Abstract
The aryl hydrocarbon receptor (AHR) is a member of the basic-helix-loop-helix/Per-ARNT-Sim (bHLH-PAS) family of transcription factors and has diverse roles in development, physiology, and environmental sensing in bilaterian animals. Studying the expression of conserved genes and function of proteins in outgroups to protostomes and deuterostomes assists in understanding the antiquity of gene function and deciphering lineage-specific differences in these bilaterian clades. We describe the developmental expression of AHR from the sea anemone Nematostella vectensis and compare its expression with three other members of the bHLH-PAS family (AHR nuclear translocator (ARNT), Cycle, and a proto-Single-Minded/Trachaeless). NvAHR expression was highest early in the larval stage with spatial expression in the basal portion of the ectoderm that became increasingly restricted to the oral pole with concentrated expression in tentacles of the juvenile polyp. The other bHLH-PAS genes showed a divergent expression pattern in later larval stages and polyps, in which gene expression was concentrated in the aboral end, with broader expression in the endoderm later in development. In co-immunoprecipitation assays, we found no evidence for heterodimerization of AHR with ARNT, contrary to the conservation of this specific interaction in all bilaterians studied to date. Similar to results with other invertebrate AHRs but in contrast to vertebrate AHRs, NvAHR failed to bind two prototypical xenobiotic AHR ligands (TCDD, BNF). Together, our data suggest that AHR's original function in Eumetazoa likely involved developmental patterning, potentially of neural tissue. The role of heterodimerization in the function of AHR may have arisen after the cnidarian-bilaterian ancestor. The absence of xenobiotic binding to NvAHR further supports a hypothesis for a derived role of this protein in chemical sensing within the chordates.
Keywords: aryl hydrocarbon receptor, development, heterodimer, Nematostella vectensis
Introduction
The recent sequencing of genomes of early diverging metazoans has revealed conservation of many of the genes that direct developmental processes in bilaterians (Putnam et al. 2007; Srivastava et al. 2008; Srivastava et al. 2010). The existence of a common genomic toolkit in widely divergent metazoans provides an opportunity to study the evolution and function of developmental regulatory genes and pathways through studies of representatives from these “basal” lineages (Erwin 2009; Technau and Steele 2011). As a member of the phylum Cnidaria, the sea anemone Nematostella vectensis has emerged as a powerful species for studying genomics and development to better characterize the ancestral eumetazoan (Darling et al. 2005; Technau and Steele 2011). Many of the studies with N. vectensis and other cnidarians have targeted gene families with roles in early developmental patterning and tissue specification; e.g., Hox (Ryan et al. 2007), Wnt (Kusserow et al. 2005), Sox (Magie et al. 2005). These and others have demonstrated that much of the diversification of transcription factor families occurred prior to the divergence of cnidarians and bilaterians. In addition, many genes in these distantly related animal groups exhibit broadly similar expression territories of mRNA, which is consistent with conserved regulatory functions. More recently, data on protein-protein and protein-DNA interactions have similarly shown that molecular interactions are conserved in cnidarians and bilaterians (Kumburegama et al. 2011; Wolenski et al. 2011), further supporting a hypothesis for ancient, conserved functions that were present in the cnidarian-bilaterian ancestor.
The basic-helix-loop-helix/Per-ARNT-Sim (bHLH-PAS) family is a large group of transcriptional regulatory proteins. The defining features of the bHLH-PAS family are two domains, which share evolutionarily conserved structures and functions among the various family members. The bHLH domain contains basic and helix-loop-helix motifs involved in protein-DNA and protein-protein interactions, respectively. The PAS domain typically contains two PAS folds—ancient protein structures that occur in bacteria, archaea, and eukarya (Gu et al. 2000); this domain forms a secondary dimerization surface for heteromeric interactions among some bHLH-PAS proteins. The bHLH-PAS family includes transcriptional activators and repressors that in bilaterian animals play key roles in developmental signaling and environmental homeostasis (Taylor and Zhulin 1999). Well-studied metazoan bHLH-PAS genes include the circadian clock regulators Period and Clock, the hypoxia-inducible factors (HIF-1α, HIF-2α, HIF-3a, Sima in Drosophila) and other hypoxia-responsive transcription factors (e.g., Trachealess (Trh)), and the aryl hydrocarbon receptor (Ahr).
bHLH-PAS proteins have been divided into α, β, and γ classes based on their functional interactions (Gu et al. 2000). α-class (“sensor”) bHLH-PAS proteins, such as AHR, HIF, and CLOCK, typically act as heterodimers with the β–class (“partner”) bHLH-PAS proteins ARNT/ARNT2 (TANGO in Drosophila, AHA-1 in C. elegans) or BMAL1/BMAL2 (CYCLE in Drosophila). Specific heterodimer combinations modulate the DNA binding characteristics for bHLH-PAS proteins; ARNT and BMAL interact with the GTG portion of a DNA recognition sequence and the sensor specifies the other half of the sequence (Powell-Coffman et al. 1998). The interactions of γ -class (“coactivator”) bHLH-PAS proteins are not as well understood but these proteins modulate the activity of transcription factors, including bHLH-PAS proteins and nuclear receptors, through protein-protein interactions.
A phylogenetic study of the diversification of the bHLH superfamily in animals showed that N. vectensis has 68 bHLH genes that represent 28-32 of the 45 classified families (Simionato et al. 2007). However, the expression and protein interactions have been reported for only a few of these genes, none of which are bHLH-PAS members. Two bHLH transcription factors, twist (Martindale et al. 2004) and COE (Pang et al. 2004), are expressed in discrete spatial domains in embryos and larvae, consistent with roles in endodermal and neural patterning, respectively. Developmental expression coupled with transient suppression and overexpression of N. vectensis members in the achaete-scute group of bHLH genes supported a role for these transcription factors in the neurogenic pathway (Layden et al. 2012). The only bHLH-PAS genes that were investigated in N. vectensis are Cycle and Clock in a study of their expression in relationship to light-dark periods (Reitzel et al. 2010). Also in this study, Reitzel et al. reported the conserved heterodimerization of these two proteins, supporting an ancient origin for a critical protein-protein interaction in the transcriptional feedback loops of the animal circadian clock. Given that the N. vectensis genome contains several bHLH-PAS genes, there is great interest in understanding their expression patterns and the possibility of conserved or partially conserved functions in comparison to their bilaterian counterparts.
The aryl hydrocarbon receptor (AHR) and AHR nuclear translocator (ARNT), both of which have orthologs in N. vectensis, are two members of the bHLH-PAS family with diverse roles in development and physiology in bilaterian animals. Recent comparative genomic studies have identified bHLH-PAS genes in early diverging animals (Larroux et al. 2008; Simionato et al. 2007) and an unicellular outgroup, Capsaspora owczarzaki (Sebé-Pedrós et al. 2011). N. vectensis, as a member of the Cnidaria, represents the first animal lineage with supported orthologs of both AHR and ARNT, while placozoans have an ortholog of AHR but not ARNT, and sponges and ctenophores (Reitzel and Peres, unpublished data) lack orthologs of either transcription factor. In bilaterians, AHR and ARNT form a protein heterodimer that regulates gene expression through activation or repression of specific target genes.
The best-characterized role of the AHR-ARNT dimer is in mediating the adaptive and maladaptive responses to toxicants in diverse vertebrates (Denison and Nagy 2003; Okey 2007). In this context, AHR regulates expression of xenobiotic- and endobiotic-metabolizing enzymes such as cytochrome P450s (CYP1A, CYP1B, CYP2S). Binding of ligand to the AHR stimulates nuclear translocation of the AHR complex and association of the ligand-bound AHR with ARNT (Hoffman et al. 1991; Reyes et al. 1992). Activation of gene transcription by AHR-ARNT complexes occurs through interactions with AHR-responsive enhancer elements (AHREs, also called DREs or XREs (Denison et al. 1989; Swanson et al. 1995), located in the promoter of dioxin-responsive genes. AHRs from diverse protostomes do not bind prototypical ligands (Butler et al. 2001); thus, the function of AHR as a sensor for environmental toxicants may be restricted to chordates.
