Abstract
A vast theoretical literature has explored the evolutionary dynamics of parasite virulence. The classic result from this modelling work is that, assuming a saturating transmission–virulence trade-off, there is a single evolutionary optimum where the parasite optimizes the epidemiological R0. However, there are an increasing number of models that have shown how ecological and epidemiological feedbacks to evolution can instead result in the creation and maintenance of multiple parasite strains. Here, we fully explore one such example, where recovered hosts have a limited ‘immune range’ resulting in partial cross-immunity to parasite strains that they have not previously encountered. Taking an adaptive dynamics approach, we show that, provided this immune range is not too wide, high levels of diversity can evolve and be maintained through multiple branching events. We argue that our model provides a more realistic picture of disease dynamics in vertebrate host populations and may be a key explanatory factor in the high levels of parasite diversity seen in natural systems.
Keywords: host–parasite, acquired immunity, virulence, evolution
1. Introduction
One of the most important applications of modelling biological evolution has been to infectious disease dynamics. As well as seeking to understand factors that select for highly virulent parasites, a key focus of theoretical work has been to explore the processes that may create and maintain the diversity of parasite strains that are seen in host populations. Classical models of host–parasite evolution, principally using the gene-for-gene and matching-allele frameworks, have found the potential for diversity through the evolutionary cycling of strains owing to the tight genetic specificities required for infection [1–5]. Since the pioneering work in the 1970s and 1980s of Anderson and May (e.g. [6]), who applied the classic ‘susceptible–infected–recovered’ (SIR) epidemiological model [7] to natural, dynamic populations, modern evolutionary studies have often focused on how ecological and epidemiological processes impact selection. These studies have largely been based upon the transmission–virulence trade-off theory, which assumes that any increase in transmission requires faster within-host growth, which in turn causes more damage to the host (see [8] for a recent review). Assuming a saturating transmission–virulence trade-off, the theory shows that the parasite is subjected to a competitive exclusion principle, with the parasite strain that maximizes the epidemiological R0 (the number of secondary infections in a nearly disease-free population) always outcompeting any other strain [9,10]. As such, it is predicted that epidemiological processes alone cannot create long-term coexistence of parasite strains. However, this prediction is based on the assumption that recovered and immune hosts are perfectly protected from all future parasite strains; hence, any mutation, no matter how large, in the parasite strain has no effect on immune hosts. In reality, we may expect that hosts will have a limited ‘immune range’ giving only partial cross-immunity to parasites with different antigenic structures; hence, new mutant (or rare) strains may be able to infect hosts which are immune to the existing resident (or common) type. Prime examples of this include influenza [11] and whooping cough [12]. Furthermore, it has been argued that cross-immunity can account for the coexistence of multiple strains (of varying virulence levels) of dengue virus in East Asia [13], where partial immunity is the cause of the varying outbreaks of different strains at different times through the year. It is therefore important to explore how partial cross-immunity, or ‘immune range’, impacts selection on parasites.
More generally, the role of ecological and epidemiological processes in shaping selection pressures in ecological populations is now central to many evolutionary studies through the framework of adaptive dynamics (e.g. [14–17]). This theoretical framework assumes that rare mutants, with small differences in phenotypic trait values, attempt to invade into an environment created by the current resident. If the mutant is successful it will grow in size to either replace or coexist with the resident. Through repeated substitutions and invasions, populations will evolve their phenotypes through trait space according to the local selection gradient. One of the most discussed outcomes from adaptive dynamics models is that of evolutionary branching. In such a case, populations are attracted to a certain ‘singular’ trait value, but once there find that it is a fitness minimum and that they can be invaded by all nearby types [17]. This gives rise to disruptive selection and the creation of two coexisting types (on either side of the singular point) that will then continue to coevolve. Such branching requires there to be more than one ‘feedback environment’, such that evolution of the trait in question alters the underlying population dynamics through at least two distinct processes [17–19]. Models have now identified a considerable range of eco-evolutionary scenarios that can create two or more coexisting strains through a process of evolutionary branching, in particular in host defence mechanisms to parasitism [20–23].
