Abstract
Growing evidence supports a role for the unfolded protein response (UPR) in carcinogenesis; however, the precise molecular mechanisms underlying this phenomenon remain elusive. Herein, we identified the circadian clock PER1 mRNA as a novel substrate of the endoribonuclease activity of the UPR sensor IRE1α. Analysis of the mechanism shows that IRE1α endoribonuclease activity decreased PER1 mRNA in tumor cells without affecting PER1 gene transcription. Inhibition of IRE1α signaling using either siRNA-mediated silencing or a dominant-negative strategy prevented PER1 mRNA decay, reduced tumorigenesis, and increased survival, features that were reversed upon PER1 silencing. Clinically, patients showing reduced survival have lower levels of PER1 mRNA expression and increased splicing of XBP1, a known IRE-α substrate, thereby pointing toward an increased IRE1α activity in these patients. Hence, we describe a novel mechanism connecting the UPR and circadian clock components in tumor cells, thereby highlighting the importance of this interplay in tumor development.
Introduction
The tumor microenvironment, and in particular, hypoxia and nutrient limitation, can lead to perturbations of endoplasmic reticulum functions, thereby resulting in the activation of an adaptive response named the unfolded protein response (UPR; refs. 1, 2). The UPR primarily provides tumor cells with the ability to cope with stress and to adapt for survival. In addition to its role in cellular adaptation, the UPR, and in particular IRE1α signaling, have been proposed to play significant roles during tumor development. This was supported by the identification of somatic mutations in the IRE1 gene (3) or the dysregulation of endoplasmic reticulum stress targets in various cancers (4–6). Moreover, the RNAse activity of IRE1α and the XBP1 transcription factor, whose mRNA is spliced by the combined action of IRE1α RNAse activity and a yet unknown ligase, have also been found to be necessary for tumor formation and growth in multiple myeloma, glioblastoma, and transformed embryonic fibroblast (7–9). Although our data have pointed toward a role for IRE1α signaling in tumor biology, IRE1α-dependent signaling pathways involved in such process still remain unclear.
In the present study, using glioblastoma as a model, we show that IRE1α endoribonuclease unexpectedly cleaves the mRNA encoded by the core circadian clock gene, PER1, thereby leading to its degradation. As PER1 is not a secretory protein but rather localizes to the cytosol/nucleus, this could therefore contribute to the regulation of a central signaling pathway and to an endoplasmic reticulum-dependent control of tumor growth. Collectively, we define a novel interplay between IRE1α and PER1 regulating tumor growth and angiogenesis, an observation consistent with the emerging role of PER1 in cancer (10, 11). Moreover, the analysis of clinical samples revealed that low PER1 mRNA expression and high XBP1 mRNA splicing correlated with poorer prognoses. These results identify IRE1α as a master regulator of cellular homeostasis in tumors, and provide the rationale for the development of IRE1α-targeted therapies in cancer cells.
Materials and Methods
Recombinant protein expression
IRE1cyto cDNA (AA 470–977) was cloned from human liver cDNAs using the Gateway technology (Invitrogen Corp.) into either pGEX-2TK or pDEST17. IRE1cyto cDNA devoid of ATG, was amplified by PCR using the Platinum Taq DNA Polymerase High Fidelity (Invitrogen Corp.) and the following amplification scheme: denaturation at 94°C for at 40 seconds, annealing at 60°C for 40 seconds, elongation at 68°C for 2 minutes, 35 cycles. The PCR products were precipitated using PEG8000 and recombined into pDONR201 using the Gateway BP clonase (Invitrogen Corp.). The plasmids were then transformed into competent DH5α cells and positive clones selected and sequenced. Positive clones were recombined into destination vectors using LR clonase (Invitrogen Corp.). Five individual colonies were selected and pooled and plasmid DNA was amplified and subsequently transformed into competent BL21 bacterial cells. Recombinant protein expression in BL21 cells was induced using 1 mmol/L IPTG for 3 hours. Bacteria were then collected by centrifugation, lysed, and recombinant proteins purified as recommended by the manufacturer (Gibco BRL). The resulting purified proteins were concentrated and dialyzed using Amicon ultra centrifugal filters (cutoff = 20,000 Da; Millipore Corp.), followed by functional testing as previously described (12, 13).
Animal experiments, intracranial injections, tumor size, and blood capillary measurements
The protocol used was as previously described (14, 15) and was approved by the local animal committee. Cell implantations (2 × 105 cells) in Nude mice were at 2 mm lateral to the bregma and 3 mm in depth using empty vector and IRE1_DN cells stably expressing pGIPZ-GFP-shPer1 or pGIPZ-GFP alone. Twenty-eight days postinjection, brain sections were observed for GFP fluorescence and stained using hematoxylin and eosin for visualization of tumor masses. Tumor volume was then estimated by measuring the length (L) and width (W) of each tumor and was calculated using the following formula (L × W2 × 0.5). CD31-positive vessels and Ki-67–positive cells were numerated after immunohistologic staining using rat antibodies against CD31 (Phar-Mingen), mouse antibody against Ki-67 (Clone MIB1, Dako), and secondary antibodies coupled to HRP (Dako). Imaging was carried out using a Nikon E600 microscope equipped with a digital camera DMX1200.
Microarray analysis
Microarray assay and preprocessing analysis were conducted in the microarray core facility of the Research Institute for Biotherapy at Montpellier using the standard Affymetrix protocol. Total RNA was extracted using TRIzol reagent (Invitrogen). RNA integrity was verified on an Agilent 2100 Bioanalyzer. For each of the samples, total RNA was reverse transcribed into cDNA, followed by in vitro transcription and biotin labeling to generate cRNA (Enzo Biochem). The fragmented, biotin-labeled cRNA was hybridized to the Human Genome U133 2.0 oligonucleotide arrays (Affymetrix) containing approximately 22,000 probes. Microarrays were stained with streptavidin antibodies and streptavidin–phycoerythrin in an Affymetrix Fluidics station. Arrays were scanned using a 3000 7G scanner. Raw data were processed into R/Bioconductor by using the Limma package (16). To determine genes whose expression increased when IRE1α is inactivated, probe set intensities were obtained by means of Gene Chip Robust Multiarray Averaging and were selected by using a corrected P value threshold of 0.05 and fold change threshold of |log2(fc)| ≥2.5 as previously described in ref. 15. The regulated genes are listed in Supplementary Table S1. Data are accessible on the NCBI Geo portal with the reference number GSE27306.
RNA isolation, reverse transcription PCR, and quantitative PCR analyses
Total RNA was prepared using the TRIzol reagent (Invitrogen Corp.). Semiquantitative analyses were carried out as previously described (17). The primers used were designed using Primer depot software (18) and are listed in Supplementary Table S4. The PCR products were resolved on 1% to 3% agarose gels. For real-time quantitative PCR (qPCR), RNA was reverse transcribed with Superscript II (Promega). All PCR reactions were carried out with a Stratagene ×4000 thermocycler (Stratagene) and the SYBR Green PCR Core Reagents Kit (Bio-Rad). Experiments were conducted in triplicate for each data point. Each sample was normalized toward the expression of the Rplp0 gene.