Although most historical studies of AHR function have focused on its role in mediating toxicant response, studies in vertebrates and a variety of invertebrate species have shown diverse developmental and physiological roles of AHR, functions that perhaps are more broadly conserved and representative of the ancestral functions of this protein (Hahn 2002; McMillan and Bradfield 2007). In vertebrates, AHR has roles in the regulation of vascular and gonadal development during fetal development and lymphocyte differentiation as part of the adaptive immune system. Studies in the nematode C. elegans revealed developmental and sensory roles for the AHR ortholog, designated AHR-1 (Powell-Coffman et al. 1998). AHR-1 is expressed in a specific subset of neurons (28 out of the 302 C. elegans neurons), including touch receptor neurons, neurons that contact the pseudocoelomic fluid, and neurons in the pharynx (Qin and Powell-Coffman 2004). AHR-1 loss-of-function mutants exhibit altered neuronal differentiation leading to defective migration and morphology of touch receptor neurons and GABAergic motor neurons (Huang et al. 2004; Qin and Powell-Coffman 2004; Smith et al. 2013; Zhang et al. 2013). In addition to its developmental roles in C. elegans, AHR-1 also continues to function in adult neurons, playing an important role in determining social feeding behavior through regulation of soluble guanylate cyclases (Qin et al. 2006). Evidence suggests that AHR-1 functions together with AHA-1, the C. elegans ortholog of ARNT (Huang et al. 2004). The Drosophila gene spineless (ss) is the ortholog of vertebrate AHR (Duncan et al. 1998). Null mutants of ss in Drosophila display defects in the development of the arista, or distal segment of the antennae (an olfactory organ), as well as deletion of the distal leg segments (Duncan et al. 1998). More recently, it was demonstrated that ss controls photoreceptor specification during development of the Drosophila compound eye; stochastic expression of ss determines the type of rhodopsin that is expressed in the photoreceptor cells, which in turn controls color sensitivity (Wernet et al. 2006). In addition, ss regulates dendrite morphology in a group of sensory neurons known as “dendritic arborization” neurons (Kim et al. 2006). Thus, Drosophila ss displays at least three distinct developmental roles, all related to sensory systems.
Collectively, the above research in vertebrates and invertebrates indicates that the fundamental molecular interactions among AHR and ARNT are evolutionarily conserved in the Bilateria and support the idea that the ancestral role of AHR is likely as a developmental regulator. By testing for conserved and divergent expression of AHR, ARNT and other bHLH-PAS genes in N. vectensis, we can begin to assess whether the expression and molecular interactions of these genes support a hypothesis for more ancient origins, and thus, provide a more complete understanding of the evolution of these transcription factors in animal evolution.
Methods
bHLH-PAS gene annotation
Previous studies have shown that N. vectensis has predicted representatives for most families in the bHLH superfamily, including bHLH-PAS. AHR was previously identified by Reitzel et al. (2008). Other bHLH-PAS genes, including ARNT, Cycle, HIF, and Sim/Trh, have also been identified (Simionato et al. 2007). These previous gene descriptions and phylogenetic analyses have relied on predicted genes with varying degrees of coverage and confirmation. We annotated four bHLH-PAS genes (AHR, ARNT, Cycle, and Sim/Trh) from N. vectensis. For each gene we searched available EST sequences at the Joint Genome Institute (JGI) and NCBI using the predicted sequences for each gene as a query. Overlapping sequences were assembled in silico to produce the most complete sequences. Large portions of each transcript – in some cases full-length transcripts (see below) - were confirmed with PCR from cDNA synthesized from RNA extracted from developmental stages using standard methods.
Developmental expression
We quantified temporal patterns of developmental expression of AHR, ARNT, Cycle, and Sim/Trh with quantitative RT-PCR (qPCR) and determined spatial expression during development with whole-mount in situ hybridization. For qPCR, we followed previously published methods (Reitzel and Tarrant 2009). Briefly, large portions of each transcript were amplified with gene specific primers (see Supplemental Table 1 for all primers), gel purified, and cloned into pGEM-T Easy (Promega). Cloned regions were confirmed by sequencing. To quantify expression, we designed primers to amplify short amplicons (76 – 102 bp) within the region cloned from each transcript with minimal predicted secondary structure (m-fold, (Zuker 2003)) that spanned one or more introns. We constructed a standard curve from serially-diluted plasmids to calculate the number of molecules per reaction. 18S ribosomal RNA was used as a control gene for expression because it represents a large proportion of total RNA and has little variation in expression in developmental stages of N. vectensis (Reitzel and Tarrant 2009). Gene expression was assayed on five developmental stages (embryo, early and late larva, juvenile, adults) with three biological replicates per stage. Expression between stages was statistically compared using one-way ANOVA with Tukey-Kramer for posthoc comparisons (JMP v.5).
Spatial expression of the four bHLH-PAS transcripts was compared with whole mount in situ hybridization using previously published methods (Martindale et al. 2004). Riboprobes were synthesized from cloned fragments (AHR: 1310 bp, ARNT: 823 bp, Cycle: 696 bp, Sim/Trh: 852 bp). Hybridization of all digoxigenin-labeled anti-sense probes was conducted at 62°C. Following NBT/BCIP staining, embryos and polyps were washed in hybridization buffer at 62°C to reduce background (Supp. Fig. 1), and were subsequently taken through a dilution series in PBS with 0.1% Tween-20, and then cleared in 80% glycerol. NBT/BCIP stained embryos and polyps were imaged under bright field Nomarski optics with a Zeiss Axiocam HR digital camera mounted on a Zeiss Axioskop 2 mot plus compound microscope. Expression was determined for embryo, larva, and early polyp developmental stages (Stefanik et al. 2013).
Co-immunoprecipitation
Using the in silico transcript annotations, we amplified full length products for AHR, ARNT, and Cycle. Full length AHR was cloned into pGEM-T Easy (Promega) and subcloned into pcDNA3.1 (Invitrogen) with EcoRI and ApaI restriction sites. Expression constructs for ARNT and Cycle labeled with a C-terminal V5 tag (pcDNA3.2/V5-DEST, Invitrogen) were previously reported (Reitzel et al. 2010). In vitro co-immunoprecipitation assays were conducted as previously described (Reitzel et al. 2010). Briefly, N. vectensis AHR, ARNT, and CYCLE proteins were synthesized by in vitro transcription and translation (IVTT; TnT quick coupled in vitro transcription/translation system, Promega) with or without [35S]-Met. [35S]-labeled AHR was combined and incubated with unlabeled ARNT and CYCLE. The mixture was precleared with normal mouse IgG and protein A agarose to reduce nonspecific binding. An antibody to V5 (Invitrogen) or IgG (negative control) was used to pull down the putative AHR/ARNT or AHR/CYCLE heterodimer complex. After several washes, the bound proteins were recovered from the beads by boiling and separated by polyacrylamide gel electrophoresis. The [35S]-labeled proteins were detected using fluorography. As a positive control, we used a similar protocol with human AHR and ARNT. [35S]-labeled human ARNT was incubated with unlabeled human AHR and the complex was immunoprecipitated with an antibody against human AHR (SA-210; Biomol, Plymouth Meeting, PA). The incubations included the addition of 1 μl of 0.2 μM (final concentration 10 nM) 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) to facilitate a more robust heterodimerization. We tested for a similar effect of TCDD on dimerization of N. vectensis AHR and ARNT.
AHR ligand binding assay by velocity sedimentation on sucrose gradients
We tested for potential ligand binding by NvAHR by using velocity sedimentation on sucrose gradients, the most sensitive assay for detecting specific binding of radioligands to AHRs (Hahn and Hestermann 2007). This method has been successfully applied to measure [3H]TCDD and [3H]β-naphthoflavone BNF) binding to AHRs from a variety of species (Butler et al. 2001; Karchner et al. 2006; Karchner et al. 1999). AHR from N. vectensis was synthesized with IVTT using the TnT in vitro transcription/translation system (Promega). IVTT-expressed AHR protein was incubated with 2 nM [3H]TCDD or 20 nM [3H]BNF, and analyzed using methods described earlier (Butler et al. 2001). The concentrations of radioligands were selected to match those shown previously to produce substantial and easily measureable levels of specific binding to vertebrate AHRs (Butler et al. 2001; Karchner et al. 1999). As positive controls, we analyzed binding by AHR1 from the Atlantic killifish Fundulus heteroclitus and AHR from mouse. Peaks corresponding to the bound ligand for each AHR protein were compared with unprogrammed lysates (UPL, TnT products produced using an empty expression construct), which served as the negative control.