That parasite evolution optimizes R0 is perhaps one of the most well-known results in evolutionary theory. However, this result has been increasingly challenged in recent years by models that include more epidemiological and ecological detail than the baseline SIR model. These have identified a number of processes which may alter selection on the parasite to prevent optimization and instead lead to evolutionary branching. These processes include:
— Superinfection. In fact, in one of the first models addressing parasite evolution [24] it was found that two strains of a parasite could stably coexist within a host population for some limited parameter range when a virulent strain is able to take over infections from an avirulent strain. This model was later extended to a fully dynamic evolutionary model [25], confirming that evolutionary branching could occur from an initially monomorphic population.
— Density-dependent host mortality. Models have traditionally assumed that density dependence acts on host reproduction, which does not impact (explicitly) on parasite fitness (in particular it does not appear in R0). However, if it is instead assumed to act on host mortality, this creates a second feedback environment for the parasite and, for certain trade-off assumptions, can allow for evolutionary branching in parasite virulence [26–28] (see also [29] for an example of branching where virulence is density dependent).
— Specialism on coexisting host strains. Gudelj et al. [30] studied a model where parasite strains could specialize on already coexisting host strains. This in effect creates two niches for parasitism, and therefore again allows for evolutionary branching. A similar effect was shown where hosts and parasites could coevolve their respective resistance and infection ‘ranges’ [31]. They found the potential for multiple branching events leading to high levels of diversity in both hosts and parasites as well as for evolutionary cycles.
— Preferential predation of infected hosts. Morozov & Best [32] analysed a model where hosts are at risk not only of parasitism but also of predation. If hosts infected with more virulent parasites are subjected to higher rates of predation, this can also promote branching in parasite virulence.
Here we shall explore, in detail, a further process which has been shown to allow for coexistence of parasite strains, namely partial cross-immunity. Adaptive immunity to infection is a vital aspect of the epidemiological dynamics of vertebrate hosts. After infection with a certain strain of disease, long-lasting memory cells are produced that are able to recognize specific antigenic configurations of previously encountered pathogens. If the host is then exposed to that strain again, it is able to launch a quick and efficient immune response to effectively prevent further infection. This is incorporated into the SIR epidemiological model through the transition from the infected to the recovered compartment, with the assumption that recovered hosts are perfectly protected from infection. From an evolutionary perspective, immune memory may not be expected to alter selection on parasite virulence because it does not impact on R0. However, this would implicitly assume that once hosts have developed immunity to a particular parasite strain, they are also immune to all other strains, suggesting a perfect ‘immune range’. In reality, however, the acquired immune memory developed by hosts may be highly specific (e.g. in dengue virus [13]). Thus, while parasite strains with very similar antigenic configurations may be recognized by the hosts’ immune systems, those emerging from larger mutations are likely to escape the hosts’ recognition systems. A number of studies have explored the role of cross-immunity in long-term epidemiological dynamics, in particular in whether immunity gained from one strain can provide immunity to a strain in the future (e.g. following year). The focus of these models has largely been to understand the population dynamics of two competing strains and the conditions for stable coexistence of strains to occur in, for example, whooping cough [12] and influenza [11]. Other studies have focused explicitly on the evolutionary turnover of influenza strains, showing that invasion of the second strain to coexistence is possible where there is partial cross-immunity [33,34]. However, more long-term evolutionary studies are needed to understand the creation and maintenance of wider diversity of parasite strains.
Here, we shall thoroughly explore the role that partial cross-immunity (through the hosts’ ‘immune range’) has on the evolution of diversity in parasite virulence through evolutionary branching. Extending the work of previous studies examining single invasion events in populations of fixed size [33,34], we examine a fully dynamic eco-evolutionary model over a much longer time scale, investigating whether a greater range of parasite strains is possible. Taking an adaptive dynamics approach, we analyse an SIR-type model which allows for varying population sizes and explore how epidemiological feedbacks may lead to the creation of diversity in parasite strains through evolutionary branching.