RNA cleavage assay
Total RNA (10 μg) from U87 or HepG2 was incubated with the cytoplasmic domain of human GST-IRE1α (5 μg) at 37°C for the indicated amounts of time in a buffer containing 250 mmol/L Tris pH 7.5, 600 mmol/L NaCl, 5 mmol/L MgCl2, 5 mmol/L MnCl2, 25 mmol/L β-mercaptoethanol, supplemented with or without 10 mmol/L ATP as previously described (12). As control, we used heat-denatured GST-IRE1α. Reverse transcriptase (RT)-PCR was then conducted using Per1 primers and Gapdh as internal control. The pcDNA3.1-hPer1 expression vector was linearized by using SspI and used as a template for in vitro transcription by using T7 polymerase (Promega) in the presence of dNTP and 32PαdUTP. In vitro transcribed radiolabeled RNA was incubated in kinase buffer (50 mmol/L Tris-HCl, pH 7.5, 150 mmol/L NaCl, 1 mmol/L MgCl2, 1 mmol/L MnCl2, 5 mmol/L 2-mercaptoethanol, and 2 mmol/L ATP) with the cytoplasmic domain of human GST-IRE1α at 37°C for increasing amounts of time. Fragments resulting from the enzymatic reaction were resolved by Tris Borate EDTA-Urea electrophoresis and visualized by radioautography on X-ray films. Secondary structure of Per1 mRNA was predicted using M-FOLD (19). For actinomycin D pulse-chase experiments, actinomycin D was applied to 50% confluent empty vector or IRE1_DN cells at a final concentration of 5 μg/mL for the indicated amounts of time. Total RNA was then extracted and reverse transcribed before qPCR analysis using the following primers targeting Per1: forward 5′-ctcagtggctgtctccttcc and reverse 5′-gagccaggagctcagagaag (fragment 898–1016) or forward 5′-gatgtgcatctggtgaagc and reverse 5′-ccttgaacgtgcctgtagca (fragment 1891–1991).
Lentiviral transduction and PER1 knockdown by shRNA
For Per1 knockdown experiments, we used the pGIPZ-GFP-lentiviral vectors expressing Per1 short-hairpin RNA (shRNA) as previously described (Open Biosystems). Lentivirus-containing supernatant was collected 48 hours after transfection in LTA-HEK293T cells, 0.2 μm filtered, and snap frozen at −80°C. U87 cells were infected with lentivirus at low multiplicity according to the manufacturer’s instructions. Cells were selected in puromycin (2.5 μg/mL) and polyclonal populations were expanded and analyzed.
Luciferase reporter gene assay
The human Per1 promoter luciferase reporter gene construct was kindly provided by U. Albrecht (Freiburg, Switzerland). The hPer1-Luc plasmid was generated as previously described (20). Following transfection, cells were incubated for 24 hours and stimulated or not with drugs for an additional 16 hours. Luciferase activity was measured using the dual luciferase kit (Promega) according the manufacturer’s instructions. Light emission was measured in a luminometer (Lumistar).
RNA interference
siRNA were designed using Greg Hannon’s webtool and listed in Supplementary Table S5. Cells were transfected by using the siRNAi Max Lipofectamine reagent (Invitrogen Corp.). Following incubation for 48 to 72 hours, total RNA was extracted and used for semi-quantitative RT-PCR.
Colony formation assay
Cells were plated at density of 104 per well in 12-well plates and cell proliferation rate was measured by cell counting (Beckman Coulter). For colony formation, 2,500 cells were seeded in 6-well plates. Twenty-four hours later, fresh medium was added and the cells were allowed to form colonies. After 2 weeks, the colonies were stained with 0.1% crystal violet and counted. The experiments were carried out at least twice in triplicate.
Antibody-based analyses
Total protein extracts and immunoblotting were conducted as previously described (14). Antibodies against PER1 and tubulin were purchased from Cogenics and Santa Cruz Biotechnology. Proteins were detected using secondary antibodies coupled to HRP (Dako) and immunoblots revealed using enhanced chemiluminescence and radioautography. For immunohistochemistry, analyses were carried out as previously described (15) using an antibody that was raised against the protein translated from Xbp1s mRNA (21).
Cell culture, transfections, and treatments
HepG2 and U87 cells were grown in Dulbecco’s Modified Eagle Medium, supplemented with 10% FBS, L-glutamine, and antibiotics. U87 and HepG2 cells were stably transfected with pcDNA3/IRE1-NCK-1, an expression vector encoding a cytoplasmic-defective IRE1α mutant (17). U87 cells were transiently transfected with pED-IRE1 WT or mutant K599A (22) expression vectors. Transfections were conducted using Lipofectamine (Invitrogen Corp.) according to the manufacturer’s recommendations. Actinomycin D were purchased from Sigma and used as indicated.
Statistical analyses
Data are presented as means ± SD. Statistical significance (P < 0.05 or lower) was determined using the Student t test 2-tailed distribution, assuming equal variance for the samples (GraphPad Prism). For in vivo studies, Kaplan–Meier curves and log-rank analysis were conducted using GraphPad 5.0.
Human samples
A total of 29 human glioblastoma samples were collected from The Bordeaux Tumor Bank and 31 + 20 samples from the Mayo Clinic. Twelve samples from normal or peritumoral brain tissues were also collected. Samples were collected according to the recommendations of the local ethics committees and informed consent was systematically obtained.
Results
IRE1α loss-of-function results in PER1 mRNA posttranscriptional upregulation
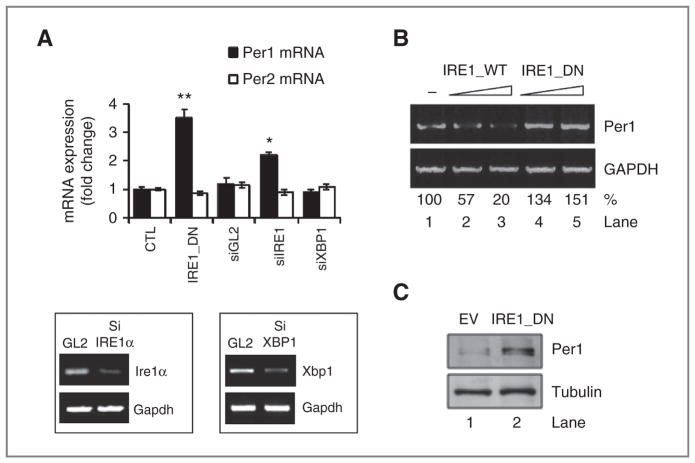
Using global expression profiles of U87 cells stably transfected with an empty vector or a well-established dominant-negative (DN)-IRE1α vector (IRE1_DN; ref. 17), we identified PER1 mRNA as a potential target of IRE1α signaling (Supplementary Table S1). PER1 mRNA expression was increased in both IRE1_DN cells and IRE1α-silenced cells, whereas XBP1 silencing had no effect (Fig. 1A). This indicates that PER1 mRNA expression regulation is dependent on IRE1α activity but not on XBP1. Similar results were obtained in additional empty vector or IRE1_DN (Supplementary Fig. S1A) HepG2 stable transfected cells (Supplementary Fig. S1B), thus ruling out any clonal or cell line-specific effects. Changes were specific of PER1, as PER2 mRNA levels were not altered in these conditions (Fig. 1A). To further investigate the relationship between IRE1α activity and PER1 mRNA expression, parental U87 cells were transiently transfected with increasing amounts of IRE1αWT or kinase dead mutant IRE1αK599A (22, 23). PER1 mRNA expression was reduced in cells overexpressing IRE1αWT in a dose-dependent manner (Fig. 1B; compare lanes 2 and 3 to lane 1). In contrast, PER1 mRNA accumulated in cells overexpressing IRE1αK599A (Fig. 1B; compare lanes 4 and 5 to lane 1). The impact of IRE1α activity inhibition on PER1 mRNA was also concomitant with an increase in PER1 protein levels in IRE1_DN cells (Fig. 1C).
Figure 1.
