
lipoprotein lipase
- SCD
stearoyl-CoA desaturase
- FADS
fatty acid desaturase
- ALA
alpha-linolenic acid
- SREBF
sterol response element binding factor
- SREBP
sterol response element binding protein
- ChREBP
Carbohydrate response element binding protein
- BRCA1
breast cancer susceptibility gene 1
- EPA
eicosapentaenoic acid
- ETA
eicosatetraenoic acid
- AA
arachidonic acid
- DGLA
dihomo-gamma-linoleic acid
- LC-PUFA
long chain polyunsaturated fatty acid
- DHA
docosahexaenoic acid
- DNA
deoxyribonucleic acid
- RNA
ribonucleic acid
- DOHaD
Developmental Origins of Health and Disease
- GWAS
genome wide association study
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abu-Elheiga L, Matzuk MM, Kordari P, Oh W, Shaikenov T, Gu Z, Wakil SJ. Mutant mice lacking acetyl-coa carboxylase 1 are embryonically lethal. Proc Natl Acad Sci U S A. 2005;102(34):12011–12016. doi: 10.1073/pnas.0505714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameur A, Enroth S, Johansson A, Zaboli G, Igl W, Johansson AC, Rivas MA, Daly MJ, Schmitz G, Hicks AA, Meitinger T, Feuk L, van Duijn C, Oostra B, Pramstaller PP, Rudan I, Wright AF, Wilson JF, Campbell H, Gyllensten U. Genetic adaptation of fatty-acid metabolism: A human-specific haplotype increasing the biosynthesis of long-chain omega-3 and omega-6 fatty acids. Am J Hum Genet. 2012;90(5):809–820. doi: 10.1016/j.ajhg.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage JA, Taylor PD, Poston L. Experimental models of developmental programming: Consequences of exposure to an energy rich diet during development. J Physiol. 2005;565(Pt 1):3–8. doi: 10.1113/jphysiol.2004.079756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslibekyan S, Jensen MK, Campos H, Linkletter CD, Loucks EB, Ordovas JM, Deka R, Rimm EB, Baylin A. Genetic variation in fatty acid elongases is not associated with intermediate cardiovascular phenotypes or myocardial infarction. Eur J Clin Nutr. 2012;66(3):353–359. doi: 10.1038/ejcn.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker D. The midwife, the coincidence, and the hypothesis. BMJ. 2003;327(7429):1428–1430. doi: 10.1136/bmj.327.7429.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxheinrich A, Stratmann B, Lee-Barkey YH, Tschoepe D, Wahrburg U. Effects of a rapeseed oil-enriched hypoenergetic diet with a high content of alpha-linolenic acid on body weight and cardiovascular risk profile in patients with the metabolic syndrome. Br J Nutr. 2012;108(4):682–691. doi: 10.1017/S0007114512002875. [DOI] [PubMed] [Google Scholar]
- Beare-Rogers J, Dieffenbacher A, JV H. Lexicon of lipid nutrition. Pure Appl Chem. 2001;73(4):685–744. [Google Scholar]
- Beigneux AP, Kosinski C, Gavino B, Horton JD, Skarnes WC, Young SG. Atp-citrate lyase deficiency in the mouse. J Biol Chem. 2004;279(10):9557–9564. doi: 10.1074/jbc.M310512200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berquin IM, Min Y, Wu R, Wu J, Perry D, Cline JM, Thomas MJ, Thornburg T, Kulik G, Smith A, Edwards IJ, D'Agostino R, Zhang H, Wu H, Kang JX, Chen YQ. Modulation of prostate cancer genetic risk by omega-3 and omega-6 fatty acids. J Clin Invest. 2007;117(7):1866–1875. doi: 10.1172/JCI31494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berquin IM, Edwards IJ, Kridel SJ, Chen YQ. Polyunsaturated fatty acid metabolism in prostate cancer. Cancer Metastasis Rev. 2011;30(3-4):295–309. doi: 10.1007/s10555-011-9299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchini JA, da Silva DF, Nardo CC, Carolino ID, Hernandes F, Junior NN. Multidisciplinary therapy reduces risk factors for metabolic syndrome in obese adolescents. 2012 doi: 10.1007/s00431-012-1865-7. [DOI] [PubMed] [Google Scholar]
- Bjermo H, Riserus U. Role of hepatic desaturases in obesity-related metabolic disorders. Curr Opin Clin Nutr Metab Care. 2010;13(6):703–708. doi: 10.1097/MCO.0b013e32833ec41b. [DOI] [PubMed] [Google Scholar]
- Bokor S, Dumont J, Spinneker A, Gonzalez-Gross M, Nova E, Widhalm K, Moschonis G, Stehle P, Amouyel P, De Henauw S, Molnar D, Moreno LA, Meirhaeghe A, Dallongeville J. Single nucleotide polymorphisms in the fads gene cluster are associated with delta-5 and delta-6 desaturase activities estimated by serum fatty acid ratios. J Lipid Res. 2010;51(8):2325–2333. doi: 10.1194/jlr.M006205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth FW, Chakravarthy MV, Spangenburg EE. Exercise and gene expression: Physiological regulation of the human genome through physical activity. J Physiol. 2002;543(Pt 2):399–411. doi: 10.1113/jphysiol.2002.019265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdge GC, Lillycrop KA. Nutrition, epigenetics, and developmental plasticity: Implications for understanding human disease. Annu Rev Nutr. 2010;30:315–339. doi: 10.1146/annurev.nutr.012809.104751. [DOI] [PubMed] [Google Scholar]