2. Modelling and results
2.1. Baseline model: perfect immune range
For comparison, we begin by first fully examining the classic case of parasite evolution within an SIRS epidemiological framework, assuming a perfect host immune range, with the resident dynamics given by
| 2.1 |
| 2.2 |
| 2.3 |
All hosts reproduce at rate a, which is reduced owing to crowding by qN, where N is the total population size. Infected hosts may be partially sterilized by factor f (where f can take values between 0 and 1). All hosts suffer background mortality rate b, with infected hosts suffering additional mortality, or virulence, at rate α. Transmission of disease is a mass-action density-dependent process with parameter β. Infected hosts recover from infection at rate γ to full immunity, but this immunity wanes at rate δ. The subscripts denote whether a trait/density relates to the resident, r, or mutant, m. These epidemiological dynamics result in a unique, stable endemic equilibrium whenever
We shall base our evolutionary analysis within the framework of adaptive dynamics [17], analysing the success of small, rare mutants attempting to invade a resident equilibrium. In this initial study, our aim is to investigate how partial cross-immunity may lead to the creation and maintenance of diversity in parasite strains within a host population. We therefore begin by assuming that each parasite type varies in its transmission–virulence strategy, following the classic trade-off theory of parasite evolution such that transmission is a saturating function of virulence, with β′(α) > 0 and β″(α) < 0. Additionally, we shall assume that each parasite type differs in its antigenicity. For analytical tractability in this initial work, we further assume that antigenicity varies along just a single axis and that there is a simple one-to-one link between antigenicity and transmission–virulence strategy, meaning that all strains can in fact be identified by their virulence rate. We acknowledge the biological simplifications made here, but these are required to gain a significant amount of analytical insight into our key question. Assuming that host immunity is perfect, such that mutant parasites are unable to infect resident immune hosts, an invading mutant parasite (at low density) would initially have dynamics
| 2.4 |
The fitness of the parasite is then defined as
| 2.5 |
Assuming small mutation steps, the parasite will evolve virulence in the direction of the local selection gradient, 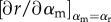 until a singular strategy, α*, is reached where
until a singular strategy, α*, is reached where
| 2.6 |
with
| 2.7 |
The stability of this singular point, α*, depends on two conditions: evolutionary stability (ES; is the point locally invadable?) and convergence stability (CS; is the point locally attracting?). The point is ES provided
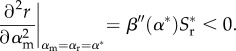 |
2.8 |
CS depends on the sum of ES and mutual invasibility (MI), which here evaluates to
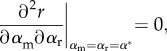 |
2.9 |
meaning that the CS condition is also simply
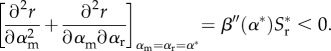 |
2.10 |
In this baseline case, then, the evolutionary behaviour depends entirely upon the trade-off curvature, β″(α). Under the standard assumption that the trade-off always has negative curvature (i.e. transmission is a saturating function of virulence), it is clear that the singular strategy will always be both ES and CS and is therefore an uninvadable attractor of evolution or a continuously stable strategy (CSS). It is straightforward to show that this singular point corresponds to where R0 is maximized. Were we to assume that the curvature could be positive, then in this case the strategy would be an invadable repeller. For evolutionary branching to occur, it is required that the singular point be attracting (CS) but invadable (not ES). In fact, it will suffice to show that the MI condition is satisfied (i.e. MI < 0) for branching to be possible, as suitable trade-offs could always be chosen to satisfy the stability conditions [31,35,36]. Here, because MI = 0, clearly branching cannot occur.
2.2. Limited immune range
We now extend the model to include a limited host immune range. We assume a one-dimensional ‘antigenic space’ [34], with the immune range purely determined by the difference between the parasite strains' virulence rates. As such, a mutant parasite strain, whether it is one of lower or higher virulence than the resident, is now able to (partially) escape the host's immune defences, which have been specifically adapted to the current resident parasite. We therefore now allow mutant parasites to gain some infections from resident immune hosts. The epidemiological model for a (monomorphic) resident parasite strain is identical to that in (2.1)–(2.3). However, the dynamics for an invading mutant parasite (at low density) would now be
| 2.11 |
The function σ(αm,αr) represents the immune range of the host and is assumed to be zero when the immune host is challenged by the parasite that it was previously infected by (i.e. at αm = αr). In other words, an immune host cannot be reinfected by the same parasite type. We assume that the host's immune range, and thus the infection success of mutant parasite types against immune hosts, is a function of how different the mutant is from the resident, using a Gaussian-type function
| 2.12 |
As shown in figure 1a, this continuous function given by (2.12) results in zero infection when the mutant and resident are identical, with the level of infection tending to β(αm) as the two strains become more different (i.e. as σ(αm, αr) → 1). Here the parameter ρ defines the host's immune range, where a higher value implies a wider range of parasite strains to which the host is immune (figure 1a).
Figure 1.