Impaired IRE1α activity leads the upregulation of PER1 mRNA. A, expression of PER1 and PER2 mRNA was measured by PCR in control (EV) and IRE1_DN U87 cells as well as U87 cells subjected to IRE1α silencing, XBP1 silencing, or luciferase silencing as control (GL2) by siRNA for 72 hours (insets). PER1 and PER2 mRNA levels were normalized to RPLP0 levels (t test; *, P < 0.05; **, P < 0.001). B, U87 cells were transiently transfected with increasing concentrations of plasmids encoding for WT-IRE1α or DN K599A IRE1α, followed by mRNA extraction. The expression of PER1 and Gapdh was assessed by RT-PCR. C, PER1 and tubulin protein levels in empty vector and IRE1_DN cells.
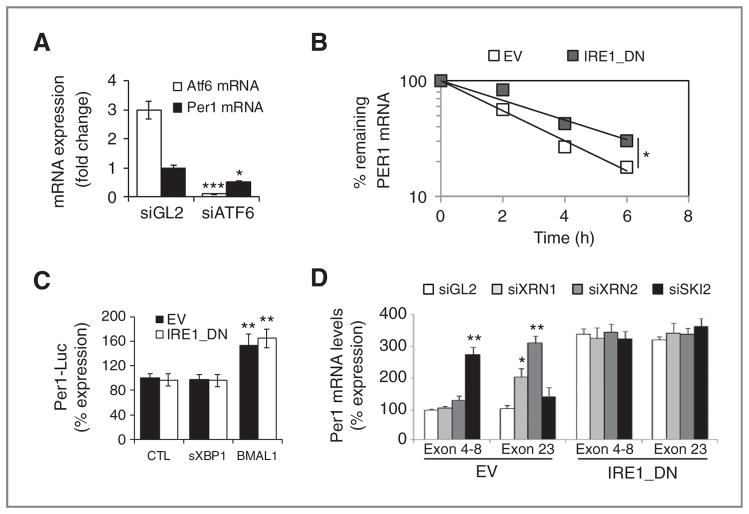
PER1 mRNA expression was previously found to be under the control of UPR-regulated transcription factor ATF6 (24). To determine whether the observed IRE1α-dependent regulation of PER1 mRNA occurred at the transcriptional level, empty vector and IRE1_DN cells were transfected with a PER1 promoter reporter construct containing the −1,500 bp upstream of the transcriptional start site. These experiments were carried out under control conditions (CTL) or upon overexpression of spliced XBP1 (XBP1) or the circadian clock regulator BMAL1 that is known to control PER1 expression (as a positive control). PER1 promoter activity remained unchanged in IRE1_DN or IRE1α-silenced cells, whereas it doubled in BMAL1-overexpressing cells and remained unchanged in cells overexpressing XBP1s (Fig. 2B). These results indicate that the increase in PER1 mRNA in the absence of functional IRE1α may occur posttranscriptionally and independently of XBP1s. We then tested whether PER1 mRNA expression increase in IRE1_DN cells was associated with an increase in PER1 mRNA half-life. This was assessed using an actinomycin D pulse-chase experiment followed by qRT-PCR (Fig. 2C). Under these conditions, PER1 mRNA half-life was significantly prolonged going from 2.3 hours in empty vector cells to 3.5 hours in IRE1_DN cells. Together, these data identify PER1 mRNA as an IRE1α-regulated target in cancer cells and provide a novel role for IRE1α activity on PER1 mRNA stability.
Figure 2.
IRE1α-mediated posttranscriptional control of PER1 mRNA in cultured cells. A, PER1 (closed) and ATF6 (open) mRNA expression as determined by quantitative RT-PCR in cells transfected with siRNA against luciferase (siGL2) and ATF6 (siATF6). Experiments were carried out in triplicate and the mean ± SD, statistical significance (Student t test) is indicated (*, P < 0.05; ***, P < 0.01). B, empty vector and IRE1_DN cells were cotransfected with control plasmid (pCMV-rL) or PER1 promoter-dependent luciferase reporter and either an empty pCDNA3 vector, a pCDNA3-sXBP1 vector, or a pCDNA3-BMAL1 vector. Cells were then lysed and lysates analyzed with the Dual-Luciferase Reporter Kit (Promega). Results were normalized against pCMV-Renilla luciferase (t test, **P < 0.05). C, actinomycin D pulse-chase was carried out as described in Materials and Methods. Total mRNA was extracted and quantitative RT-PCR experiments were conducted using PER1 mRNA-specific primer pairs. The experiment was repeated 3 times and data are presented as mean ± SD. Statistical significance was determined using Student t test, *, P < 0.03. D, empty vector and IRE1_DN cells were transfected with siRNA against XRN1/2 or SKI2. RNA was isolated after 48 hours and was used to amplify different regions of PER1 mRNA. Experiments were carried out in triplicate and the mean ± SD, statistical significance (Student t test) is indicated (*, P < 0.05; **, P < 0.01).
PER1 mRNA is cleaved by IRE1α
Next, as IRE1α was shown to control mRNA levels through direct cleavage (25), we examine whether PER1 mRNA was a direct target of IRE1α endoribonuclease activity. We studied the effects of IRE1α activity on PER1 mRNA regulation through internal cleavage sites. PER1 mRNA potential cleavage fragments amounts were measured upon siRNA-mediated silencing of the ribonucleases XRN1/2 and SKI2, which respectively contribute to RNA degradation 5′-3′ and 3′-5′, as previously described (26). We confirmed that treatment with XRN1/2 or SKI2 siRNAs specifically reduced the expression of target mRNAs without affecting the expression of endogenous Ire1α mRNA (Supplementary Fig. S2). Treatment with XRN1/2 or SKI2 siRNA did not affect PER1 mRNA sequences corresponding to 5′ (exons 4–8) and 3′ (exon 23) mRNA ends in IRE1_DN cells (Fig. 2D), showing that no PER1 mRNA cleavage fragments were present in these cells. In empty vector cells, SKI2 knockdown led to the accumulation of the 5′ PER1 mRNA sequence corresponding to the exons 4 to 8, and thus located upstream of potential IRE1α cleavage sites. In contrast, the fragment located downstream of these sites (exon 23) did not accumulate when compared with control irrelevant siRNA (GL2; Fig. 2D). Conversely, treatment with siRNA targeting the 5′ to 3′ exonucleases XRN1/2 only led to the increase of the fragment, downstream of this site (exon 23; Fig. 2D).