- Burkhardt R, Toh SA, Lagor WR, Birkeland A, Levin M, Li X, Robblee M, Fedorov VD, Yamamoto M, Satoh T, Akira S, Kathiresan S, Breslow JL, Rader DJ. Trib1 is a lipid- and myocardial infarction-associated gene that regulates hepatic lipogenesis and vldl production in mice. J Clin Invest. 2010;120(12):4410–4414. doi: 10.1172/JCI44213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron DJ, Tong Z, Yang Z, Kaminoh J, Kamiyah S, Chen H, Zeng J, Chen Y, Luo L, Zhang K. Essential role of elovl4 in very long chain fatty acid synthesis, skin permeability barrier function, and neonatal survival. Int J Biol Sci. 2007;3(2):111–119. doi: 10.7150/ijbs.3.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campa D, McKay J, Sinilnikova O, Husing A, Vogel U, Hansen RD, Overvad K, Witt PM, Clavel-Chapelon F, Boutron-Ruault MC, Chajes V, Rohrmann S, Chang-Claude J, Boeing H, Fisher E, Trichopoulou A, Trichopoulos D, Palli D, Villarini A, Sacerdote C, Mattiello A, Tumino R, Peeters PH, van Gils CH, Bas Bueno-de-Mesquita H, Lund E, Chirlaque MD, Sala N, Suarez LR, Barricarte A, Dorronsoro M, Sanchez MJ, Lenner P, Hallmans G, Tsilidis K, Bingham S, Khaw KT, Gallo V, Norat T, Riboli E, Rinaldi S, Lenoir G, Tavtigian SV, Canzian F, Kaaks R. Genetic variation in genes of the fatty acid synthesis pathway and breast cancer risk. Breast Cancer Res Treat. 2009;118(3):565–574. doi: 10.1007/s10549-009-0347-8. [DOI] [PubMed] [Google Scholar]
- Campa D, Husing A, Chang-Claude J, Dostal L, Boeing H, Kroger J, Tjonneland A, Roswall N, Overvad K, Dahm CC, Rodriguez L, Sala N, Perez MJ, Larranaga N, Chirlaque MD, Ardanaz E, Khaw KT, Wareham N, Allen NE, Travis RC, Trichopoulou A, Naska A, Bamia C, Palli D, Sieri S, Tumino R, Sacerdote C, van Kranen HJ, Bas Bueno-de-Mesquita H, Stattin P, Johansson M, Chajes V, Rinaldi S, Romieu I, Siddiq A, Norat T, Riboli E, Kaaks R, Canzian F. Genetic variability of the fatty acid synthase pathway is not associated with prostate cancer risk in the european prospective investigation on cancer (epic) Eur J Cancer. 2011;47(3):420–427. doi: 10.1016/j.ejca.2010.09.029. [DOI] [PubMed] [Google Scholar]
- Casper ML, Barnett E, Williams GIJ, Halverson JA, Braham VE, Greenlund KJ. Atlas of stroke mortality: Racial, ethnic, and geographic disparities in the united states. Atlanta, GA: Department of Health and Human Services, Centers for Disease Control and Prevention; 2003. [Google Scholar]
- Castro LF, Monroig O, Leaver MJ, Wilson J, Cunha I, Tocher DR. Functional desaturase fads1 (delta5) and fads2 (delta6) orthologues evolved before the origin of jawed vertebrates. PLoS One. 2012;7(2):e31950. doi: 10.1371/journal.pone.0031950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy MV, Pan Z, Zhu Y, Tordjman K, Schneider JG, Coleman T, Turk J, Semenkovich CF. “New” Hepatic fat activates pparalpha to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab. 2005;1(5):309–322. doi: 10.1016/j.cmet.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Chakravarthy MV, Zhu Y, Lopez M, Yin L, Wozniak DF, Coleman T, Hu Z, Wolfgang M, Vidal-Puig A, Lane MD, Semenkovich CF. Brain fatty acid synthase activates pparalpha to maintain energy homeostasis. J Clin Invest. 2007;117(9):2539–2552. doi: 10.1172/JCI31183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy MV, Lodhi IJ, Yin L, Malapaka RR, Xu HE, Turk J, Semenkovich CF. Identification of a physiologically relevant endogenous ligand for pparalpha in liver. Cell. 2009a;138(3):476–488. doi: 10.1016/j.cell.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy MV, Zhu Y, Yin L, Coleman T, Pappan KL, Marshall CA, McDaniel ML, Semenkovich CF. Inactivation of hypothalamic fas protects mice from diet-induced obesity and inflammation. J Lipid Res. 2009b;50(4):630–640. doi: 10.1194/jlr.M800379-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, Miriami E, Karczewski KJ, Hariharan M, Dewey FE, Cheng Y, Clark MJ, Im H, Habegger L, Balasubramanian S, O'Huallachain M, Dudley JT, Hillenmeyer S, Haraksingh R, Sharon D, Euskirchen G, Lacroute P, Bettinger K, Boyle AP, Kasowski M, Grubert F, Seki S, Garcia M, Whirl-Carrillo M, Gallardo M, Blasco MA, Greenberg PL, Snyder P, Klein TE, Altman RB, Butte AJ, Ashley EA, Gerstein M, Nadeau KC, Tang H, Snyder M. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148(6):1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirala SS, Chang H, Matzuk M, Abu-Elheiga L, Mao J, Mahon K, Finegold M, Wakil SJ. Fatty acid synthesis is essential in embryonic development: Fatty acid synthase null mutants and most of the heterozygotes die in utero. Proc Natl Acad Sci U S A. 2003;100(11):6358–6363. doi: 10.1073/pnas.0931394100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P, Ntambi JM, Friedman JM. Stearoyl-coa desaturase-1 and the metabolic syndrome. Curr Drug Targets Immune Endocr Metabol Disord. 2003;3(4):271–280. doi: 10.2174/1568008033340117. [DOI] [PubMed] [Google Scholar]