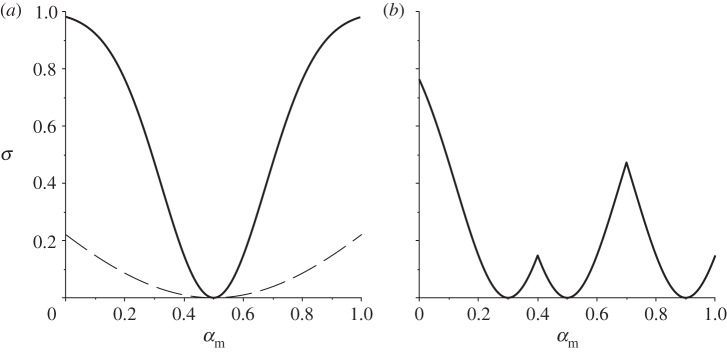
Examples of the ‘immune range’ function. (a) Different shapes of σ(αm, αr), as in equation (2.12), for varying ρ (αr = 0.5; solid line: ρ = 0.25; dashed line: ρ = 1). (b) An example immune range of a host that has immunity to multiple parasite strains, with σ(αm, αA) = min{σ(αm, α1),…,σ(αm, αn)} as in equation (2.29) (α1 = 0.3, α2 = 0.5, α3 = 0.9, ρ = 0.25).
Before continuing, we should mention here some notational details. For convenience, we shall assume the following notation when dealing with derivatives of σ(αm, αr):
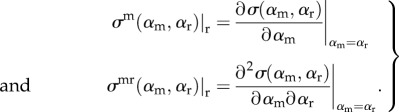 |
2.13 |
We also collect together some important aspects of σ(αm, αr) here
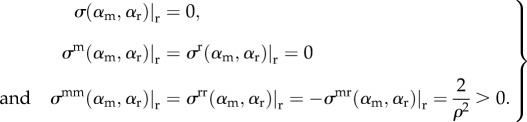 |
2.14 |
The fitness for this model becomes
| 2.15 |
However, when evaluated at αm = αr, owing to the relations in (2.14) we find again that
| 2.16 |
with Sr still given by (2.7). Thus, the selection gradient at any point, and therefore the location of any singular points, is unaffected by σ(αm,αr), the ability of mutant parasites to infect resident immunes. This is owing to the assumption of small mutations, required by the framework of adaptive dynamics, as the amount of infection from immune hosts remains negligible.
While the location of the singular point is not affected by this alteration, the stability conditions at that point are. In particular, the ES condition becomes
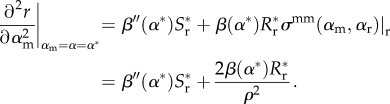 |
2.17 |
This term is larger than that in the previous model (equation (2.8)), and hence depending on the immune range the ES status of the singular strategy could be changed. In particular, we note that for narrow ranges (ρ small) the immune range term will dominate and the singular strategy would always be invadable by any mutant; this is further emphasized by the fact that decreasing ρ increases fitness (2.15) of any invader.
The MI condition is no longer zero, but now becomes
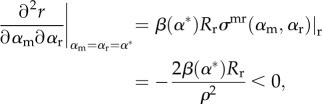 |
2.18 |
which, being negative, gives the potential for evolutionary branching.
Finally, the full CS condition remains
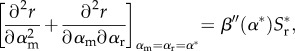 |
2.19 |
as in the previous model (equation (2.10)), and we particularly draw attention to the fact that it still does not depend on immune range or indeed immune period, unlike the ES and MI conditions. It is important to note, then, that the transmission–virulence trade-off must still be saturating (i.e. β″(α) < 0) for a singular point to be attracting but that so long as the immune range is narrow enough (ρ small) any saturating trade-off may yield a branching point.