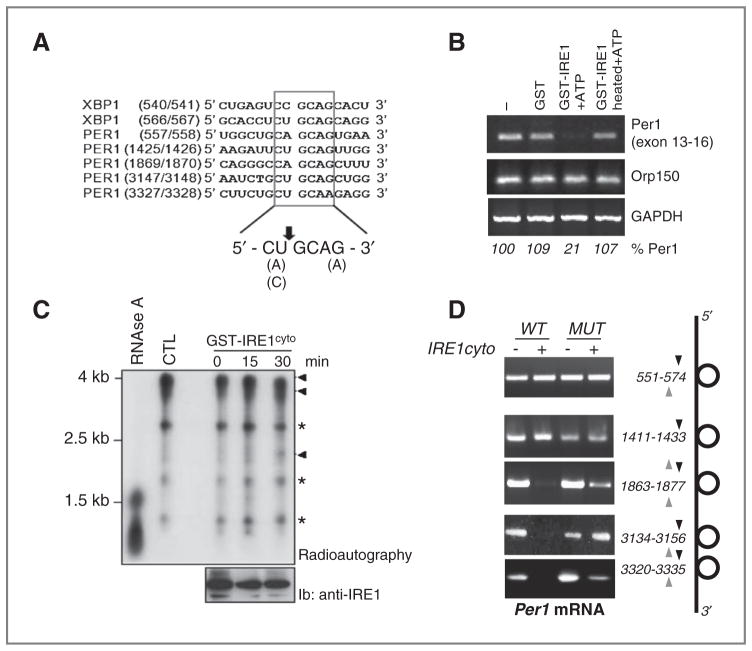
Sequence analysis revealed that five IRE1α consensus cleavage sites were present on human PER1 mRNA (Fig. 3A). Moreover, these cleavage sites were associated with P-loops structures, thereby creating potential cleavage sites for IRE1α endoribonuclease (Supplementary Fig. S3A). We then tested whether IRE1α could directly cleave PER1 mRNA. Total RNA from U87 cells was subjected to an in vitro IRE1α-mediated cleavage assay (12). This reaction was followed by RT-PCR to monitor PER1 mRNA levels (Fig. 3B). A strong decrease in PER1 mRNA level was observed when total RNA was incubated with IRE1α and ATP, whereas mRNA levels of the housekeeping genes ORP150 or GAPDH were unchanged (Fig. 3B). A positive control for IRE1α endoribonuclease activity was obtained using XBP1 mRNA as a substrate (Supplementary Fig. S3B). These results show that IRE1α cleaves PER1 mRNA in vitro. Then, to identify the cleavage products resulting from PER1 mRNA, in vitro transcribed and radiolabeled PER1 mRNA was subjected to IRE1α cleavage as described above. These experiments showed a major radiolabeled fragment of approximately 4 kb corresponding to the mRNA transcribed from the PER1 cDNA. In addition, three bands corresponding to entities of, respectively, 2.7, 1.7, and 1 kb were also present in the original transcription reaction (Fig. 3C, lane 2). All the radiolabeled material was RNA as shown by RNase A-mediated degradation (Fig. 3C, lane 1). In the presence of GST-IRE1α-cyto, a band of approximately 2 kb (Fig. 3C, lanes 3–5) appeared across time and could correspond to the product generated following IRE1α-mediated cleavage at nucleotide 1920 (Site 3, Supplementary Table S2), thus suggesting that IRE1α cleaves PER1 mRNA at least at the cleavage site #3. To determine whether IRE1α can also cleave PER1 mRNA at others sites, the five putative sites were mutated by insertion of a single mutation with the site CUGCAC where G was replaced by A. Mutated cDNA were in vitro transcribed and subjected to IRE1-mediated cleavage as above. Site-specific PCR amplification was then carried out for each reaction (Fig. 3D). This revealed that out of the 5 potential cleavage sites identified, only 3, namely 1920, 3197, and 3378 were cleaved by IRE1α in vitro (Fig. 3D). Taken together, these data are consistent with an IRE1α-dependent cleavage of PER1 mRNA.
Figure 3.
IRE1α-mediated posttranscriptional control of PER1 mRNA in vitro. A, sequence alignment of XBP1 mRNA IRE1α-mediated cleavage sites with similar regions in PER1 mRNAs. B, in vitro RNA cleavage assay. Total RNA extracted from U87 cells was incubated with GST or GST-IRE1α-cyto in the presence of ATP for 2 hours at 37°C. RT-PCR was then conducted to determine PER1, ORP150, and GAPDH mRNA levels. C, PER1 cDNA sequence cloned into the pCDNA3 vector was used as template for in vitro transcription using the T7 Ribomax kit (Promega) in the presence of 32P-UTP. The resulting radiolabeled riboprobe was then incubated or not with dephosphorylated GST-hIRE1cyto for the indicated periods of time or with RNase A for 15 minutes at room temperature. The reaction products were resolved by PAGE and revealed by radioautography on X-ray films. The amount of recombinant GST-IRE1cyto added to the reaction is shown in the bottom blot using immunoblot with anti-IRE1 antibodies. *, nonspecific bands; Arrowheads, full and cleaved PER1 mRNA products. D, PER1 mRNA wild-type and mutated on each potential IRE1α cleavage sites were transcribed in vitro and subjected to in vitro cleavage with GST-hIRE1cyto as in F. Reaction products were then subjected to RT-PCR with specific primers flanking each cleavage site.
IRE-dependent PER1 degradation modulates cancer cell survival and tumor progression in vivo
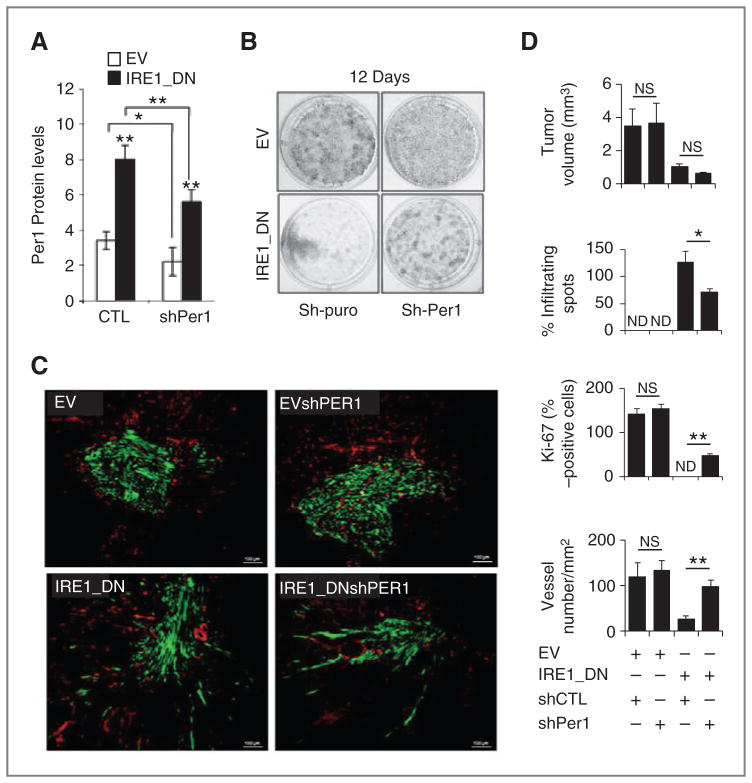
To determine the biologic significance of PER1 mRNA cleavage by IRE1α on tumor cell growth, PER1 mRNA expression was attenuated using lentiviral-mediated delivery of GFP-shPER1 in empty vector and IRE1_DN cells (or GFP empty vector as control). We first confirmed that pGIPZ-GFP-shPER1 viral particles effectively reduced PER1 mRNA and protein expression in U87 cells using RT-PCR and immunoblotting (Fig. 4A and S4). Using these cells, the impact of PER1 regulation by IRE1α anchorage-independent cell growth was investigated. After 2 weeks, the IRE1_DN cells showed a reduced ability to form colonies compared with empty vector cells (Fig. 4B). We then investigated the effects of PER1 silencing on tumor growth using our previously described in vivo orthotopic glioblastoma model (14). Fluorescence microscopy analysis of the tumors revealed GFP expression in tumor cells, thereby further confirming successful and stable lentiviral transduction in tumor cells up to 28 days postinjection (Fig. 4C). This also revealed that low PER1 expression in an IRE1α wild-type background neither impacted on tumor volume (Fig. 4C and D), tumor shape (Fig. 4C and D), nor on the number of tumor proliferating cells (Fig. 3D). IRE1_DN cell-derived tumors were smaller (P < 0.001) with extensive tumor cell infiltration in surrounding parenchyma (Fig. 4D; P < 0.001). Interestingly, at 28 days postinjection, the size of IRE1_DNsh-PER1 cell-derived tumors was comparable with that of IRE1_DN cell-derived tumors (Fig. 4D), however, with reduced tumor cell infiltrates (Fig. 4D) and better-delimited perimeters as compared with empty vector-derived tumors. This phenotype was accompanied by a marked restoration of proliferation within the tumor (Fig. 4D). These results confirm a role of IRE1α signaling in tumor growth in vivo and show the involvement of the IRE1α/PER1 axis in this process. Moreover, tumor angiogenesis, which is abnormal in IRE1_DN-derived tumors (14, 15), was investigated in PER1-silenced cells using CD31 immunostaining (Fig. 4C). High vascular density was apparent in empty vector and EVshPER1 cell-derived tumors (Fig. 4C). Tumor vascularization was partially restored in IRE1_DNshPER1-derived tumors (Fig. 4C and D). These results establish that the loss of cancer cell proliferation and tumor vascularization due to impairment of IRE1α activity is in part mediated by increased PER1 expression and suggest a potential role for PER1 in tumor angiogenesis. As the IRE1α/PER1 axis impacts on tumor growth capacity and angiogenesis, we then measured the consequences of its alteration on mouse survival following orthotopic injection (Fig. 5A). IRE1α signaling inhibition (IRE1_DN cell-derived tumors) increased the survival of tumor-bearing mice compared with those bearing empty vector cell-derived tumors. This survival advantage was lost in PER1 knocked-down tumors (Fig. 5A), thereby reinforcing the existence of functional interplay between IRE1α and PER1 underlying IREα.