- Cohen P, Friedman JM. Leptin and the control of metabolism: Role for stearoyl-coa desaturase-1 (scd-1) J Nutr. 2004;134(9):2455S–2463S. doi: 10.1093/jn/134.9.2455S. [DOI] [PubMed] [Google Scholar]
- Corella D, Ordovas JM. Interactions between dietary n-3 fatty acids and genetic variants and risk of disease. Br J Nutr. 2012;107(Suppl 2):S271–283. doi: 10.1017/S0007114512001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier H, Rudkowska I, Paradis AM, Thifault E, Garneau V, Lemieux S, Couture P, Vohl MC. Association between polymorphisms in the fatty acid desaturase gene cluster and the plasma triacylglycerol response to an n-3 pufa supplementation. Nutrients. 2012;4(8):1026–1041. doi: 10.3390/nu4081026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous-Bou M, Rennert G, Salazar R, Rodriguez-Moranta F, Rennert HS, Lejbkowicz F, Kopelovich L, Lipkin SM, Gruber SB, Moreno V. Genetic polymorphisms in fatty acid metabolism genes and colorectal cancer. Mutagenesis. 2012;27(2):169–176. doi: 10.1093/mutage/ger066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CL, Pollock NK, Waller JL, Allison JD, Dennis BA, Bassali R, Melendez A, Boyle CA, Gower BA. Exercise dose and diabetes risk in overweight and obese children: A randomized controlled trial. JAMA. 2012;308(11):1103–1112. doi: 10.1001/2012.jama.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi S. Getting to the root of america's racial health inequalities. Lancet. 2012;380(9847):1043. doi: 10.1016/s0140-6736(12)61584-0. [DOI] [PubMed] [Google Scholar]
- Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F. Srebp transcription factors: Master regulators of lipid homeostasis. Biochimie. 2004;86(11):839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Ellison PT. Evolutionary perspectives on the fetal origins hypothesis. Am J Hum Biol. 2005;17(1):113–118. doi: 10.1002/ajhb.20097. [DOI] [PubMed] [Google Scholar]
- Fiorentino M, Zadra G, Palescandolo E, Fedele G, Bailey D, Fiore C, Nguyen PL, Migita T, Zamponi R, Di Vizio D, Priolo C, Sharma C, Xie W, Hemler ME, Mucci L, Giovannucci E, Finn S, Loda M. Overexpression of fatty acid synthase is associated with palmitoylation of wnt1 and cytoplasmic stabilization of beta-catenin in prostate cancer. Lab Invest. 2008;88(12):1340–1348. doi: 10.1038/labinvest.2008.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rios A, Delgado-Lista J, Perez-Martinez P, Phillips CM, Ferguson JF, Gjelstad IM, Williams CM, Karlstrom B, Kiec-Wilk B, Blaak EE, Lairon D, Planells R, Malczewska-Malec M, Defoort C, Riserus U, Saris WH, Lovegrove JA, Drevon CA, Roche HM, Lopez-Miranda J. Genetic variations at the lipoprotein lipase gene influence plasma lipid concentrations and interact with plasma n-6 polyunsaturated fatty acids to modulate lipid metabolism. Atherosclerosis. 2011;218(2):416–422. doi: 10.1016/j.atherosclerosis.2011.07.092. [DOI] [PubMed] [Google Scholar]
- Glaser C, Lattka E, Rzehak P, Steer C, Koletzko B. Genetic variation in polyunsaturated fatty acid metabolism and its potential relevance for human development and health. Matern Child Nutr. 2011;7(Suppl 2):27–40. doi: 10.1111/j.1740-8709.2011.00319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA. Living with the past: Evolution, development, and patterns of disease. Science. 2004;305(5691):1733–1736. doi: 10.1126/science.1095292. [DOI] [PubMed] [Google Scholar]
- Gong J, Campos H, McGarvey S, Wu Z, Goldberg R, Baylin A. Genetic variation in stearoyl-coa desaturase 1 is associated with metabolic syndrome prevalence in costa rican adults. J Nutr. 2011;141(12):2211–2218. doi: 10.3945/jn.111.143503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene ER, Huang S, Serhan CN, Panigrahy D. Regulation of inflammation in cancer by eicosanoids. Prostaglandins Other Lipid Mediat. 2011;96(1-4):27–36. doi: 10.1016/j.prostaglandins.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremeaux V, Drigny J, Nigam A, Juneau M, Guilbeault V, Latour E, Gayda M. Long-term lifestyle intervention with optimized high-intensity interval training improves body composition, cardiometabolic risk, and exercise parameters in patients with abdominal obesity. Am J Phys Med Rehabil. 2012;91(11):941–950. doi: 10.1097/PHM.0b013e3182643ce0. [DOI] [PubMed] [Google Scholar]