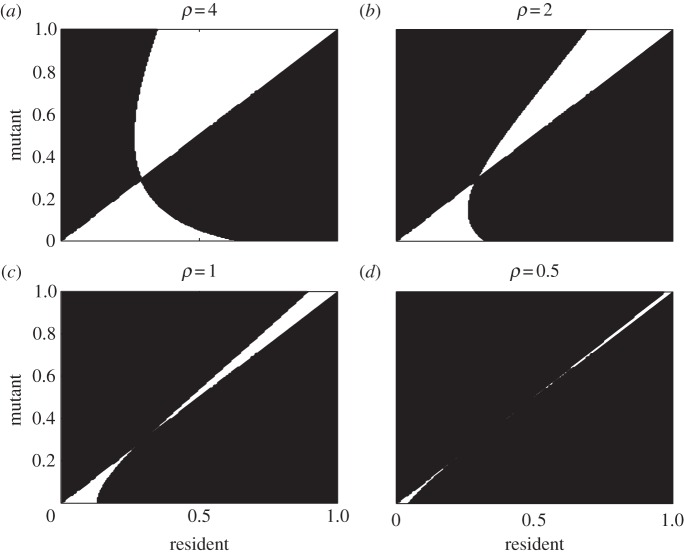
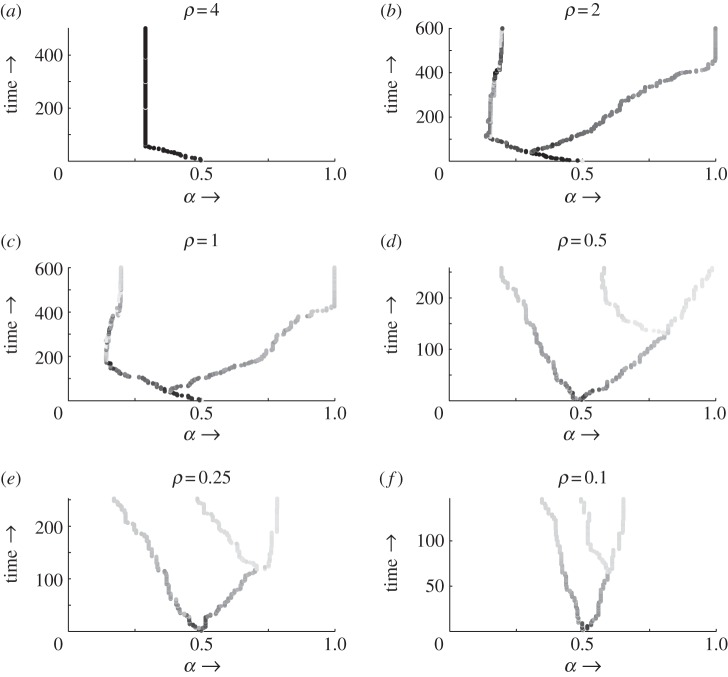
It is clear from these conditions, therefore, that while immune range does not affect the location of a parasite's singular strategy, it does change the stability properties. As we show in the pairwise invasion plots (PIPs) in figure 2, by altering the immune range, and the parameter ρ in particular, we may therefore transform a CSS into a branching point (A → B). Reducing the range still further (B → C → D) results in a larger proportion of mutants being able to invade their residents. As ρ → 0, all mutants are able to fully infect immune hosts; however, all resident hosts are protected from being replaced and going extinct (as they will also have positive fitness when rare). Assuming non-negligible mutations, we therefore reach a point where any mutant will always invade its resident, resulting in immediate coexistence. These differing behaviours can also be seen from the outcomes of evolutionary simulations (see appendix A for a description of the simulation procedure). Figure 3a corresponds to the PIP of figure 2a and shows the population evolving to the CSS and then remaining there. Figure 3b–d corresponds to the PIPs of figure 2b–d and shows that the population evolves to the singular strategy but then branches into two coexisting strains that then continue to coevolve. For reasons discussed above, as ρ decreases, coexistence between the mutant and resident strains becomes more likely, and hence the branching event occurs sooner (before α reaches the singular point), and eventually, as in three dimensions, it occurs immediately.
Figure 2.
Pairwise invasion plots for parasite evolution for varying ρ. Resident trait values are along the horizontal axis and mutant values are along the vertical axis. For each resident–mutant pair, if the mutant's fitness is positive there is black shading and if it is negative there is white shading. Evolution will proceed by small mutational steps up or down the main diagonal until a zero-isocline is crossed, and there is an evolutionarily singular strategy. (a) A CSS at the singular point; (b–d) evolutionary branching points. Parameter values: a = 2, q = 0.1, f = 0.1, b = 0.5, γ = 0.5, δ = 0.1 and β(α) = 4 − 2(1 − α)/(1 + 2α).
Figure 3.
Output from evolutionary simulations (see appendix A for a description of the simulation procedure). The shading represents the density of strains as (evolutionary) time progresses. (a) The CSS from figure 2a, and (b–d) the branching point from figure 2b–d, with the two coexisting strains continuing to evolve. (d–f) A second branching point occurring, with (d) relating to figure 4a–c. Parameter values are as of figure 2, with the initial α = 0.5 and mutation size given by a normal distribution with standard deviation of 0.01.