Results
bHLH-PAS gene annotation
Nematostella vectensis AHR is encoded by a transcript whose coding sequence is 2,370 bp in length composed of 13 exons, which span a total of 12,637 bp of genomic DNA. Using the in silico assembly of the longest gene product, we annotated 230 bp of 5′ UTR and 479 bp of 3′ UTR, including a predicted polyadenylation signal sequence located 391 bp from the stop codon.
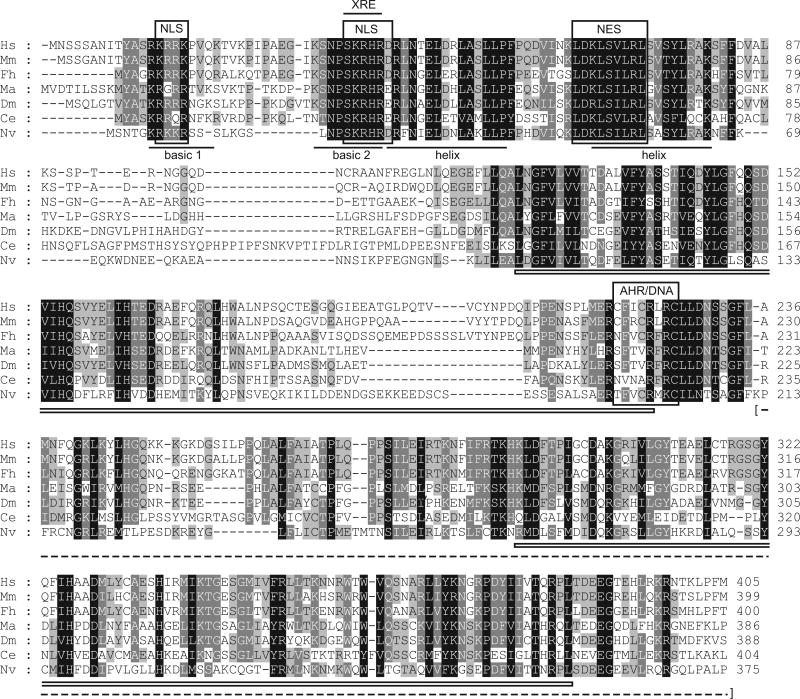
NvAHR protein contains well-conserved bHLH and PAS domains with high similarity to those of bilaterian AHR orthologs (Fig. 1). The least conserved region in these domains was the segment between the two basic regions that is reduced in NvAHR. NvAHR contains the diagnostic AHR motif SKRHR, involved in binding the XRE half site, one strongly (SKRHRD) and one moderately (RKKR) conserved nuclear localization signal, and a putative nuclear export signal (LDKLSILRL). AHR from N. vectensis also contains a moderately conserved domain that controls AHR/DNA interactions (TFVCRMKC), similar to other invertebrates, but the critical cysteine at position 198 (216 in mouse) is conserved, unlike AHRs from the protostomes D. melanogaster, C. elegans, and M. arenaria (Butler et al. 2001). The region of NvAHR corresponding to the ligand binding domain identified in vertebrate AHRs shows equal similarity with protostome and deuterostome orthologs (identity: ∼32-33%, similarity: ∼55%), with the exception of C. elegans which was considerably lower (identity: 18%, similarity: 45%). NvAHR lacks a Q-rich region, common to the transactivation domain of AHRs from other species, but the C-terminal end of the protein contains many prolines and serines, a molecular signature consistent with transactivation function in AHRs from other animals.
Fig. 1.
Alignment of N-terminal portion of Nematostella vectensis AHR with AHR proteins from diverse bilaterians. The basic and helix motifs of the bHLH domain are underlined, the two PAS domains are indicated by open boxes, and the presumed ligand-binding region is indicated by a dashed line and bracketed. Protein domains: NLS: nuclear localization signal; NES: nuclear export signal; AHR/DNA: AHR/DNA interaction domain. Taxa abbreviations: Hs: Homo sapiens; Mm: Mus musculus; Fh: Fundulus heteroclitus (AHR1); Ma: Mya arenaria; Dm: Drosophila melanogaster; Ce: Caenorhabditis elegans; Nv: Nematostella vectensis.
Developmental expression
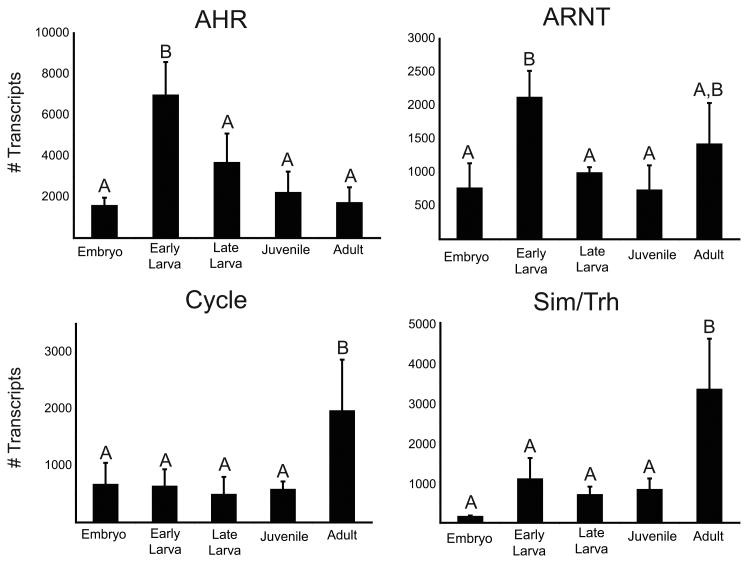
All four bHLH-PAS genes showed statistical differences in quantitative expression over the five developmental stages we measured (Fig. 2). AHR expression peaked in the early larval stage, at which time expression was significantly higher than at all other stages (F4,19 = 15.44, p < 0.0001). Expression decreased throughout the remainder of pre-adult stages to levels similar to those in embryos. ARNT showed a temporal pattern of expression similar to that of AHR, with highest expression in early larvae (F4,19 = 8.06, p = 0.0011); however, ARNT expression showed a second peak in the adult stage. Cycle and Sim/Trh had similar expression profiles, with highest expression in the adult stage (F4,19 = 6.39, p = 0.0033; F4,19 = 14.89, p < 0.0001, respectively) and overall low expression in developmental stages. The control gene, 18S, showed no statistical difference in expression over the sampled stages (not shown, p = 0.542).
Fig. 2.
Quantitative transcript expression analysis of four Nematostella vectensis bHLH-PAS genes during a developmental time series. Developmental stages assayed are indicated on the x-axis and gene expression in molecules per μl is indicated on the y-axis. Bars indicated the mean ± standard deviation. Letters above the bars indicate significantly different expression at p = 0.05.
All four N. vectensis bHLH-PAS genes showed specific spatial expression patterns over development (Fig. 3, subsequent letter references refer to panels in this figure). Expression of AHR was absent from the cleavage (a) and blastula (b) stages but AHR was expressed throughout the ectoderm in the gastrula, with the strongest signal near the blastopore. In later developmental stages, AHR was expressed in oral and aboral basal ectoderm in the planula (d), forming a ring of expression around the apical end (inset of panel d). AHR expression was also located in the oral and aboral ectoderm in tentacle bud (e) stages. In the juvenile polyp (f), AHR expression was restricted to the tentacle ectoderm. Expression of ARNT was absent from the cleavage (g) and blastula (h) stages. ARNT was expressed in oral and aboral ectoderm at gastrula (i) and planula (j) stages, but, by the tentacle bud stage ARNT expression was restricted to the aboral ectoderm (k). In the juvenile polyp (l), ARNT was broadly expressed in endodermal tissues of the mesenteries and body wall. Cycle expression was not detected until the tentacle bud stage, at which expression was restricted to the aboral endoderm with strong expression in a subset of cells in the mesenteries (q-r). Sim/Trh expression was not detected in development until the planula stage (compare s-w), at which time expression was in the basal ectoderm, predominately in the oral and aboral ends. Expression of Sim/Trh later became restricted to the aboral ectoderm of the tentacle bud stage. Sim/Trh was expressed throughout the endoderm of the juvenile polyp (x).