- Guillou H, Zadravec D, Martin PG, Jacobsson A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog Lipid Res. 2010;49(2):186–199. doi: 10.1016/j.plipres.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Harkewicz R, Du H, Tong Z, Alkuraya H, Bedell M, Sun W, Wang X, Hsu YH, Esteve-Rudd J, Hughes G, Su Z, Zhang M, Lopes VS, Molday RS, Williams DS, Dennis EA, Zhang K. Essential role of elovl4 protein in very long chain fatty acid synthesis and retinal function. J Biol Chem. 2012;287(14):11469–11480. doi: 10.1074/jbc.M111.256073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyert DL, Xu J. Deaths: Preliminary data for 2011. National Vital Statistics Reports. 2012;61(6) [PubMed] [Google Scholar]
- Illig T, Gieger C, Zhai G, Romisch-Margl W, Wang-Sattler R, Prehn C, Altmaier E, Kastenmuller G, Kato BS, Mewes HW, Meitinger T, de Angelis MH, Kronenberg F, Soranzo N, Wichmann HE, Spector TD, Adamski J, Suhre K. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42(2):137–141. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabuchi K, Nakayama H, Iwahara C, Takamori K. Significance of glycosphingolipid fatty acid chain length on membrane microdomain-mediated signal transduction. FEBS Lett. 2010;584(9):1642–1652. doi: 10.1016/j.febslet.2009.10.043. [DOI] [PubMed] [Google Scholar]
- Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural basis of wnt recognition by frizzled. Science. 2012;337(6090):59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakumar A, Tai MH, Huang WY, al-Feel W, Hsu M, Abu-Elheiga L, Chirala SS, Wakil SJ. Human fatty acid synthase: Properties and molecular cloning. Proc Natl Acad Sci U S A. 1995;92(19):8695–8699. doi: 10.1073/pnas.92.19.8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, Voight BF, Bonnycastle LL, Jackson AU, Crawford G, Surti A, Guiducci C, Burtt NP, Parish S, Clarke R, Zelenika D, Kubalanza KA, Morken MA, Scott LJ, Stringham HM, Galan P, Swift AJ, Kuusisto J, Bergman RN, Sundvall J, Laakso M, Ferrucci L, Scheet P, Sanna S, Uda M, Yang Q, Lunetta KL, Dupuis J, de Bakker PI, O'Donnell CJ, Chambers JC, Kooner JS, Hercberg S, Meneton P, Lakatta EG, Scuteri A, Schlessinger D, Tuomilehto J, Collins FS, Groop L, Altshuler D, Collins R, Lathrop GM, Melander O, Salomaa V, Peltonen L, Orho-Melander M, Ordovas JM, Boehnke M, Abecasis GR, Mohlke KL, Cupples LA. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41(1):56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A. Very long-chain fatty acids: Elongation, physiology and related disorders. J Biochem. 2012;152(5):387–395. doi: 10.1093/jb/mvs105. [DOI] [PubMed] [Google Scholar]
- Kim OY, Lim HH, Yang LI, Chae JS, Lee JH. Fatty acid desaturase (fads) gene polymorphisms and insulin resistance in association with serum phospholipid polyunsaturated fatty acid composition in healthy korean men: Cross-sectional study. Nutr Metab (Lond) 2011;8(1):24. doi: 10.1186/1743-7075-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korner A, Ma L, Franks PW, Kiess W, Baier LJ, Stumvoll M, Kovacs P. Sex-specific effect of the val1483ile polymorphism in the fatty acid synthase gene (fas) on body mass index and lipid profile in caucasian children. Int J Obes (Lond) 2007;31(2):353–358. doi: 10.1038/sj.ijo.0803428. [DOI] [PubMed] [Google Scholar]
- Kovacs P, Harper I, Hanson RL, Infante AM, Bogardus C, Tataranni PA, Baier LJ. A novel missense substitution (val1483ile) in the fatty acid synthase gene (fas) is associated with percentage of body fat and substrate oxidation rates in nondiabetic pima indians. Diabetes. 2004;53(7):1915–1919. doi: 10.2337/diabetes.53.7.1915. [DOI] [PubMed] [Google Scholar]
- Kridel SJ, Axelrod F, Rozenkrantz N, Smith JW. Orlistat is a novel inhibitor of fatty acid synthase with antitumor activity. Cancer Res. 2004;64(6):2070–2075. doi: 10.1158/0008-5472.can-03-3645. [DOI] [PubMed] [Google Scholar]