Figure 4.
Impact of PER1 mRNA expression levels on IRE1_DN cell-derived tumors. A, the expression of PER1 was monitored using immunoblot analysis in empty vector and IRE1_DN cells silenced or not for PER1 (shPER1). Quantification of 3 independent experiments is represented as the mean ± SD. B, empty vector (EV) and IRE1_DN cells and their shPER1 counterparts were seeded in 6-well plates at equal densities. Cells were allowed to form colonies for 12 days. The colonies were stained with crystal violet 0.1%. C, intracranial implantation of U87 cells expressing either the IRE1_DN or the empty vector in the presence of pGIPZ-GFP-shPER1 or pGIPZ-GFP lentiviral vector was done in nude mice (n = 16). Immunohistochemical staining of tumor and surrounding tissue was done using anti-CD31 antibodies (red). Scale bar, 100 μm. D, quantification of implanted tumors’ features. Intracerebral tumor volume was determined. Four independent tumors were measured for each clone. Infiltrating spots were estimated by counting tumor field at ×5 magnification for each condition (t test, ns, nonsignificant; *, P < 0.05). The percentage of dividing cells (Ki-67 positive) in the 4 types of tumors was estimated by counting 5 different fields at ×40 magnification for each experiment. The mean Ki-67 intensity per condition is plotted with error bars representing SD. Significant differences are indicated between each empty vector and IRE1_DN pairs, and between empty vector compared with IRE1_DN. Vascular density was quantified by counting vessels from 5 randomly chosen fields per animal (n = 4 animals per conditions) and normalized to the tumor surface. Significant differences are indicated between each empty vector and IRE1_DN pairs, and between empty vector compared with IRE1_DN (t test, NS, nonsignificant; ***, P < 0.0005; **, P < 0.001; *, P < 0.05).
Figure 5.
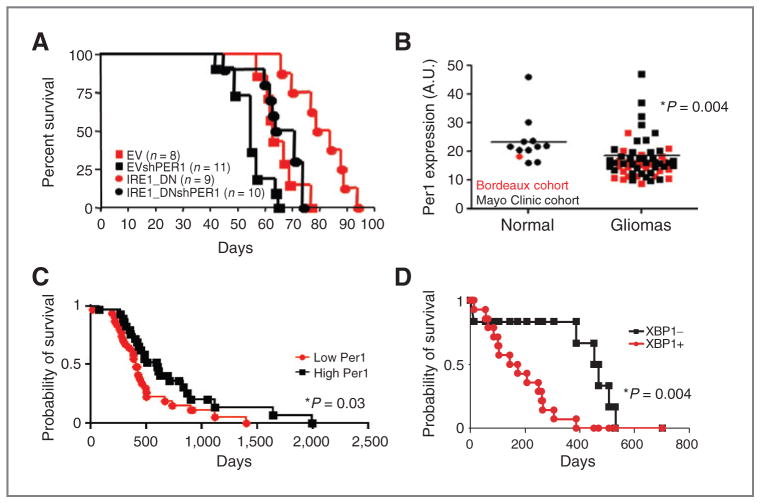
IRE1/PER1 signaling axis in tumor growth. A, overall survival of mice subjected to intracranial implantation of empty vector and IRE1_DN cells and their shPER1 counterparts was reported in Kaplan–Meier survival curves. (EV vs. IRE1_DN, P < 0.001; IRE1_DN vs. IRE1_DNshPER1, P < 0.001; EV vs. IRE1_DNshPER1, P = NS; log-rank test). B, qPCR analysis of PER1 mRNA expression in 60 glioblastoma cancer samples and 12 normal brain tissues. Bordeaux cohort is indicated in red, Mayo Clinic cohort in black. The results are expressed in arbitrary units as a ratio of PER1 transcripts to Rplp0 transcripts. The P value is indicated. C, high (n = 31) and low (n = 29) PER1 mRNA level correlates with patient survival. Values were plotted in Kaplan–Meier survival curves. Statistical difference between the 2 groups is indicated. Statistical difference between the 2 groups in indicated P = 0.03; log-rank test. D, Kaplan–Meier survival curves of patients displaying negative sXBP1 staining (6; XBP1−) or positive sXBP1 staining (14; XBP1+). P = 0.004; log-rank test.
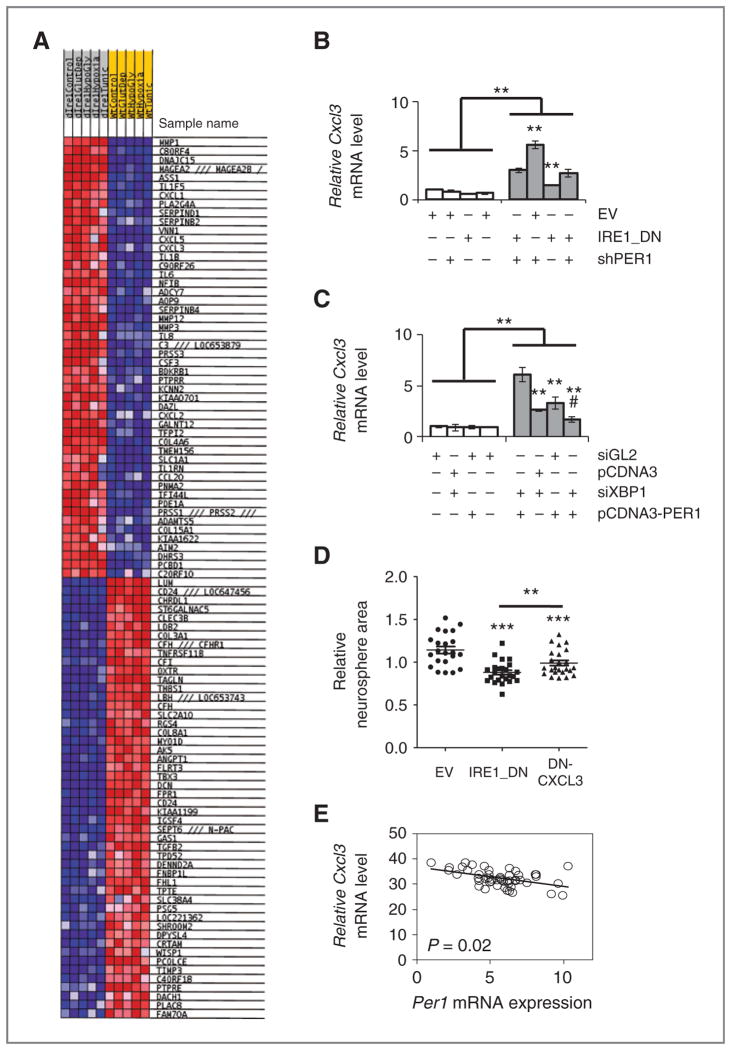
Next, we sought to define mediating IRE1α/PER1 axis, to this end we identified the genes controlled by this axis and potentially involved in the control of tumor growth/angiogenesis. mRNA expression profiles in IRE1_DN and subjected or not to known endoplasmic reticulum stress inducers such as glucose or glutamine deprivation, hypoxia or Tun exposure were compared with those obtained in empty vector cells. These experimental conditions are also known to recapitulate microenvironmental stresses. We identified the top 50 genes up and downregulated all conditions included (Fig. 6A). As PER1 has been defined as a transcriptional repressor, we focused our attention on the genes downregulated in IRE1_DN cells, which were found to be enriched in both cytokine–cytokine receptor interaction and chemokine signaling pathways. To further explore the IRE1α/PER1–dependent cytokine and chemokine regulatory networks upon endoplasmic reticulum stress, validation of potential target genes was carried out using RT-qPCR. The proangiogenic chemokine CXCL3 was the most significant gene that showed restoration of its expression in IRE1_DNsh-PER1 cells upon glucose deprivation (Fig. 6B). We postulated that this might be due to the coordinated regulation of CXCL3 expression by XBP1 that was already proposed by Gargalovic and colleagues (27) and the downregulation of PER1, which in this context would play a repressor role. As anticipated from our model, U87 cells transiently silenced for XBP1 and/or overexpressing PER1 and exposed or not to endoplasmic reticulum stress induced by Glc deprivation showed an attenuation of CXCL3 mRNA expression increase mediated by Glc deprivation (Fig. 6C). This shows that XBP1s increase and PER1 down-regulation both contribute to the regulation of CXCL3 mRNA expression. Hence, integrated IRE1α signaling specifically controls chemokine expression upon stress.