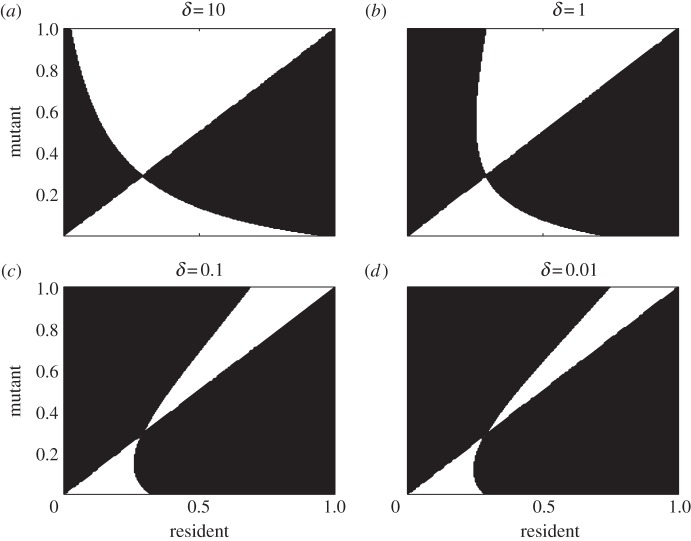
Clearly, the MI and ES terms also depend on the size of the resident immune population, with greater densities of R (recovered hosts) leading to a greater range of trade-off shapes yielding evolutionary branching. This will depend to varying extents on all of the parameters in our model, with a key dependence being on the rate of waning immunity, δ. Intuitively, we would expect R to be a decreasing function of δ (if immunity wanes quickly, few hosts will ever be immune), and although we may note that Rr = γIr/(b + δ), because I is itself a function of δ this is of limited analytical insight. Numerical investigation does indeed suggest that R is a decreasing function of δ, meaning that lower values of δ will make the ES term more positive and the MI term more negative, increasing the potential for evolutionary branching, as seen in the PIPs in figure 5. We also note that because S (susceptible hosts) is independent of δ, the location of the singular point is again unaffected.
Figure 5.
Pairwise invasion plots for varying rates of waning immunity, δ. Parameter values are as of figure 2 with ρ = 2.
2.3. Two-strain system
If the parasite evolves to an evolutionary branching point, it will undergo disruptive selection and emerge as two coexisting strains on either side of the singular point. The two-strain resident dynamics of this system then become
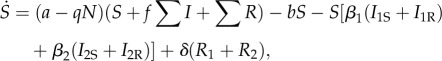 |
2.20 |
| 2.21 |
| 2.22 |
| 2.23 |
| 2.24 |
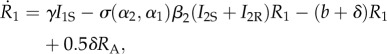 |
2.25 |
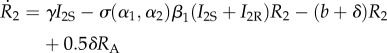 |
2.26 |
| 2.27 |
The parameters retain their meanings from the previous model. There are now four infected types, where I1S is a host infected with parasite strain 1 that was susceptible to both strains, while I1R was recovered and immune from strain 2. There are two recovered and immune types to each strain and RA denotes a recovered host immune to both (all) parasite strains. We note that we have assumed that fully immune hosts are equally likely to lose their immunity to either strain. We also note here that σmi(αm, αj)|i = 0, for i ≠ j,m as this should lead to cancellations in the derivatives.
The fitness of a mutant from either strain is given by
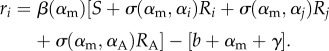 |
2.28 |
The term, σ(αm, αA)RA, represents a mutant parasite strain attempting to infect a host that is immune to both existing resident strains. We shall assume that in an n-strain system
| 2.29 |
such that the host's local immune range always holds true (figure 1b). We note that other forms for this function could be used, though our simulation results suggest that this choice has a very minor quantitative effect on the outcomes.