- Kuhajda FP, Jenner K, Wood FD, Hennigar RA, Jacobs LB, Dick JD, Pasternack GR. Fatty acid synthesis: A potential selective target for antineoplastic therapy. Proc Natl Acad Sci U S A. 1994;91(14):6379–6383. doi: 10.1073/pnas.91.14.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunesova M, Hlavaty P, Tvrzicka E, Stankova B, Kalouskova P, Viguerie N, Larsen TM, Van Baak MA, Jebb SA, Martinez JA, Pfeiffer AF, Kafatos A, Handjieva-Darlenska T, Hill M, Langin D, Zak A, Astrup A, Saris WH. Fatty acid composition of adipose tissue triglycerides after weight loss and weight maintenance. The diogenes study. 2012 doi: 10.33549/physiolres.932414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JH, Paik JK, Kim OY, Jang Y, Lee SH, Ordovas JM, Lee JH. Fads gene polymorphisms in koreans: Association with omega6 polyunsaturated fatty acids in serum phospholipids, lipid peroxides, and coronary artery disease. Atherosclerosis. 2011;214(1):94–100. doi: 10.1016/j.atherosclerosis.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Lattka E, Illig T, Heinrich J, Koletzko B. Do fads genotypes enhance our knowledge about fatty acid related phenotypes? Clin Nutr. 2010;29(3):277–287. doi: 10.1016/j.clnu.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Li C, Aldai N, Vinsky M, Dugan ME, McAllister TA. Association analyses of single nucleotide polymorphisms in bovine stearoyl-coa desaturase and fatty acid synthase genes with fatty acid composition in commercial cross-bred beef steers. Anim Genet. 2012;43(1):93–97. doi: 10.1111/j.1365-2052.2011.02217.x. [DOI] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319(5866):1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Li W, Sandhoff R, Kono M, Zerfas P, Hoffmann V, Ding BC, Proia RL, Deng CX. Depletion of ceramides with very long chain fatty acids causes defective skin permeability barrier function, and neonatal lethality in elovl4 deficient mice. Int J Biol Sci. 2007;3(2):120–128. doi: 10.7150/ijbs.3.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Mondal AK, Murea M, Sharma NK, Tonjes A, Langberg KA, Das SK, Franks PW, Kovacs P, Antinozzi PA, Stumvoll M, Parks JS, Elbein SC, Freedman BI. The effect of acacb cis-variants on gene expression and metabolic traits. PLoS One. 2011;6(8):e23860. doi: 10.1371/journal.pone.0023860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald ML, Singaraja RR, Bissada N, Ruddle P, Watts R, Karasinska JM, Gibson WT, Fievet C, Vance JE, Staels B, Hayden MR. Absence of stearoyl-coa desaturase-1 ameliorates features of the metabolic syndrome in ldlr-deficient mice. J Lipid Res. 2008;49(1):217–229. doi: 10.1194/jlr.M700478-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnard C, Bachelier R, Vincent A, Jaquinod M, Kieffer S, Lenoir GM, Venezia ND. Brca1 interacts with acetyl-coa carboxylase through its tandem of brct domains. Oncogene. 2002;21(44):6729–6739. doi: 10.1038/sj.onc.1205915. [DOI] [PubMed] [Google Scholar]
- Mannen H. Identification and utilization of genes associated with beef qualities. Anim Sci J. 2011;82(1):1–7. doi: 10.1111/j.1740-0929.2010.00845.x. [DOI] [PubMed] [Google Scholar]
- Marmot M, Allen J, Bell R, Bloomer E, Goldblatt P. Who european review of social determinants of health and the health divide. Lancet. 2012;380(9846):1011–1029. doi: 10.1016/S0140-6736(12)61228-8. [DOI] [PubMed] [Google Scholar]
- Martinez JA, Cordero P, Campion J, Milagro FI. Interplay of early-life nutritional programming on obesity, inflammation and epigenetic outcomes. Proc Nutr Soc. 2012;71(2):276–283. doi: 10.1017/S0029665112000055. [DOI] [PubMed] [Google Scholar]
- Mathias RA, Fu W, Akey JM, Ainsworth HC, Torgerson DG, Ruczinski I, Sergeant S, Barnes KC, Chilton FH. Adaptive evolution of the fads gene cluster within africa. PLoS One. 2012;7(9):e44926. doi: 10.1371/journal.pone.0044926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaka T, Shimano H, Yahagi N, Kato T, Atsumi A, Yamamoto T, Inoue N, Ishikawa M, Okada S, Ishigaki N, Iwasaki H, Iwasaki Y, Karasawa T, Kumadaki S, Matsui T, Sekiya M, Ohashi K, Hasty AH, Nakagawa Y, Takahashi A, Suzuki H, Yatoh S, Sone H, Toyoshima H, Osuga J, Yamada N. Crucial role of a long-chain fatty acid elongase, elovl6, in obesity-induced insulin resistance. Nat Med. 2007;13(10):1193–1202. doi: 10.1038/nm1662. [DOI] [PubMed] [Google Scholar]
- Mead JR, Irvine SA, Ramji DP. Lipoprotein lipase: Structure, function, regulation, and role in disease. J Mol Med (Berl) 2002;80(12):753–769. doi: 10.1007/s00109-002-0384-9. [DOI] [PubMed] [Google Scholar]
- Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7(10):763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- Merino DM, Ma DW, Mutch DM. Genetic variation in lipid desaturases and its impact on the development of human disease. Lipids Health Dis. 2010;9:63. doi: 10.1186/1476-511X-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino DM, Johnston H, Clarke S, Roke K, Nielsen D, Badawi A, El-Sohemy A, Ma DW, Mutch DM. Polymorphisms in fads1 and fads2 alter desaturase activity in young caucasian and asian adults. Mol Genet Metab. 2011;103(2):171–178. doi: 10.1016/j.ymgme.2011.02.012. [DOI] [PubMed] [Google Scholar]