Figure 6.
Relevance of IRE1 signaling in cancer. A, heatmap for microarray results. Blue, upregulation; Red, downregulation. B, U87 cells-expressing or not functional IRE1α and silenced or not for PER1 were starved or not of glucose for 16 hours in presence of dialyzed serum; CXCL3 mRNA abundance was measured by qPCR. Messenger RNA levels were normalized to those of RPLP0 and to untreated control. Error bars represent the SDs of at least 3 independent experiments. (t test, NS, nonsignificant; *, P < 0.05). C, U87 cells were transfected with siXBP1 for 72 hours and/or with pcDNA3PER1 for 24 hours before glucose deprivation for additional 16 hours. Total RNA was purified from these cells and analyzed by qRT-PCR for CXCL3 expression (using GAPDH as internal reference). The experiment was carried out in triplicate and is presented as the mean ± SD. Statistical significance was determined using the Student t test. *, P < 0.06; ***, P < 0.01; #, P < 0.03. D, sixty-two human glioblastoma samples (24 Bordeaux cohort; 38 Mayo cohort) were analyzed for Per1 and Cxcl3 mRNA expression using qRT-PCR. Three technical replicates were conducted. Data indicate a negative correlation (slope = −0.75) with statistical significance (P = 0.0208).
Low levels of PER1 gene correlates with poor survival in patients
To investigate whether the IRE1α/PER1 axis was of clinical relevance, human glioblastoma samples from 2 independent sources (CHU Bordeaux and Mayo Clinic) were analyzed for PER1 mRNA expression by qPCR. This revealed that both cohorts presented an expression of PER1 mRNA lower in tumors than that observed in normal or nontumoral tissues (red/Bordeaux; black/Mayo, Fig. 5B). To evaluate the impact of low PER1 expression on prognosis, postsurgery survival of 60 patients with glioblastoma was followed (Fig. 5C). These patients were classified into 2 groups in which PER1 expression was either lower or higher than average PER1 mRNA. The low PER1 group contained 31 patients and the high PER1 group contained 29 patients. Interestingly, high PER1 expression significantly correlated with increased survival with a median of 599 days compared with 411 days in the low PER1 group (P = 0.03; Fig. 5C). This result is in agreement with those obtained in in vitro and in vivo tumor models showing that PER1 differentially affects patient outcome and strongly supports a specific role for the IRE1α/PER1 axis in the pathogenesis of glioblastoma. To confirm that low PER1 levels are associated with high IRE1α activity, we investigated whether the tumors contained the translation product of Xbp1s mRNA (XBP1s protein). To this end, paraffin-embedded specimens from 20 other glioblastoma samples (Supplementary Table S3) were analyzed for the presence of XBP1s protein using immunohistochemistry with a monoclonal antibody specific to XBP1s (Supplementary Fig. S5 and Supplementary Table S3; 21). XBP1s protein expression was then correlated with patient survival. The data were represented as Kaplan–Meier plots discriminating between 2 populations that exhibited either a positive or a negative XBP1s staining (Fig. 5D). This showed that the absence of/low XBP1s staining significantly correlated with enhanced survival. Together with data obtained with the expression of PER1 mRNA, these results suggested that IRE1α signaling activation in glioblastoma correlated with low patient survival. Finally, we tested the relevance of CXCL3 to glioblastoma outcome using the Rembrandt database (28). High expression of CXCL3 mRNA seemed to correlate with low patient survival (not shown). Interestingly and as expected, in our cohort of human tumors, expression of CXCL3 mRNA negatively correlated with that of PER1 mRNA (Fig. 6D). This further reinforced the instrumental role of an IRE1α-dependent pathway in tumor aggressiveness. Taken together, these data further support a specific and important role for IRE1α signaling in human glioblastoma and show that PER1 is a genuine signaling intermediate in glioblastoma progression. Moreover, these results suggest that IRE1α may be a suitable therapeutic target for patients with this disease.
Discussion
Our results identify PER1 mRNA as a novel and atypical substrate (coding for a cytosolic/nuclear protein) of RIDD contributing to cancer development. As PER1 is a core gene of the circadian clock, our observation could also be placed in the perspective of a previous report (29) that shows the significance of the UPR/circadian clock connection in the control of hepatic metabolism. In addition, another study showed that a connection between the eIF2α-dependent transcription factor ATF4 and the circadian clock transcription systems plays an important role in multidrug resistance in tumor cells (30). In this context, IRE1α-mediated PER1 mRNA decay could also represent another pathway in the well-described posttranscriptional regulation mechanisms of the circadian clock (31), whose relevance to cancer still remains to be investigated.
The findings included in this report show a direct clinical relevance of this newly identified IREα/Per1 axis as we have determined that PER1 mRNA may be a useful marker for predicting patient survival (Fig. 5). In addition to its role in the control of the circadian clock, PER1 has also directly been involved in cell stress response, through interactions with ATM and Chk2 to participate in γ-irradiation–induced apoptosis (11). Moreover, low PER1 mRNA expression was observed in a variety of cancers (10, 11, 32, 33), thereby suggesting its involvement in cancer development. Our data show that low PER1 together with high XBP1s expression are significantly associated with lower glioma patient survival. These observations point toward an instrumental role of IRE1α in glioma development. This was also supported by the suggested driver role of IRE1α mutations in cancer including glioma (3), however, the functional implication of these mutations remains to be shown.
IRE1α RNAse-dependent signaling in tumor could on the one hand regulate XBP1 mRNA splicing thereby leading to previously shown induction of proinflammatory cytokines as reported in many instances (27, 34–36) and in parallel enhance this effect by repressing PER1 expression, which in turn could act as derepression of cytokine expression, as illustrated for other core circadian genes (37–39). Mutually exclusive phenotypic changes in glioma observed upon impairement of IRE1α signaling from massive/angiogenic to diffuse/avascular could either result from the IRE1α-dependent activation of a cell-autonomous proinflammatory/angiogenic phenotype or from the coordinated posttranscriptional stabilization of specific mRNA (RIDD substrates), thereby leading to major changes in tumor cell–microenvironment interactions.