The selection gradients are therefore
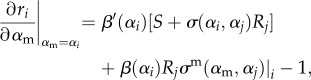 |
2.30 |
for each strain i. (Here, we make the reasonable assumption that given a mutation arising from resident strain i then locally min{σ(αm, α1),(αm, α2),…,σ(αm, αn)} = σ(αm, αi).) This produces a two-dimensional coevolutionary system, with co-singular strategies occurring where the selection gradients of both strains are simultaneously zero. The stability of such a co-singular point requires similar conditions to above, namely (a) Is the strategy ES for each strain? (b) Is the strategy MI for each strain? (c) Is the strategy coevolutionary convergent? Condition (c) now depends on the eigenvalues from a 2 × 2 Jacobian matrix. We note that MI is now a stability condition in its own right. As for either strain to be able to branch again it is required that it must have MI < 0, and that the trade-off curvature can always be chosen to satisfy the other conditions [31,35,36], we therefore limit our analysis to determining the sign of this condition. After some work, we find this condition reduces to (appendix B)
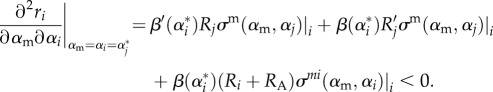 |
2.31 |
The sign of the first two terms will depend on whether strain j has higher or lower virulence than the mutant of strain i. However, the final term is again a multiple of −2β(α*)/ρ2, which will dominate for ρ small. Once more, then, wherever an attracting co-singular point exists, by choosing the curvature of the immune range to be high enough it will be possible for MI < 0, and therefore to see further branching.
We can consider the coevolutionary behaviour of the phase space of the two strains by building upon our PIPs from figure 2. Firstly, we can identify the region of coexistence for the two strains by ‘folding’ the PIP along its main diagonal [17]. Coexistence can occur where regions of positive invasion fitness overlap upon this folding (as here both strains could invade the other when rare). In figure 4a, we have performed this on the PIP from figure 2d, with shaded regions denoting coexistence and the white circle identifying the initial branching point. After branching, we can examine the coevolutionary trajectory through this coexistence region (we can focus on just one side of the main diagonal, as the two cases are symmetric). We can then find the isoclines for each strain by solving equations (2.30) numerically for each strain pair and finding the zero contours. These isoclines are overlaid as the lines on figure 4a. Here we see a very large coexistence region, with the two strains initially diverging away after branching (i.e. trajectories head northwest, with strain 1 to lower values and strain 2 to higher values). The two isoclines are shown to cross at (α1, α2) ≈ (0.21, 0.84), indicating a co-singular point. We demonstrate the behaviour in the insets by numerically plotting PIPs for each strain nearby the co-singular point, assuming the other is at its co-singular point. These show that the point is attracting for both strains, forming a CSS for strain 1 but a further branching point for strain 2. We would therefore expect the parasite to branch once more to form a three-strain system. This is confirmed by the evolutionary simulations in figure 3d. Extending the model to greater numbers of strains becomes computationally expensive. However, from the results of previous work we can conclude that, by showing the potential to branch to three coexisting strains, we expect there to be no limit to the level of diversity that can evolve [31,37,38].
Figure 4.
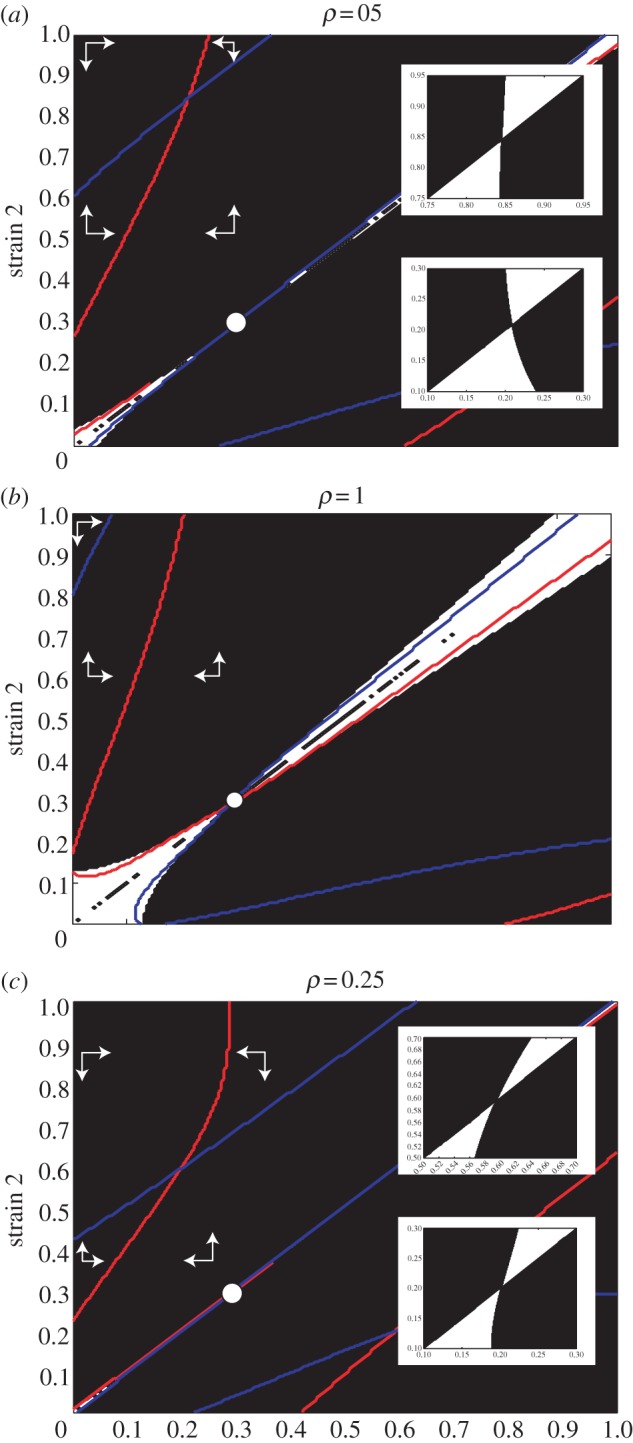