- Merkel M, Eckel RH, Goldberg IJ. Lipoprotein lipase: Genetics, lipid uptake, and regulation. J Lipid Res. 2002;43(12):1997–2006. doi: 10.1194/jlr.r200015-jlr200. [DOI] [PubMed] [Google Scholar]
- Metcalf B, Henley W, Wilkin T. Effectiveness of intervention on physical activity of children: Systematic review and meta-analysis of controlled trials with objectively measured outcomes (earlybird 54) BMJ. 2012;345:e5888. doi: 10.1136/bmj.e5888. [DOI] [PubMed] [Google Scholar]
- Migita T, Ruiz S, Fornari A, Fiorentino M, Priolo C, Zadra G, Inazuka F, Grisanzio C, Palescandolo E, Shin E, Fiore C, Xie W, Kung AL, Febbo PG, Subramanian A, Mucci L, Ma J, Signoretti S, Stampfer M, Hahn WC, Finn S, Loda M. Fatty acid synthase: A metabolic enzyme and candidate oncogene in prostate cancer. J Natl Cancer Inst. 2009;101(7):519–532. doi: 10.1093/jnci/djp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milagro FI, Mansego ML, De Miguel C, Martinez JA. Dietary factors, epigenetic modifications and obesity outcomes: Progresses and perspectives. 2012 doi: 10.1016/j.mam.2012.06.010. [DOI] [PubMed] [Google Scholar]
- Miura GI, Treisman JE. Lipid modification of secreted signaling proteins. Cell Cycle. 2006;5(11):1184–1188. doi: 10.4161/cc.5.11.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M, Man WC, Ntambi JM. Targeted disruption of stearoyl-coa desaturase1 gene in mice causes atrophy of sebaceous and meibomian glands and depletion of wax esters in the eyelid. J Nutr. 2001;131(9):2260–2268. doi: 10.1093/jn/131.9.2260. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Dobrzyn A, Elias PM, Ntambi JM. Stearoyl-coa desaturase-2 gene expression is required for lipid synthesis during early skin and liver development. Proc Natl Acad Sci U S A. 2005;102(35):12501–12506. doi: 10.1073/pnas.0503132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon YA, Hammer RE, Horton JD. Deletion of elovl5 leads to fatty liver through activation of srebp-1c in mice. J Lipid Res. 2009;50(3):412–423. doi: 10.1194/jlr.M800383-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcillo S, Martin-Nunez GM, Rojo-Martinez G, Almaraz MC, Garcia-Escobar E, Mansego ML, de Marco G, Chaves FJ, Soriguer F. Elovl6 genetic variation is related to insulin sensitivity: A new candidate gene in energy metabolism. PLoS One. 2011;6(6):e21198. doi: 10.1371/journal.pone.0021198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau K, Dizin E, Ray H, Luquain C, Lefai E, Foufelle F, Billaud M, Lenoir GM, Venezia ND. Brca1 affects lipid synthesis through its interaction with acetyl-coa carboxylase. J Biol Chem. 2006;281(6):3172–3181. doi: 10.1074/jbc.M504652200. [DOI] [PubMed] [Google Scholar]
- Moreno-Navarrete JM, Botas P, Valdes S, Ortega FJ, Delgado E, Vazquez-Martin A, Bassols J, Pardo G, Ricart W, Menendez JA, Fernandez-Real JM. Val1483ile in fasn gene is linked to central obesity and insulin sensitivity in adult white men. Obesity (Silver Spring) 2009;17(9):1755–1761. doi: 10.1038/oby.2009.65. [DOI] [PubMed] [Google Scholar]
- Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, Li X, Li H, Kuperwasser N, Ruda VM, Pirruccello JP, Muchmore B, Prokunina-Olsson L, Hall JL, Schadt EE, Morales CR, Lund-Katz S, Phillips MC, Wong J, Cantley W, Racie T, Ejebe KG, Orho-Melander M, Melander O, Koteliansky V, Fitzgerald K, Krauss RM, Cowan CA, Kathiresan S, Rader DJ. From noncoding variant to phenotype via sort1 at the 1p13 cholesterol locus. Nature. 2010;466(7307):714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasseri K, Moulton LH. Patterns of death in the first and second generation immigrants from selected middle eastern countries in california. J Immigr Minor Health. 2011;13(2):361–370. doi: 10.1007/s10903-009-9270-7. [DOI] [PubMed] [Google Scholar]
- Neel JV. Diabetes mellitus: A “Thrifty” Genotype rendered detrimental by “Progress”? Am J Hum Genet. 1962;14:353–362. [PMC free article] [PubMed] [Google Scholar]
- Nguyen PL, Ma J, Chavarro JE, Freedman ML, Lis R, Fedele G, Fiore C, Qiu W, Fiorentino M, Finn S, Penney KL, Eisenstein A, Schumacher FR, Mucci LA, Stampfer MJ, Giovannucci E, Loda M. Fatty acid synthase polymorphisms, tumor expression, body mass index, prostate cancer risk, and survival. J Clin Oncol. 2010;28(25):3958–3964. doi: 10.1200/JCO.2009.27.0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D, Lee Y, La B, Yeo J, Chung E, Kim Y, Lee C. Fatty acid composition of beef is associated with exonic nucleotide variants of the gene encoding fasn. Mol Biol Rep. 2012;39(4):4083–4090. doi: 10.1007/s11033-011-1190-7. [DOI] [PubMed] [Google Scholar]