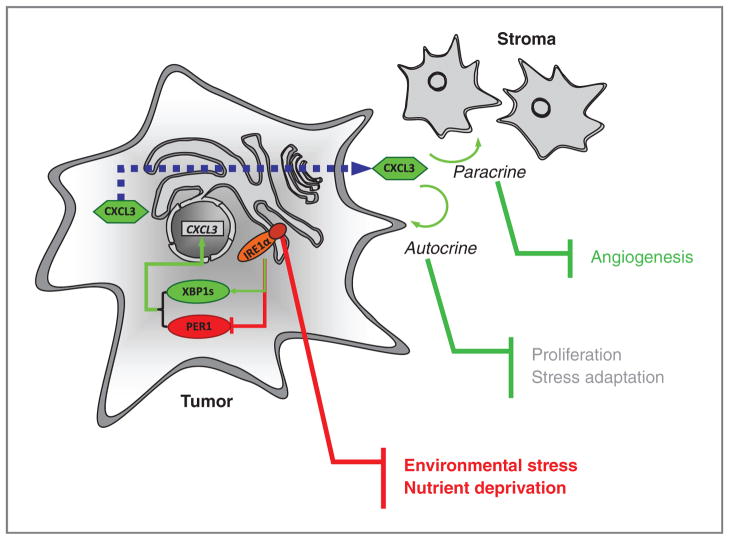
As such, our study shows that CXCL3 mRNA is per se an IRE1α/XBP1s/PER1-dependent target in our model as determined in Fig. 6. These data are also consistent with the emerging role of CXCL3 as a key player in cancer development (27, 40) that also applies to glioblastoma (41, 42). Consequently, we identify here an IRE1α-dependent mechanism that coincidentally activates XBP1 mRNA splicing and PER1 mRNA decay. This provides a molecular link between IRE1α activation and tumor cell adaptation, and directly links IRE1α activity to proinflammatory/angiogenic phenotypes (Fig. 7).
Figure 7.
Schematic representation of the IRE1α-dependent activation loop that controls tumor cell adaptation. Tumor cell is presented in light gray and stromal cells in dark gray. Proteins are represented by circles; green, upregulation; red, downregulation. Connections following stress-mediated activation of IRE1α are presented in green for activation and red for inhibition. The dashed blue line represents the traffic of CXCL3 protein through the secretory pathway.
Taken together, these data further support a specific and important role for IRE1α signaling in human glioblastoma and show that PER1 is a genuine signaling intermediate in glioblastoma progression. Moreover, these results suggest that IRE1α may constitute a suitable therapeutic target for patients with this disease. As a consequence, this suggests that controlling the interplay between UPR signaling and the circadian clock component might also be a suitable strategy to slow down cancer progression; our results may consequently define a model for novel therapeutic option for cancers.
Supplementary Material
Acknowledgments
The authors thank M. Moenner (Université Bordeaux 1, Bordeaux, France) for precious help and fruitful discussions, S. Manié (UMR CNRS 5286, INSERM 1052, Cancer Research Center of Lyon, Lyon, France), and the Chevet lab for critical reading of the manuscript. The authors also thank Dr S. Gery (University of California, Los Angeles, CA) for the gift of pcDNA3.1-hPER1 expression vector and Dr U. Albrecht (Freiburg, Switzerland) for providing us with the pPER1-Luc vector.
Grant Support
This work was supported by an Avenir program (INSERM), grants from the Institut National du Cancer (INCa), Ligue contre le cancer, a Marie Curie International Reintegration Grant (E. Chevet), a grant from the Mayo Clinic Cancer Centre (M.E. Fernandez-Zapico), a grant from Institut Fédératif de Recherche 66 (O. Pluquet). O. Pluquet was supported by fellowships from INSERM and Association pour la Recherche contre le Cancer. M. Bouchecareilh was supported from a fellowship from le Conseil Régional d’Aquitaine and la Fondation pour la Recherche Française (FRM). Human glioblastoma samples were collected through the Bordeaux Tumor Bank (JP Merlio, CHU Bordeaux, France) funded by the Cancéropôle Grand Sud-Ouest and by a CEREPEG project grant (PHRC 2003; H. Loiseau) or through the Mayo Clinic Department of Clinical Pathology and funded by the Mayo Clinic SPORE in Brain Cancer P50 CA108961 (Rochester).
Footnotes
Disclosure of Potential Conflicts of Interest
J.N. Sarkaria has a commercial research grant from Genentech, Basilea, Sanofi, and Merck. E. Chevet has a commercial research grant from Servier. No potential conflicts of interest were disclosed by the other authors.
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors’ Contributions
Conception and design: O. Pluquet, N. Dejeans, M.E. Fernandez-Zapico, E. Chevet
Development of methodology: O. Pluquet, N. Dejeans, M. Bouchecareilh, R. Pineau, A. Higa, S. Loriot, S. Taouji, F. Saltel, E. Chevet
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): O. Pluquet, S. Lhomond, R. Pineau, A. Higa, C. Combe, N. Dugot-Senant, A. Vital, H. Loiseau, J.N. Sarkaria, F.J. Rodriguez, E. Chevet
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): O. Pluquet, N. Dejeans, A. Vital, S.J.C. Gosline, S. Taouji, F. Saltel, M. Hallett, K. Anderson, W. Wu, E. Chevet
Writing, review, and/or revision of the manuscript: O. Pluquet, N. Dejeans, M. Bouchecareilh, A. Higa, A. Vital, J.N. Sarkaria, W. Wu, F.J. Rodriguez, J. Rosenbaum, M.E. Fernandez-Zapico, E. Chevet
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): M. Bouchecareilh, R. Pineau, M. Delugin, G. Cubel, F.J. Rodriguez
Study supervision: N. Dejeans, E. Chevet
References
- 1.Moenner M, Pluquet O, Bouchecareilh M, Chevet E. Integrated endoplasmic reticulum stress responses in cancer. Cancer Res. 2007;67:10631–4. doi: 10.1158/0008-5472.CAN-07-1705. [DOI] [PubMed] [Google Scholar]
- 2.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–89. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 3.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bifulco G, Miele C, Di Jeso B, Beguinot F, Nappi C, Di Carlo C, et al. Endoplasmic reticulum stress is activated in endometrial adenocarcinoma. Gynecol Oncol. 2012;125:220–5. doi: 10.1016/j.ygyno.2011.11.045. [DOI] [PubMed] [Google Scholar]
- 5.Davies MP, Barraclough DL, Stewart C, Joyce KA, Eccles RM, Barraclough R, et al. Expression and splicing of the unfolded protein response gene XBP-1 are significantly associated with clinical outcome of endocrine-treated breast cancer. Int J Cancer. 2008;123:85–8. doi: 10.1002/ijc.23479. [DOI] [PubMed] [Google Scholar]
- 6.Dejeans N, Glorieux C, Guenin S, Beck R, Sid B, Rousseau R, et al. Overexpression of GRP94 in breast cancer cells resistant to oxidative stress promotes high levels of cancer cell proliferation and migration: implications for tumor recurrence. Free Radic Biol Med. 2012;52:993–1002. doi: 10.1016/j.freeradbiomed.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 7.Papandreou I, Denko NC, Olson M, Van Melckebeke H, Lust S, Tam A, et al. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117:1311–4. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romero-Ramirez L, Cao H, Nelson D, Hammond E, Lee AH, Yoshida H, et al. XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res. 2004;64:5943–7. doi: 10.1158/0008-5472.CAN-04-1606. [DOI] [PubMed] [Google Scholar]
- 9.Romero-Ramirez L, Cao H, Regalado MP, Kambham N, Siemann D, Kim JJ, et al. X box-binding protein 1 regulates angiogenesis in human pancreatic adenocarcinomas. Transl Oncol. 2009;2:31–8. doi: 10.1593/tlo.08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ, Chang JG. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26:1241–6. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- 11.Gery S, Koeffler HP. The role of circadian regulation in cancer. Cold Spring Harb Symp Quant Biol. 2007;72:459–64. doi: 10.1101/sqb.2007.72.004. [DOI] [PubMed] [Google Scholar]