Fig. 3.
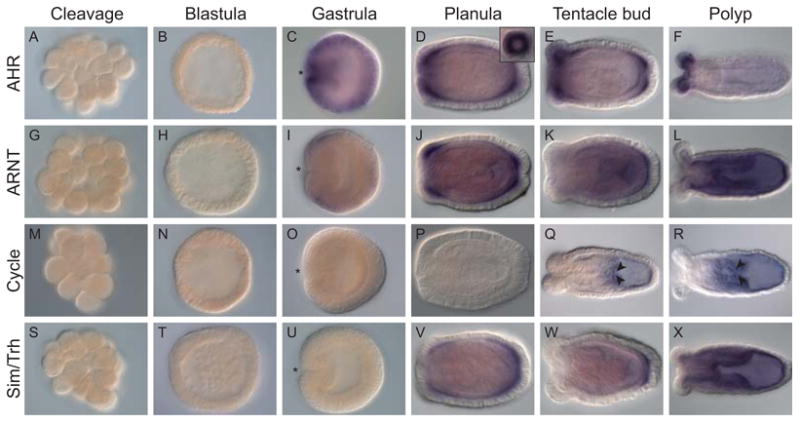
Spatial expression of bHLH-PAS gene transcripts in Nematostella vectensis embryos and polyps. Whole mount in situ hybridization of (a-f) NvAHR (g-l) NvARNT, (m-r) NvCycle, and (s-x) NvSim/Trh. NvAHR expression was not detectable in the cleavage (a) and blastula (b) stages. NvAHR was expressed throughout the ectoderm at the gastrula stage (c), with the strongest signal at the blastopore (*). NvAHR was expressed in oral and aboral ectoderm at planula (d), localized to a ring around the apical tuft (inset in panel d), and tentacle bud (e) stages. In the juvenile polyp (f), NvAHR expression was restricted to the tentacle ectoderm. NvARNT expression was not detectable in the cleavage (g) and blastula (h) stages. NvARNT was expressed in oral and aboral ectoderm at gastrula (i) and planula (j) stages. In the tentacle bud stage (k) NvARNT expression was restricted to the aboral ectoderm. In the juvenile polyp (l) NvARNT was expressed in the endoderm. NvCycle expression was not detectable in the cleavage (m), blastula (n), gastrula (o), or planula (p) stages. NvCycle expression was restricted to the aboral endoderm of the tentacle bud (q) and juvenile polyp (r) stages, with strong expression in a subset of cells in the mesenteries (arrowheads). NvSim/Trh expression was not detectable in the cleavage (s), blastula (t), or gastrula (u) stages. NvSim/Trh was expressed in the oral and aboral ectoderm of the planula (v), and in the aboral ectoderm of the tentacle bud stage (w). NvSim/Trh was expressed throughout the endoderm of the juvenile polyp (x).
Co-immunoprecipitation
We tested for protein-protein interactions of NvAHR with NvARNT and NvCYCLE. All were expressed well by in vitro transcription-translation (IVTT, Fig. 4a, left). However, we detected no evidence for dimerization of AHR with either of these proteins (Fig. 4a, right). Identical results were observed when TCDD (10 nM) was present during incubation of NvAHR and NvARNT (data not shown). Positive control reactions using human AHR and ARNT resulted in a strong, specific band (Fig. 4b).
Fig. 4.
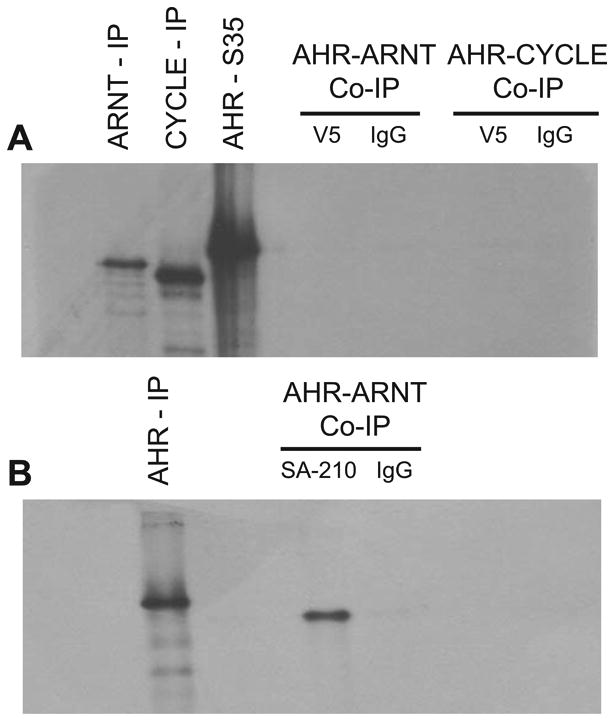
Protein-protein interactions of Nematostella vectensis AHR compared with human AHR. (a) Co-immunoprecipitation showing no evidence for dimerization of NvAHR protein with either NvARNT or NvCYCLE. Full length constructs of NvAHR were synthesized with [35S]methionine, incubated with unlabeled NvARNT and NvCYCLE, and then isolated using an antibody to a V5-epitope (V5) or mouse IgG. Both NvARNT and NvCYCLE were successfully immunoprecipitated with the anti-V5 antibody. (b) Positive control co-immunoprecipitation of human AHR and ARNT with addition of TCDD. For further details, refer to Methods, Co-immunoprecipitation. Hs AHR-ARNT complexes were immunoprecipitated using SA-210, a polyclonal antibody for human AHR.
NvAHR ligand binding
We assessed the potential for ligand binding of in vitro-expressed AHR from N. vectensis, using AHRs from the fish Fundulus heteroclitus (AHR1) and mouse as positive controls. The two ligands, TCDD and BNF, are prototypical halogenated and non-halogenated ligands for vertebrate AHRs that exhibit specific binding peaks when analyzed with velocity sedimentation on sucrose gradients. As expected, both of the vertebrate AHRs showed specific binding to TCDD and BNF (Fig. 5a and 5b, respectively), with mouse AHR exhibiting greater binding of TCDD under these conditions. In contrast, N. vectensis AHR showed no evidence for specific binding of either TCDD or BNF. Similar to our observations in co-immunoprecipitation, NvAHR protein showed strong in vitro expression (inset within Fig. 5a).
Fig. 5.
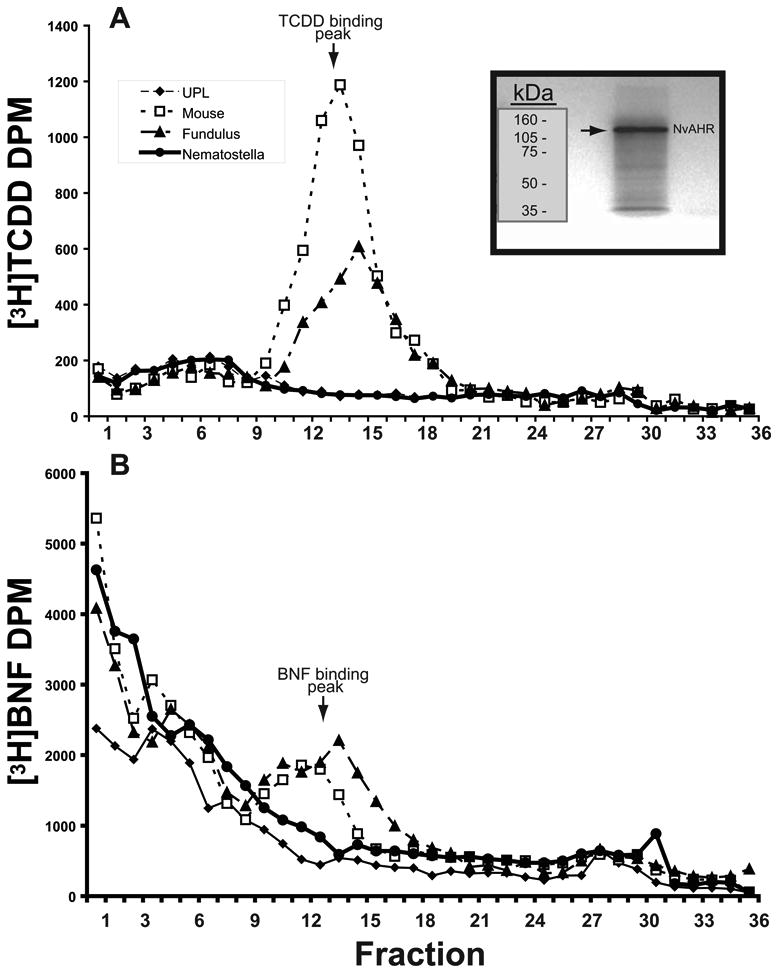
Absence of TCDD and BNF binding by Nematostella vectensis AHR. (a) Specific binding of [3H]TCDD to AHR from mouse and Fundulus (AHR1). NvAHR showed a profile similar to that of the unprogrammed TnT lysate (no AHR) negative controls. (b) Specific binding of [3H]BNF to the same AHRs as in (A). Mouse and Fundulus AHRs show a specific binding peak; however, NvAHR had no evidence for binding. DPM = disintegrations per minute. Inset: SDS-PAGE and fluorography of in vitro transcription/translation reaction of NvAHR showing a single product.