- Perez-Martinez P, Perez-Caballero AI, Garcia-Rios A, Yubero-Serrano EM, Camargo A, Gomez-Luna MJ, Marin C, Gomez-Luna P, Dembinska-Kiec A, Rodriguez-Cantalejo F, Tinahones FJ, Roche HM, Perez-Jimenez F, Lopez-Miranda J, Delgado-Lista J. Effects of rs7903146 variation in the tcf7l2 gene in the lipid metabolism of three different populations. PLoS One. 2012;7(8):e43390. doi: 10.1371/journal.pone.0043390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Goumidi L, Bertrais S, Field MR, Cupples LA, Ordovas JM, McMonagle J, Defoort C, Lovegrove JA, Drevon CA, Blaak EE, Kiec-Wilk B, Riserus U, Lopez-Miranda J, McManus R, Hercberg S, Lairon D, Planells R, Roche HM. Acc2 gene polymorphisms, metabolic syndrome, and gene-nutrient interactions with dietary fat. J Lipid Res. 2010;51(12):3500–3507. doi: 10.1194/jlr.M008474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razani B, Zhang H, Schulze PC, Schilling JD, Verbsky J, Lodhi IJ, Topkara VK, Feng C, Coleman T, Kovacs A, Kelly DP, Saffitz JE, Dorn GW, 2nd, Nichols CG, Semenkovich CF. Fatty acid synthase modulates homeostatic responses to myocardial stress. J Biol Chem. 2011;286(35):30949–30961. doi: 10.1074/jbc.M111.230508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riancho JA, Vazquez L, Garcia-Perez MA, Sainz J, Olmos JM, Hernandez JL, Perez-Lopez J, Amado JA, Zarrabeitia MT, Cano A, Rodriguez-Rey JC. Association of acacb polymorphisms with obesity and diabetes. Mol Genet Metab. 2011;104(4):670–676. doi: 10.1016/j.ymgme.2011.08.013. [DOI] [PubMed] [Google Scholar]
- Ringseis R, Eder K. Regulation of genes involved in lipid metabolism by dietary oxidized fat. Mol Nutr Food Res. 2011;55(1):109–121. doi: 10.1002/mnfr.201000424. [DOI] [PubMed] [Google Scholar]
- Rysman E, Brusselmans K, Scheys K, Timmermans L, Derua R, Munck S, Van Veldhoven PP, Waltregny D, Daniels VW, Machiels J, Vanderhoydonc F, Smans K, Waelkens E, Verhoeven G, Swinnen JV. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res. 2010;70(20):8117–8126. doi: 10.1158/0008-5472.CAN-09-3871. [DOI] [PubMed] [Google Scholar]
- Sassa T, Suto S, Okayasu Y, Kihara A. A shift in sphingolipid composition from c24 to c16 increases susceptibility to apoptosis in hela cells. Biochim Biophys Acta. 2012;1821(7):1031–1037. doi: 10.1016/j.bbalip.2012.04.008. [DOI] [PubMed] [Google Scholar]
- Schleinitz D, Kloting N, Korner A, Berndt J, Reichenbacher M, Tonjes A, Ruschke K, Bottcher Y, Dietrich K, Enigk B, Filz M, Schon MR, Jenkner J, Kiess W, Stumvoll M, Bluher M, Kovacs P. Effect of genetic variation in the human fatty acid synthase gene (fasn) on obesity and fat depot-specific mrna expression. Obesity (Silver Spring) 2010;18(6):1218–1225. doi: 10.1038/oby.2009.392. [DOI] [PubMed] [Google Scholar]
- Schneider JG, Yang Z, Chakravarthy MV, Lodhi IJ, Wei X, Turk J, Semenkovich CF. Macrophage fatty-acid synthase deficiency decreases diet-induced atherosclerosis. J Biol Chem. 2010;285(30):23398–23409. doi: 10.1074/jbc.M110.100321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeff LC, McKenna MT. Cervical cancer mortality among foreign-born women living in the united states, 1985 to 1996. Cancer Detect Prev. 2003;27(3):203–208. doi: 10.1016/s0361-090x(03)00062-x. [DOI] [PubMed] [Google Scholar]
- Sergeant S, Hugenschmidt CE, Rudock ME, Ziegler JT, Ivester P, Ainsworth HC, Vaidya D, Case LD, Langefeld CD, Freedman BI, Bowden DW, Mathias RA, Chilton FH. Differences in arachidonic acid levels and fatty acid desaturase (fads) gene variants in african americans and european americans with diabetes or the metabolic syndrome. Br J Nutr. 2012;107(4):547–555. doi: 10.1017/S0007114511003230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan CN, Petasis NA. Resolvins and protectins in inflammation resolution. Chem Rev. 2011;111(10):5922–5943. doi: 10.1021/cr100396c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SH, Kraus WE, Newgard CB. Metabolomic profiling for the identification of novel biomarkers and mechanisms related to common cardiovascular diseases: Form and function. Circulation. 2012;126(9):1110–1120. doi: 10.1161/CIRCULATIONAHA.111.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw DI, Tierney AC, McCarthy S, Upritchard J, Vermunt S, Gulseth HL, Drevon CA, Blaak EE, Saris WH, Karlstrom B, Helal O, Defoort C, Gallego R, Lopez-Miranda J, Siedlecka D, Malczewska-Malec M, Roche HM, Lovegrove JA. Lipgene food-exchange model for alteration of dietary fat quantity and quality in free-living participants from eight european countries. Br J Nutr. 2009;101(5):750–759. doi: 10.1017/S0007114508039962. [DOI] [PubMed] [Google Scholar]
