Abstract
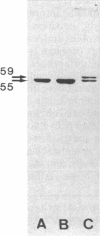
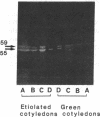
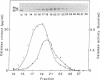
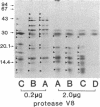
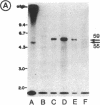
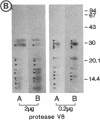
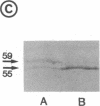
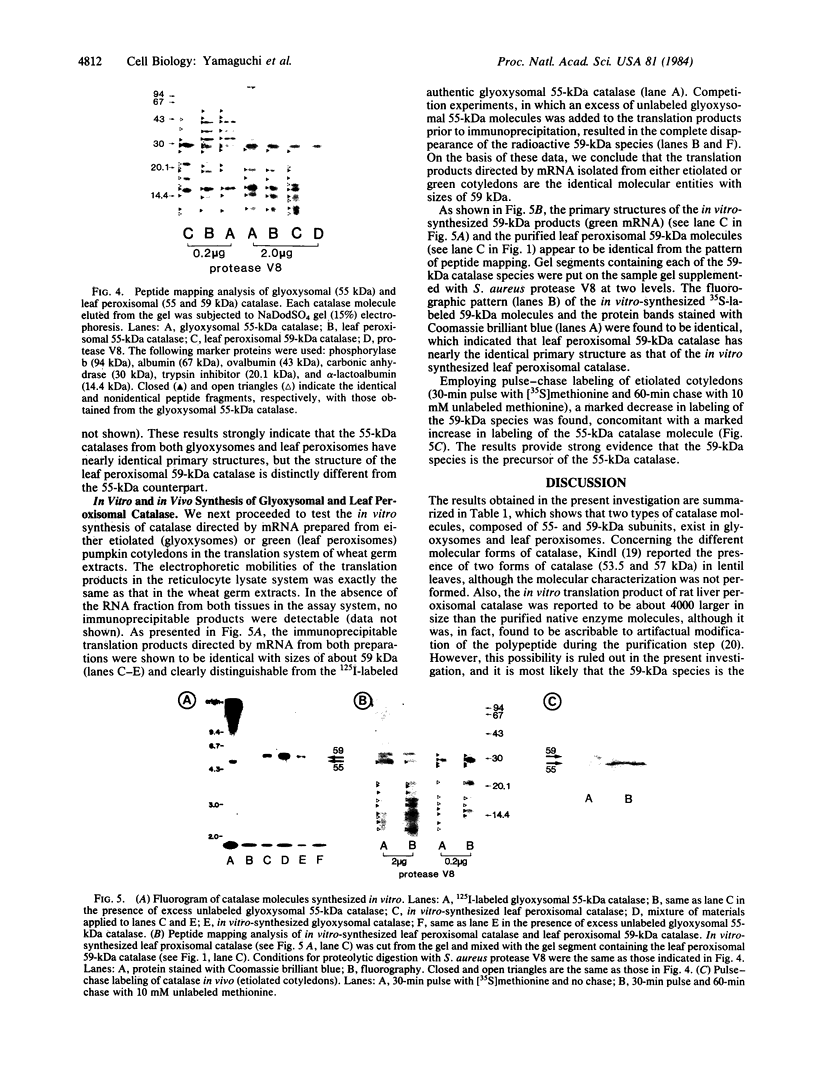
As an approach to study the mechanism of the microbody transition (glyoxysomes to leaf peroxisomes) in greening pumpkin cotyledons, catalase molecules were purified from the two different types of microbody and their structural properties were compared. The purified glyoxysomal catalase was found to consist of four identical subunits (55 kDa), whereas the leaf peroxisomal catalase contains two different forms of monomeric subunit (55 and 59 kDa). These different catalase species cross-reacted with the rabbit antibody raised against the glyoxysomal enzyme. During gel filtration on an Ultrogel AcA 34 column, the leaf peroxisomal 55-kDa polypeptide eluted slightly faster than the leaf peroxisomal 59-kDa polypeptide. The profile of catalase activities exactly paralleled the elution pattern of the 55-kDa molecules, which indicated that the 59-kDa polypeptide was enzymically inactive. Peptide mapping analysis using Staphylococcus aureus protease V8 showed that the glyoxysomal 55-kDa polypeptide was identical to the leaf peroxisomal 55-kDa species, whereas the leaf peroxisomal 59-kDa polypeptide had a different primary structure from the 55-kDa polypeptide. In an in vitro translation system directed by mRNA isolated from etiolated and green cotyledons, glyoxysomal and leaf peroxisomal catalases were synthesized as the identical 59-kDa polypeptide. From peptide mapping analysis, the in vitro-translated 59-kDa polypeptide was found to have a nearly identical primary structure to that of the leaf peroxisomal 59-kDa species. In vivo pulse-chase labeling experiments using etiolated cotyledons showed the conversion of the 59-kDa polypeptide to the 55-kDa molecular species. The overall results strongly indicate that the 59-kDa polypeptide is a precursor form of catalase in pumpkin cotyledons.