Discussion
The evolution of bHLH-PAS family can be traced to a single founder outside of the animal kingdom (Sebé-Pedrós et al. 2011); however, the radiation of this family occurred within animals (Simionato et al. 2007). Cnidarians, represented in this case by the sea anemone Nematostella vectensis, are the first branch from the animal stem that contain both a bona fide AHR and its specific dimerization partner ARNT. N. vectensis also has orthologs of most other bHLH-PAS genes conserved in protostomes and deuterostomes, providing strong evidence that this family diversified prior to the cnidarian-bilaterian ancestor. However, the conservation of nucleotide sequence does not necessarily imply conservation of expression or function. Thus, comparative characterization of specific genes is necessary for inferring their shared and divergent roles during animal evolution. Like other bHLH-PAS genes, AHR plays diverse roles in bilaterian animals, so studying species representative of outgroups (e.g., cnidarians) can provide insight into the evolution of these genes.
Previous phylogenetic analyses have shown that AHR is likely a result of a gene duplication event of a proto-AHR/HIF/Sim/TRH gene, sometime after the divergence of sponges from the animal stem but prior to the Placozoan-Cnidarian-Bilaterian ancestor (Larroux et al. 2008; Sebé-Pedrós et al. 2011; Simionato et al. 2007). Indeed, a reconstruction of the diversification of the bHLH-PAS family from these previous phylogenetic analyses provides a defined hypothesis for the expansion of the family (Fig. 6). Despite the ancient divergence of AHR, N. vectensis AHR shows high sequence similarity to bilaterian AHRs in the core bHLH and PAS domains. NvAHR lacks a Q-rich domain in the C-terminal portion of the protein present in AHR from many bilaterians; however, this region is also absent from the AHR ortholog in C. elegans as well as some AHR paralogs in vertebrates (Hahn 2002).
Fig. 6.
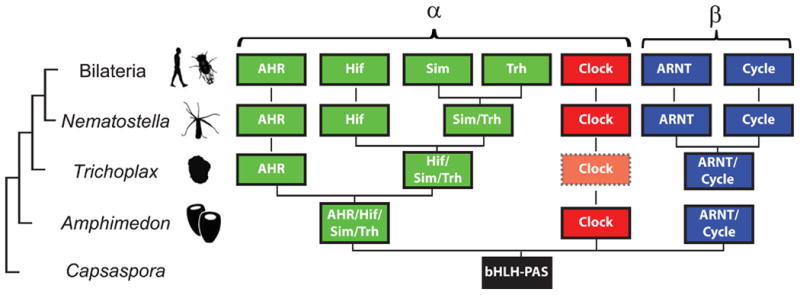
Early evolutionary emergence of the bHLH-PAS family. The cladogram at left represents inferred phylogenetic relationships of these taxa. Previous phylogenetic analyses in early diverging animals and the unicellular Capsaspora have identified the diversification of the bHLH superfamily, including the bHLH-PAS family. The summarized distribution supports the hypothesis that the sponge Amphimedon has three genes that represent an ortholog of Clock, an ancestral gene that later duplicated and diverged into the β class (ARNT and Cycle), and an ancestral gene that diversified into numerous members of the α class, including AHR, Hif, Trh, and Sim. The next branch of the tree, represented by the placozoan Trichoplax, appears to have lost Clock (indicated by dotted line and shading) and duplicated the sponge-type α class gene into a well-supported AHR and a proto-Hif/Sim/Trh. At the cnidarian-bilaterian ancestor, these members of the bHLH-PAS family appear to have fully diversified with the potential exception of Sim and Trh. We have indicated this gene as a proto-Sim/Trh gene in Nematostella. However, in phylogenetic analyses this gene has been shown to be part of a well-supported group with Trh orthologs from bilaterians, leading to two hypotheses: a proto-Sim/Trh that duplicated and diverged prior to the bilaterian ancestor (as shown) or a Sim gene loss in cnidarians, as suggested by the strong support for the Nv “proto-Sim/Trh” gene as an ortholog of Trh. In the Bilateria, particularly in vertebrates, many bHLH-PAS genes have duplicated one or numerous times giving rise to multiple paralogs (Hahn 2002, not depicted in figure).
The temporal and spatial patterns of AHR, ARNT, Cycle, and Sim/Trh expression in N. vectensis are consistent with a role in development of this sea anemone. All four genes showed differential expression between the developmental stages assayed, with highest expression either in early larvae (AHR, ARNT) or adults (Cycle, Sim/Trh). Analysis of spatial expression by ISH showed that expression was largely restricted to the basal portion of the ectoderm for all genes in early developmental stages; however, the regions varied among genes and across developmental stages. NvAHR showed the most distinct pattern, in which expression was increasingly restricted to the oral end during development, specifically in the tentacle buds. By contrast, ARNT and Sim/Trh displayed concentrated expression in the aboral end in later stage larvae and, later, a shift to broad endodermal expression in early juvenile polyps.
Previous research comparing the function of AHR orthologs in protostomes and deuterostomes has suggested that a potential conserved role for AHR is in the development and specification of neuronal sensory systems (e.g., Huang et al. 2004; Kim et al. 2006; Qin and Powell-Coffman 2004; Smith et al. 2013; Zhang et al. 2013). The cnidarians represent an early diverging phylum containing true neurons that are diffusely organized in what is traditionally termed a nerve net. Despite lacking a centralized nervous system, cnidarians do have concentrations of neurons around sensory areas of the tentacles and body column. Marlow et al. (2009) and Nakanishi et al. (2012) provided a description of the development and distribution of neuronal cells in N. vectensis. Subsequently, Sinigaglia et al. (2013) described the expression and function of a suite of genes involved in patterning bilaterian anterior development that are expressed in the aboral region of N. vectensis during development. Larvae sense their environment primarily with the apical organ positioned at the base of a bundle of cilia (apical tuft) on the aboral end, which is a component of the larval sensory system. The aboral expression of NvAHR is consistent with a possible role in localization of apical sensory system of the planula larva because expression forms a ring around this structure and borders expression with genes specific to the apical organ, including FGFs, FGF receptor, and COE (Rentzsch et al. 2008). These AHR-positive cells correspond to the location of ganglion cells located in the basal ectoderm of the developing larva and cells that surround the apical organ with centrally located nuclei compared to neurons in the apical organ (Chia and Koss 1979; Nakanishi et al. 2012).
In the juvenile stage, N. vectensis contains FMRFamide-expressing and, to a lesser extent, GABAergic neurons scattered throughout the ectoderm, with a large concentration in the developing tentacles. NvAHR was expressed in the basal portion of the oral ectoderm that contains some of the neuron cell bodies and ganglion cells (Marlow et al. 2009; Nakanishi et al. 2012). Sensory cells span the ectoderm tissue layer where they terminate on the outer surface, but there are other neuronal cell types distributed in the ectoderm. Thus, we could hypothesize that NvAHR may have some function in specification of a set of these neurons specific to the tentacles. However, previous expression of numerous presumptive neuronal markers (Layden et al. 2012; Marlow et al. 2009; Sinigaglia et al. 2013) showed punctuate expression patterns corresponding to the specification of individual nerve cells, not the broad expression we observed here for NvAHR. The precise role of NvAHR in larval and juvenile neurogenesis and specification will require gene knockdown studies coupled with histological characterization of abnormal phenotypes to discern how disruption of AHR signaling impacts development.