- Shurbaji MS, Kalbfleisch JH, Thurmond TS. Immunohistochemical detection of a fatty acid synthase (oa-519) as a predictor of progression of prostate cancer. Hum Pathol. 1996;27(9):917–921. doi: 10.1016/s0046-8177(96)90218-x. [DOI] [PubMed] [Google Scholar]
- Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: The impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61(4):212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- Simopoulos AP. Genetic variants in the metabolism of omega-6 and omega-3 fatty acids: Their role in the determination of nutritional requirements and chronic disease risk. Exp Biol Med (Maywood) 2010;235(7):785–795. doi: 10.1258/ebm.2010.009298. [DOI] [PubMed] [Google Scholar]
- Sinilnikova OM, Ginolhac SM, Magnard C, Leone M, Anczukow O, Hughes D, Moreau K, Thompson D, Coutanson C, Hall J, Romestaing P, Gerard JP, Bonadona V, Lasset C, Goldgar DE, Joulin V, Venezia ND, Lenoir GM. Acetyl-coa carboxylase alpha gene and breast cancer susceptibility. Carcinogenesis. 2004;25(12):2417–2424. doi: 10.1093/carcin/bgh273. [DOI] [PubMed] [Google Scholar]
- Speakman JR. Thrifty genes for obesity, an attractive but flawed idea, and an alternative perspective: The ‘drifty gene’ hypothesis. Int J Obes (Lond) 2008;32(11):1611–1617. doi: 10.1038/ijo.2008.161. [DOI] [PubMed] [Google Scholar]
- Stoffel W, Holz B, Jenke B, Binczek E, Gunter RH, Kiss C, Karakesisoglou I, Thevis M, Weber AA, Arnhold S, Addicks K. Delta6-desaturase (fads2) deficiency unveils the role of omega3- and omega6-polyunsaturated fatty acids. EMBO J. 2008;27(17):2281–2292. doi: 10.1038/emboj.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud CK, Nara TY, Roqueta-Rivera M, Radlowski EC, Lawrence P, Zhang Y, Cho BH, Segre M, Hess RA, Brenna JT, Haschek WM, Nakamura MT. Disruption of fads2 gene in mice impairs male reproduction and causes dermal and intestinal ulceration. J Lipid Res. 2009;50(9):1870–1880. doi: 10.1194/jlr.M900039-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryjecki C, Roke K, Clarke S, Nielsen D, Badawi A, El-Sohemy A, Ma DW, Mutch DM. Enzymatic activity and genetic variation in scd1 modulate the relationship between fatty acids and inflammation. Mol Genet Metab. 2012;105(3):421–427. doi: 10.1016/j.ymgme.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Suburu J, Chen YQ. Lipids and prostate cancer. Prostaglandins Other Lipid Mediat. 2012;98(1-2):1–10. doi: 10.1016/j.prostaglandins.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K, Shin SY, Petersen AK, Mohney RP, Meredith D, Wagele B, Altmaier E, Deloukas P, Erdmann J, Grundberg E, Hammond CJ, de Angelis MH, Kastenmuller G, Kottgen A, Kronenberg F, Mangino M, Meisinger C, Meitinger T, Mewes HW, Milburn MV, Prehn C, Raffler J, Ried JS, Romisch-Margl W, Samani NJ, Small KS, Wichmann HE, Zhai G, Illig T, Spector TD, Adamski J, Soranzo N, Gieger C. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477(7362):54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinnen JV, Van Veldhoven PP, Timmermans L, De Schrijver E, Brusselmans K, Vanderhoydonc F, Van de Sande T, Heemers H, Heyns W, Verhoeven G. Fatty acid synthase drives the synthesis of phospholipids partitioning into detergent-resistant membrane microdomains. Biochem Biophys Res Commun. 2003;302(4):898–903. doi: 10.1016/s0006-291x(03)00265-1. [DOI] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O'Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, Konig IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AC, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Doring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D, Karpe F, Lafontan M, Frayn K. Physical activity and exercise in the regulation of human adipose tissue physiology. Physiol Rev. 2012;92(1):157–191. doi: 10.1152/physrev.00012.2011. [DOI] [PubMed] [Google Scholar]
- Walker CL, Ho SM. Developmental reprogramming of cancer susceptibility. Nat Rev Cancer. 2012;12(7):479–486. doi: 10.1038/nrc3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Dubois RN. Eicosanoids and cancer. Nat Rev Cancer. 2010;10(3):181–193. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr. 2007;27:363–388. doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- Westerberg R, Tvrdik P, Unden AB, Mansson JE, Norlen L, Jakobsson A, Holleran WH, Elias PM, Asadi A, Flodby P, Toftgard R, Capecchi MR, Jacobsson A. Role for elovl3 and fatty acid chain length in development of hair and skin function. J Biol Chem. 2004;279(7):5621–5629. doi: 10.1074/jbc.M310529200. [DOI] [PubMed] [Google Scholar]