Keywords: inactive enzyme, microbody transition, protein synthesis
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cleveland D. W., Fischer S. G., Kirschner M. W., Laemmli U. K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977 Feb 10;252(3):1102–1106. [PubMed] [Google Scholar]
- Furuta S., Hashimoto T., Miura S., Mori M., Tatibana M. Cell-free synthesis of the enzymes of peroxisomal beta-oxidation. Biochem Biophys Res Commun. 1982 Mar 30;105(2):639–646. doi: 10.1016/0006-291x(82)91482-6. [DOI] [PubMed] [Google Scholar]
- Gietl C., Hock B. Organelle-bound malate dehydrogenase isoenzymes are synthesized as higher molecular weight precursors. Plant Physiol. 1982 Aug;70(2):483–487. doi: 10.1104/pp.70.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman B. M., Blobel G. Biogenesis of peroxisomes: intracellular site of synthesis of catalase and uricase. Proc Natl Acad Sci U S A. 1978 Oct;75(10):5066–5070. doi: 10.1073/pnas.75.10.5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A. H., Beevers H. Isolation of microbodies from plant tissues. Plant Physiol. 1971 Nov;48(5):637–641. doi: 10.1104/pp.48.5.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa T., Beevers H. The development of microbodies (glyoxysomes and leaf peroxisomes) in cotyledons of germinating watermelon seedlings. Plant Physiol. 1975 Feb;55(2):258–264. doi: 10.1104/pp.55.2.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa T., McGregor D. I., Beevers H. Development of enzymes in the cotyledons of watermelon seedlings. Plant Physiol. 1973 Jan;51(1):66–71. doi: 10.1104/pp.51.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Miyata S., Okamoto K., Watanabe A., Akazawa T. Enzymic Mechanism of Starch Breakdown in Germinating Rice Seeds: 10. IN VIVO AND IN VITRO SYNTHESIS OF alpha-AMYLASE IN RICE SEED SCUTELLUM. Plant Physiol. 1981 Dec;68(6):1314–1318. doi: 10.1104/pp.68.6.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M., Bhusawang P., Strzalka K., Akazawa T. Developmental formation of glutamine synthetase in greening pumpkin cotyledons and its subcellular localization. Plant Physiol. 1982 Aug;70(2):353–356. doi: 10.1104/pp.70.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roa M., Blobel G. Biosynthesis of peroxisomal enzymes in the methylotrophic yeast Hansenula polymorpha. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6872–6876. doi: 10.1073/pnas.80.22.6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbi M., Lazarow P. B. Peptide mapping of peroxisomal catalase and its precursor. Comparison to the primary wheat germ translation product. J Biol Chem. 1982 Jan 25;257(2):964–970. [PubMed] [Google Scholar]
- Robbi M., Lazarow P. B. Synthesis of catalase in two cell-free protein-synthesizing systems and in rat liver. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4344–4348. doi: 10.1073/pnas.75.9.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B. E., Paterson B. M. Efficient translation of tobacco mosaic virus RNA and rabbit globin 9S RNA in a cell-free system from commercial wheat germ. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2330–2334. doi: 10.1073/pnas.70.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefer S., Teifel W., Kindl H. Plant microbody proteins, I. Purification and characterization of catalase from leaves of lens culinaris. Hoppe Seylers Z Physiol Chem. 1976 Feb;357(2):163–175. doi: 10.1515/bchm2.1976.357.1.163. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelease R. N., Becker W. M., Gruber P. J., Newcomb E. H. Microbodies (Glyoxysomes and Peroxisomes) in Cucumber Cotyledons: Correlative Biochemical and Ultrastructural Study in Light- and Dark-grown Seedlings. Plant Physiol. 1971 Oct;48(4):461–475. doi: 10.1104/pp.48.4.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaessen R. T., Kreike J., Groot G. S. Protein transfer to nitrocellulose filters. A simple method for quantitation of single proteins in complex mixtures. FEBS Lett. 1981 Feb 23;124(2):193–196. doi: 10.1016/0014-5793(81)80134-2. [DOI] [PubMed] [Google Scholar]
- Yamada T., Tanaka A., Horikawa S., Numa S., Fukui S. Cell-free translation and regulation of Candida tropicalis catalase messenger RNA. Eur J Biochem. 1982 Dec 15;129(2):251–255. doi: 10.1111/j.1432-1033.1982.tb07046.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi J., Nishimura M. Purification of glyoxysomal catalase and immunochemical comparison of glyoxysomal and leaf peroxisomal catalase in germinating pumpkin cotyledons. Plant Physiol. 1984 Feb;74(2):261–267. doi: 10.1104/pp.74.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]