For bilaterian animals studied to date, AHR is a specific dimerization partner of ARNT; however, ARNT is a more promiscuous partner, forming heterodimers with other bHLH-PAS proteins. In addition to serving as a dimerization partner for AHR, ARNT (and its invertebrate orthologs TANGO and AHA-1) also functions as the partner for HIF, SIM, and TRH, and may also act as a homodimer (Sogawa et al. 1995). The importance of ARNT as a dimerization partner for multiple bHLH-PAS proteins is reflected in the lethal phenotype of null-mutants in model vertebrates (Maltepe et al. 1997) and invertebrates (Emmons et al. 1999). Using co-immunoprecipitation with in vitro proteins, we found no evidence for dimerization of NvAHR with NvARNT. Although the absence of heterodimerization may result from shortcomings of an in vitro system, previous work using identical methods showed conserved dimerization between two other N. vectensis bHLH-PAS proteins, CLOCK and CYCLE (Reitzel et al. 2010). Thus, it is possible that these proteins are not partners in the sea anemone. Consistent with this observation, NvAHR and NvARNT have discrete spatial expression domains during later stage development, but overlap with the other bHLH-PAS genes reported here earlier in development. Numerous ARNT partners identified in bilaterians are present in N. vectensis and at least one, Sim/TRH reported here, has an expression domain that consistently overlaps with that of ARNT. Which, if any, of the other N. vectensis bHLH-PAS protein form a dimer with ARNT awaits further research.
Recent research has identified critical residues mediating protein-protein interactions of AHR and ARNT from mouse and human, respectively (Hao et al. 2011). Using a bacterial two-hybrid screen for identification of impaired protein interactions, Hao et al. (2011) identified 22 amino acids in ARNT and 10 in AHR with a significant effect on specific heterodimerization of these two proteins. We compared the N. vectensis residues at these 32 homologous positions and found either identical or chemically-similar amino acids at all 22 positions in ARNT but at only 7 of the 10 positions in AHR. The three variant positions in AHR correspond to two sites N-terminal to the PAS-A domain (position 103 and 106 in mAHR), a region of low similarity across species, and one residue in the second beta-fold of the PAS-A domain (position 131 in mAHR). Whether these, or other, amino acids are critical for dimerization in N. vectensis await future research utilizing targeted mutagenesis. The variant amino acids in N. vectensis AHR and ARNT represent sites of interest for understanding specific residues for dimerization surfaces facilitating protein-protein interactions during animal evolution.
Accumulating evidence has shown that AHR may function alone as a participant in other protein-protein interactions that regulate gene expression or other cellular processes. AHR has also been shown to interact with other cellular regulatory proteins, including the retinoblastoma tumor suppressor protein (Puga et al. 2002), Src kinase (Matsumura 2009), KLF6 (Wilson et al. 2013), and the NF-κB components RelA and RelB (Tian 2009). The significance of these interactions with respect to AHR function are not yet clear, but these examples suggest that AHR participates in a rich set of protein-protein interactions in the absence of ARNT that may explain its varied roles in development.
Similar to AHR from protostomes (D. melanogaster, C. elegans, M. arenaria reported in (Butler et al. 2001)), N. vectensis AHR failed to bind TCDD and BNF, two ligands bound with high affinity by vertebrate AHRs. Our results showing that the AHR from this sea anemone, representative of a group of organisms that form an outgroup to the Bilateria, also does not bind these prototypical ligands provides additional evidence that the evolution of AHR binding to xenobiotics represents a novel function that evolved in the chordate lineage (Hahn 2002). A plethora of recent work has identified a diverse suite of potential endogenous AHR ligands (reviewed in Denison et al. 2011; Nguyen and Bradfield 2008). These ligands include metabolites of tryptophan, heme, and arachidonic acid. Of these, arachidonic acid metabolites, including prostaglandins, are particularly of interest due to the rich diversity of these compounds in cnidarians (Rowley et al. 2005). Identification of endogenous ligand(s) for a cnidarian AHR would provide a useful insight into the evolution of ligand specificity and how this specificity has been conserved throughout animal evolution.
Recently, a characterization of the bHLH complement, including bHLH-PAS members, of the coral Acropora digitifera (Gyoja et al. 2012), identified two candidate AHR genes in this coral species as well as in N. vectensis. In phylogenetic analyses, the second N. vectensis AHR (which we refer to here as NvAHRb1; JGI gene model 130163) grouped with the AHR described in this study (AHRa) as well as two coral AHR genes. The tree topology suggested a gene duplication event in the anthozoan lineage. The annotation of NvAHRb for that study was solely based on a gene prediction in the N. vectensis genome and lacked confirmation. Using gene specific primers, we have successfully amplified a small fragment of NvAHRb from cDNA synthesized from RNA from various developmental stages. We used this fragment for similarity searches of a transcriptome assembled by Tulin et al. (2013), where we identified a full length transcript for NvAHRb (comp2049_c0_seq1). Contrary to NvAHRa, NvAHRb lacks a conserved PAS-B domain and has generally lower sequence conservation (Supp. Fig. 2). Based on the observed sequence divergence, this second confirmed cnidarian AHR elicits new questions about the function of AHR paralogs in development and physiology of N. vectensis and the potential for subfunctionalization of duplicated genes. Previous research in teleost fishes, where AHR paralogs are common, has shown subfunctionalization of AHR paralogs in development and in mediating effects of toxicants (Karchner et al. 2005; Powell et al. 2000). Future research could test for similar functional diversification of this second cnidarian AHR.
Conclusion
The bHLH-PAS family expanded and diversified early in animal evolution, where most previously classified bilaterian members, including AHR and ARNT, diverged prior to the cnidarian-bilaterian ancestor. NvAHR expression was broadly similar to that of other N. vectensis bHLH-PAS genes early in development but later diverged with predominant expression restricted to the developing tentacles. Expression of NvAHR is consistent with a potential function in specification of neurons or other cells surrounding the apical organ during larval development and correlates with tentacle-specific neurons, as indicated by expression that overlaps with the distribution of neuronal cell bodies. The absence of protein-protein interactions between NvAHR and NvARNT suggests that the evolution of this heterodimerization reaction may have evolved after the cnidarian-bilaterian ancestor and that ARNT may dimerize with other bHLH-PAS proteins in N. vectensis. NvAHR failed to bind prototypical AHR ligands, providing additional support for a role of AHR as a mediator of toxicant response evolved in the chordate lineage.
Supplementary Material
Acknowledgments
This research was supported by Award Number F32HD062178 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development (NICHD) to AMR, the Tropical Research Initiative of the Woods Hole Oceanographic Institution to AMT, and by Award Number R01ES006272 from the National Institute of Environmental Health Sciences to MEH. AMR was also supported by incentive funds provided by the University of North Carolina at Charlotte.
Footnotes
N. vectensis AHR paralogs are annotated as NvAHRa and NvAHRb to avoid confusion with the numerical annotation of vertebrate AHR paralogs (e.g., AHR1 and AHR2), which resulted from independent duplication events.