- 12.Bouchecareilh M, Caruso ME, Roby P, Parent S, Rouleau N, Taouji S, et al. AlphaScreen-based characterization of the bifunctional kinase/RNase IRE1alpha: a novel and atypical drug target. J Biomol Screen. 2010;15:406–17. doi: 10.1177/1087057110363823. [DOI] [PubMed] [Google Scholar]
- 13.Bouchecareilh M, Higa A, Fribourg S, Moenner M, Chevet E. Peptides derived from the bifunctional kinase/RNase enzyme IRE1{alpha} modulate IRE1{alpha} activity and protect cells from endoplasmic reticulum stress. FASEB J. 2011;25:3115–29. doi: 10.1096/fj.11-182931. [DOI] [PubMed] [Google Scholar]
- 14.Drogat B, Auguste P, Nguyen DT, Bouchecareilh M, Pineau R, Nalbantoglu J, et al. IRE1 signaling is essential for ischemia-induced vascular endothelial growth factor-A expression and contributes to angiogenesis and tumor growth in vivo. Cancer Res. 2007;67:6700–7. doi: 10.1158/0008-5472.CAN-06-3235. [DOI] [PubMed] [Google Scholar]
- 15.Auf G, Jabouille A, Guerit S, Pineau R, Delugin M, Bouchecareilh M, et al. Inositol-requiring enzyme 1alpha is a key regulator of angiogenesis and invasion in malignant glioma. Proc Natl Acad Sci U S A. 2010;107:15553–8. doi: 10.1073/pnas.0914072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen DT, Kebache S, Fazel A, Wong HN, Jenna S, Emadali A, et al. Nck-dependent activation of extracellular signal-regulated kinase-1 and regulation of cell survival during endoplasmic reticulum stress. Mol Biol Cell. 2004;15:4248–60. doi: 10.1091/mbc.E03-11-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui W, Taub DD, Gardner K. qPrimerDepot: a primer database for quantitative real time PCR. Nucleic Acids Res. 2007;35:D805–9. doi: 10.1093/nar/gkl767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Motzkus D, Maronde E, Grunenberg U, Lee CC, Forssmann W, Albrecht U. The human PER1 gene is transcriptionally regulated by multiple signaling pathways. FEBS Lett. 2000;486:315–9. doi: 10.1016/s0014-5793(00)02315-2. [DOI] [PubMed] [Google Scholar]
- 21.Bouchecareilh M, Marza E, Caruso ME, Chevet E. Small GTPase signaling and the unfolded protein response. Methods Enzymol. 2011;49:343–60. doi: 10.1016/B978-0-12-385928-0.00019-5. [DOI] [PubMed] [Google Scholar]
- 22.Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–24. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welihinda AA, Tirasophon W, Green SR, Kaufman RJ. Protein serine/threonine phosphatase Ptc2p negatively regulates the unfolded-protein response by dephosphorylating Ire1p kinase. Mol Cell Biol. 1998;18:1967–77. doi: 10.1128/mcb.18.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, et al. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–64. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–31. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iqbal J, Dai K, Seimon T, Jungreis R, Oyadomari M, Kuriakose G, et al. IRE1beta inhibits chylomicron production by selectively degrading MTP mRNA. Cell Metab. 2008;7:445–55. doi: 10.1016/j.cmet.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gargalovic PS, Gharavi NM, Clark MJ, Pagnon J, Yang WP, He A, et al. The unfolded protein response is an important regulator of inflammatory genes in endothelial cells. Arterioscler Thromb Vasc Biol. 2006;26:2490–6. doi: 10.1161/01.ATV.0000242903.41158.a1. [DOI] [PubMed] [Google Scholar]
- 28.Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7:157–67. doi: 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cretenet G, Le Clech M, Gachon F. Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab. 2010;11:47–57. doi: 10.1016/j.cmet.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 30.Igarashi T, Izumi H, Uchiumi T, Nishio K, Arao T, Tanabe M, et al. Clock and ATF4 transcription system regulates drug resistance in human cancer cell lines. Oncogene. 2007;26:4749–60. doi: 10.1038/sj.onc.1210289. [DOI] [PubMed] [Google Scholar]
- 31.Kojima S, Shingle DL, Green CB. Post-transcriptional control of circadian rhythms. J Cell Sci. 2011;124:311–20. doi: 10.1242/jcs.065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin YM, Chang JH, Yeh KT, Yang MY, Liu TC, Lin SF, et al. Disturbance of circadian gene expression in hepatocellular carcinoma. Mol Carcinog. 2008;47:925–33. doi: 10.1002/mc.20446. [DOI] [PubMed] [Google Scholar]
- 33.Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–64. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 34.Kaser A, Martinez-Naves E, Blumberg RS. Endoplasmic reticulum stress: implications for inflammatory bowel disease pathogenesis. Curr Opin Gastroenterol. 2012;26:318–26. doi: 10.1097/MOG.0b013e32833a9ff1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miani M, Colli ML, Ladriere L, Cnop M, Eizirik DL. Mild endoplasmic reticulum stress augments the proinflammatory effect of IL-1beta in pancreatic rat beta-cells via the IRE1alpha/XBP1s pathway. Endocrinology. 2010;153:3017–28. doi: 10.1210/en.2011-2090. [DOI] [PubMed] [Google Scholar]
- 36.Villeneuve J, Lepreux S, Mulot A, Berard AM, Higa-Nishiyama A, Costet P, et al. A protective role for CD154 in hepatic steatosis in mice. Hepatology. 2010;52:1968–79. doi: 10.1002/hep.23935. [DOI] [PubMed] [Google Scholar]
- 37.Hashiramoto A, Yamane T, Tsumiyama K, Yoshida K, Komai K, Yamada H, et al. Mammalian clock gene Cryptochrome regulates arthritis via proinflammatory cytokine TNF-alpha. J Immunol. 2011;184:1560–5. doi: 10.4049/jimmunol.0903284. [DOI] [PubMed] [Google Scholar]
- 38.Lee JH, Sancar A. Regulation of apoptosis by the circadian clock through NF-kappaB signaling. Proc Natl Acad Sci U S A. 2012;108:12036–41. doi: 10.1073/pnas.1108125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narasimamurthy R, Hatori M, Nayak SK, Liu F, Panda S, Verma IM. Circadian clock protein cryptochrome regulates the expression of proinflammatory cytokines. Proc Natl Acad Sci U S A. 2012;109:12662–7. doi: 10.1073/pnas.1209965109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44CD24 stem cell-like breast cancer cells in human tumors. J Clin Invest. 2011;121:2723–35. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bruyere C, Mijatovic T, Lonez C, Spiegl-Kreinecker S, Berger W, Kast RE, et al. Temozolomide-induced modification of the CXC chemokine network in experimental gliomas. Int J Oncol. 2011;38:1453–64. doi: 10.3892/ijo.2011.964. [DOI] [PubMed] [Google Scholar]
- 42.Kammerer R, Buchner A, Palluch P, Pongratz T, Oboukhovskij K, Beyer W, et al. Induction of immune mediators in glioma and prostate cancer cells by non-lethal photodynamic therapy. PLoS ONE. 2011;6:e21834. doi: 10.1371/journal.pone.0021834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.