References
- Butler RB, Kelley ML, Powell WH, Hahn ME, Van Beneden RJ. An aryl hydrocarbon receptor homologue from the soft-shell clam, Mya arenaria: Evidence that invertebrate AHR homologues lack TCDD and BNF binding. Gene. 2001;278:223–234. doi: 10.1016/s0378-1119(01)00724-7. [DOI] [PubMed] [Google Scholar]
- Chia FS, Koss R. Fine structural studies of the nervous system and the apical organ in the planula larva of the sea anemone Anthopleura elegantissima. J Morphol. 1979;160:275–297. doi: 10.1002/jmor.1051600303. [DOI] [PubMed] [Google Scholar]
- Darling JA, Reitzel AM, Burton PM, Mazza ME, Ryan JF, Sullivan JC, Finnerty JR. Rising starlet: the starlet sea anemone, Nematostella vectensis. BioEssays. 2005;27:211–221. doi: 10.1002/bies.20181. [DOI] [PubMed] [Google Scholar]
- Denison MS, Fisher JM, Whitlock JP. Protein-DNA interactions at recognition sites for the dioxin-Ah receptor complex. J Biol Chem. 1989;264:16478–16482. [PubMed] [Google Scholar]
- Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- Denison MS, Soshilov AA, He G, Degroot DE, Zhao B. Exactly the same but different: Promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol Sci. 2011;124:1–22. doi: 10.1093/toxsci/kfr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan DM, Burgess EA, Duncan I. Control of distal antennal identity and tarsal development in Drosophila by spineless-aristapedia, a homolog of the mammalian dioxin receptor. Gene Dev. 1998;12:1290–1303. doi: 10.1101/gad.12.9.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons RB, Duncan D, Estes PA, Kiefel P, Mosher JT, Sonnenfeld M, Ward MP, Duncan I, Crews ST. The spineless-aristapedia and tango bHLH-PAS proteins interact to control antennal and tarsal development in Drosophila. Development. 1999;126:3937–3945. doi: 10.1242/dev.126.17.3937. [DOI] [PubMed] [Google Scholar]
- Erwin DH. Early origin of the bilaterian developmental toolkit. Philos Trans R Soc Lond B Biol Sci. 2009;364:2253–2261. doi: 10.1098/rstb.2009.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YZ, Hogenesch JB, Bradfield CA. The PAS superfamily: sensors of environmental and developmental signals. Annu Rev Pharmacol Toxicol. 2000;40:519–561. doi: 10.1146/annurev.pharmtox.40.1.519. [DOI] [PubMed] [Google Scholar]
- Gyoja F, Kawashima T, Satoh N. A genomewide survey of bHLH transcription factors in the coral Acropora digitifera identifies three novel orthologous families, pearl, amber, and peridot. Dev Genes Evol. 2012;222:63–76. doi: 10.1007/s00427-012-0388-6. [DOI] [PubMed] [Google Scholar]
- Hahn ME. Aryl hydrocarbon receptors: Diversity and evolution. Chem Biol Interact. 2002;141:131–160. doi: 10.1016/s0009-2797(02)00070-4. [DOI] [PubMed] [Google Scholar]
- Hahn ME, Hestermann EV. Receptor-mediated Mechanisms of Toxicity. In: Di Giulio RT, Hinton DE, editors. The Toxicology of Fishes. Taylor & Francis; 2007. pp. 235–272. Chapter 5. [Google Scholar]
- Hao N, Whitelaw ML, Shearwin KE, Dodd IB, Chapman-Smith A. Identification of residues in the N-terminal PAS domains important for dimerization of Arnt and AhR. Nucleic Acids Res. 2011;39:3695–3709. doi: 10.1093/nar/gkq1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman EC, Reyes H, Chu FF, Sander F, Conley LH, Brooks BA, Hankinson O. Cloning of a factor required for activity of the Ah (dioxin) receptor. Science. 1991;252:954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- Huang X, Powell-Coffman JA, Jin Y. The AHR-1 aryl hydrocarbon receptor and its co-factor the AHA-1 aryl hydrocarbon receptor nuclear translocator specify GABAergic neuron cell fate in C. elegans. Development. 2004;131:819–828. doi: 10.1242/dev.00959. [DOI] [PubMed] [Google Scholar]
- Karchner SI, Franks DG, Hahn ME. AHR1B, a new functional aryl hydrocarbon receptor in zebrafish: tandem arrangement of ahr1b and ahr2 genes. Biochem J. 2005;392:153–161. doi: 10.1042/BJ20050713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchner SI, Franks DG, Kennedy SW, Hahn ME. The molecular basis for differential dioxin sensitivity in birds: Role of the aryl hydrocarbon receptor. Proceedings of the National Academy of Sciences. 2006;103:6252–6257. doi: 10.1073/pnas.0509950103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchner SI, Powell WH, Hahn ME. Identification and functional characterization of two highly divergent aryl hydrocarbon receptors (AHR1 and AHR2) in the teleost Fundulus heteroclitus : Evidence for a novel subfamily of ligand-binding basic helix loop helix-PER-ARNT-SIM (bHLH-PAS) factors. J Biol Chem. 1999;274:33814–33824. doi: 10.1074/jbc.274.47.33814. [DOI] [PubMed] [Google Scholar]
- Kim MD, Jan LY, Jan YN. The bHLH-PAS protein Spineless is necessary for the diversification of dendrite morphology of Drosophila dendritic arborization neurons. Gene Dev. 2006;20:2806–2819. doi: 10.1101/gad.1459706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumburegama S, Wijesena N, Xu R, Wikramanayake A. Strabismus-mediated primary archenteron invagination is uncoupled from Wnt/beta-catenin-dependent endoderm cell fate specification in Nematostella vectensis (Anthozoa, Cnidaria): Implications for the evolution of gastrulation. EvoDevo. 2011;2:2. doi: 10.1186/2041-9139-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, Technau U, Von Haeseler A, Hobmayer B, Martindale MQ, Holstein TW. Unexpected complexity of the Wnt gene family in a sea anemone. Nature. 2005;433:156–160. doi: 10.1038/nature03158. [DOI] [PubMed] [Google Scholar]
- Larroux C, Luke GN, Koopman P, Rokhsar DS, Shimeld SM, Degnan BM. Genesis and expansion of metazoan transcription factor gene classes. Mol Biol Evol. 2008;25:980–996. doi: 10.1093/molbev/msn047. [DOI] [PubMed] [Google Scholar]
- Layden MJ, Boekhout M, Martindale MQ. Nematostella vectensis achaete-scute homolog NvashA regulates embryonic ectodermal neurogenesis and represents an ancient component of the metazoan neural specification pathway. Development. 2012;139:1013–1022. doi: 10.1242/dev.073221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magie CR, Pang K, Martindale MQ. Genomic inventory and expression of Sox and Fox genes in the cnidarian Nematostella vectensis. Dev Genes Evol. 2005;215:618–630. doi: 10.1007/s00427-005-0022-y. [DOI] [PubMed] [Google Scholar]
- Maltepe E, Schmidt JV, Baunoch D, Bradfield CA, Simon MC. Abnormal angiogenesis and responses to glucose and oxygen deprivation in mice lacking the protein ARNT. Nature. 1997;386:403–407. doi: 10.1038/386403a0. [DOI] [PubMed] [Google Scholar]
- Marlow HQ, Srivastava M, Matus DQ, Rokhsar D, Martindale MQ. Anatomy and development of the nervous system of Nematostella vectensis, an anthozoan cnidarian. Dev Neurobiol. 2009;69:235–254. doi: 10.1002/dneu.20698. [DOI] [PubMed] [Google Scholar]
- Martindale MQ, Pang K, Finnerty JR. Investigating the origins of triploblasty: “mesodermal” gene expression in a diploblastic animal, the sea anemone, Nematostella vectensis (Phylum Cnidaria; Class Anthozoa) Development. 2004;131:2463–2474. doi: 10.1242/dev.01119. [DOI] [PubMed] [Google Scholar]
- Matsumura F. The significance of the nongenomic pathway in mediating inflammatory signaling of the dioxin-activated Ah receptor to cause toxic effects. Biochem Pharmacol. 2009;77:608–626. doi: 10.1016/j.bcp.2008.10.013. [DOI] [PubMed] [Google Scholar]
- McMillan BJ, Bradfield CA. The aryl hydrocarbon receptor sans xenobiotics: endogenous function in genetic model systems. Mol Pharmacol. 2007;72:487–498. doi: 10.1124/mol.107.037259. [DOI] [PubMed] [Google Scholar]
- Nakanishi N, Renfer E, Technau U, Rentzsch F. Nervous systems of the sea anemone Nematostella vectensis are generated by ectoderm and endoderm and shaped by distinct mechanisms. Development. 2012;139:347–357. doi: 10.1242/dev.071902. [DOI] [PubMed] [Google Scholar]
- Nguyen LP, Bradfield CA. The search for endogenous activators of the aryl hydrocarbon receptor. Chem Res Toxicol. 2008;21:102–116. doi: 10.1021/tx7001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okey AB. An aryl hydrocarbon receptor odyssey to the shores of toxicology. Toxicol Sci. 2007;98:5–38. doi: 10.1093/toxsci/kfm096. [DOI] [PubMed] [Google Scholar]
- Pang K, Matus DQ, Martindale MQ. The ancestral role of COE genes may have been in chemoreception: evidence from the development of the sea anemone, Nematostella vectensis (Phylum Cnidaria; Class Anthozoa) Dev Genes Evol. 2004;214:134–138. doi: 10.1007/s00427-004-0383-7. [DOI] [PubMed] [Google Scholar]
- Powell-Coffman JA, Bradfield CA, Wood WB. Caenorhabditis elegans orthologs of the aryl hydrocarbon receptor and its heterodimerization partner the aryl hydrocarbon receptor nuclear translocator. Proc Natl Acad Sci USA. 1998;95:2844–2849. doi: 10.1073/pnas.95.6.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell WH, Bright R, Bello SM, Hahn ME. Developmental and tissue-specific expression of AHR1, AHR2, and ARNT2 in dioxin-sensitive and-resistant populations of the marine fish Fundulus heteroclitus. Toxicol Sci. 2000;57:229–239. doi: 10.1093/toxsci/57.2.229. [DOI] [PubMed] [Google Scholar]
- Puga A, Xia Y, Elferink C. Role of the aryl hydrocarbon receptor in cell cycle regulation. Chem Biol Interact. 2002;141:117. doi: 10.1016/s0009-2797(02)00069-8. [DOI] [PubMed] [Google Scholar]
- Putnam N, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov V, Jurka J, Genikhovich G, Grigoriev I, Lucas S, Steele R, Finnerty J, Technau U, Martindale M, Rokhsar D. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Qin H, Powell-Coffman JA. The Caenorhabditis elegans aryl hydrocarbon receptor, AHR-1, regulates neuronal development. Dev Biol. 2004;270:64–75. doi: 10.1016/j.ydbio.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Qin H, Zhai Z, Powell-Coffman JA. The Caenorhabditis elegans AHR-1 transcription complex controls expression of soluble guanylate cyclase genes in the URX neurons and regulates aggregation behavior. Dev Biol. 2006;298:606–615. doi: 10.1016/j.ydbio.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Reitzel AM, Behrendt L, Tarrant AM. Light entrained rhythmic gene expression in the sea anemone Nematostella vectensis: the evolution of the animal circadian clock. PLoS ONE. 2010;5:e12805. doi: 10.1371/journal.pone.0012805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel AM, Sullivan JC, Traylor-Knowles N, Finnerty JR. Genomic survey of candidate stress-response genes in the estuarine anemone Nematostella vectensis. Biol Bull. 2008;214:233–254. doi: 10.2307/25470666. [DOI] [PubMed] [Google Scholar]
- Reitzel AM, Tarrant AM. Nuclear receptor complement of the cnidarian Nematostella vectensis: phylogenetic relationships and developmental expression patterns. BMC Evol Biol. 2009;9:230. doi: 10.1186/1471-2148-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch F, Fritzenwanker JH, Scholz CB, Technau U. FGF signalling controls formation of the apical sensory organ in the cnidarian Nematostella vectensis. Development. 2008;135:1761–1769. doi: 10.1242/dev.020784. [DOI] [PubMed] [Google Scholar]
- Reyes H, Reisz-Porszasz S, Hankinson O. Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science. 1992;256:1193–1195. doi: 10.1126/science.256.5060.1193. [DOI] [PubMed] [Google Scholar]
- Rowley AF, Vogan CL, Taylor GW, Clare AS. Prostaglandins in non-insectan invertebrates: recent insights and unsolved problems. J Expt Biol. 2005;208:3–14. doi: 10.1242/jeb.01275. [DOI] [PubMed] [Google Scholar]
- Ryan JF, Mazza ME, Pang K, Matus DQ, Baxevanis AD, Martindale MQ, Finnerty JR. Pre-Bilaterian origins of the Hox cluster and the Hox code: Evidence from the sea anemone, Nematostella vectensis. PLoS ONE. 2007;2:e153. doi: 10.1371/journal.pone.0000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A, De Mendoza A, Lang BF, Degnan BM, Ruiz-Trillo I. Unexpected repertoire of metazoan transcription factors in the unicellular holozoan Capsaspora owczarzaki. Mol Biol Evol. 2011;28:1241–1254. doi: 10.1093/molbev/msq309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, Degnan BM, Vervoort M. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol Biol. 2007;7:33. doi: 10.1186/1471-2148-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinigaglia C, Busengdal H, Leclère L, Technau U, Rentzsch F. The bilaterian head patterning gene six3/6 controls aboral domain development in a cnidarian. PLoS Biol. 2013;11:e1001488. doi: 10.1371/journal.pbio.1001488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Cody j, O'brien T, Chatzigeorgiou M, Spencer WC, Feingold-Link E, Husson Steven j, Hori S, Mitani S, Gottschalk A, Schafer William r, Miller David m., Iii Sensory neuron fates are distinguished by a transcriptional switch that regulates dendrite branch stabilization. Neuron. 2013;79:266–280. doi: 10.1016/j.neuron.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogawa K, Nakano R, Kobayashi A, Kikuchi Y, Ohe N, Matsushita N, Fujii-Kuriyama Y. Possible function of Ah receptor nuclear translocator (Arnt) homodimer in transcriptional regulation. Proc Natl Acad Sci USA. 1995;92:1936–1940. doi: 10.1073/pnas.92.6.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, Signorovitch AY, Moreno MA, Kamm K, Grimwood J, Schmutz J, Shapiro H, Grigoriev IV, Buss LW, Schierwater B, Dellaporta SL, Rokhsar DS. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier MEA, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, Larroux C, Putnam NH, Stanke M, Adamska M, Darling A, Degnan SM, Oakley TH, Plachetzki DC, Zhai Y, Adamski M, Calcino A, Cummins SF, Goodstein DM, Harris C, Jackson DJ, Leys SP, Shu S, Woodcroft BJ, Vervoort M, Kosik KS, Manning G, Degnan BM, Rokhsar DS. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanik DJ, Friedman LE, Finnerty JR. Collecting, rearing, spawning and inducing regeneration of the starlet sea anemone, Nematostella vectensis. Nat Protocols. 2013;8:916–923. doi: 10.1038/nprot.2013.044. [DOI] [PubMed] [Google Scholar]
- Swanson HI, Chan WK, Bradfield CA. DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J Biol Chem. 1995;270:26292–26302. doi: 10.1074/jbc.270.44.26292. [DOI] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB. PAS Domains: Internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau U, Steele RE. Evolutionary crossroads in developmental biology: Cnidaria. Development. 2011;138:1447–1458. doi: 10.1242/dev.048959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y. Ah receptor and NF-κB interplay on the stage of epigenome. Biochem Pharmacol. 2009;77:670–680. doi: 10.1016/j.bcp.2008.10.023. [DOI] [PubMed] [Google Scholar]
- Tulin S, Aguiar D, Istrail S, Smith J. A quantitative reference transcriptome for Nematostella vectensis early embryonic development: a pipeline for de novo assembly in emerging model systems. EvoDevo. 2013;4:16. doi: 10.1186/2041-9139-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernet MF, Mazzoni EO, Celik A, Duncan DM, Duncan I, Desplan C. Stochastic spineless expression creates the retinal mosaic for colour vision. Nature. 2006;440:174–180. doi: 10.1038/nature04615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SR, Joshi AD, Elferink CJ. The tumor suppressor Kruppel-like factor 6 is a novel aryl hydrocarbon receptor DNA binding partner. J Pharm Exp Ther. 2013;345:419–429. doi: 10.1124/jpet.113.203786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolenski FS, Garbati MR, Lubinski TJ, Traylor-Knowles N, Dresselhaus E, Stefanik DJ, Goucher H, Finnerty JR, Gilmore TD. Characterization of the core elements of the NF-kB signaling pathway of the sea anemone Nematostella vectensis. Mol Cell Biol. 2011;31:1076–1087. doi: 10.1128/MCB.00927-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li X, Jevince AR, Guan L, Wang J, Hall DH, Huang X, Ding M. Neuronal target identification requires AHA-1-mediated fine-tuning of Wnt signaling in C. elegans. PLoS Genet. 2013;9:e1003618. doi: 10.1371/journal.pgen.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.