Dynamics of the two-strain system for varying ρ. The main plots show the coexistence region after branching, created by folding the PIPs from figure 2 over their main diagonal and shading areas of MI. The red lines show the isoclines of strain 1 and the blue lines show the isoclines of strain 2, with the arrows indicating the direction of selection. Where the two isoclines cross there is a coevolutionary singular strategy, with the independent behaviour of each strain shown by the inset PIPs (in (a) and (c)). Parameter values are as of figure 2, with ρ as marked.
As suggested above, we find that the potential for multiple branching in this system depends strongly on the immune range function (i.e. ρ). In figure 4b,c, we explore the impact of varying ρ. Figure 4b shows the coexistence plot for higher ρ (ρ = 1; cf. figure 2c), which shows that no further co-singular point occurs within the parameter range. In this case, we would expect to see strain 2 maximize virulence and strain 1 adopt a CSS at α1 ≈ 0.2. This is demonstrated by the simulation in figure 3c. Figure 4c shows the coexistence plot for lower ρ (ρ = 0.25), which now shows a co-singular point, which is a branching point for both strains (insets figure 4c; we would expect here just one of the two strains to branch, as demonstrated by the simulation in figure 3e). This ties to previous work [37] on branching in competition models, and [31] on branching in host–parasite coevolutionary models, with the steepness of such a function increasing the degree of branching and eventual coexistence. Furthermore, in the case of the first branching, as ρ decreases there is an increase in the possibility of coexistence, and hence branching occurred earlier (figure 3b–d); this is echoed for the second branching event, in that as ρ decreases the second branching event takes place sooner (figure 3d–f) and we would expect this to be true for the third branching event and beyond. We also note that the process of branching ‘slows down’ the overall evolutionary dynamics, such that we see less extreme parasite types at a set time point when more than one branching has occurred (in fact, evolution will necessarily slow down as it approaches a singular point as the selection gradient approaches zero).
3. Discussion
The classic result from the evolutionary theory of parasite virulence is that, assuming a saturating transmission–virulence trade-off, there is a single evolutionary optimum where R0 is maximized [9,10]. However, the theory has long identified instances where coexistence of parasite strains within a host population owing to ecological/epidemiological feedbacks can occur. Here, we have thoroughly explored a model of partial cross-immunity, where a host's immune memory is less effective against parasite strains that are dissimilar to those it has previously been infected by. We have shown that, provided the immune range of the host is sufficiently limited, high levels of coexistence can, and are very likely to, emerge through multiple evolutionary branching events.
Specificity is one of the most fundamental aspects of the vertebrate adaptive immune system. It is therefore inappropriate to assume that hosts infected with any parasite strain will be immune to all future mutant variants that may arise and partial cross-immunity should be considered. Previous studies have often found that parasite genotypes will cycle when there is cross-immunity, but it has also been shown that an invading strain can grow to coexist with a pre-existing strain [33,34]. We have extended these results to a fully dynamic framework, showing that, subject to the assumptions of adaptive dynamics [17], high levels of diversity may evolve. We note that our results show that evolutionary branching occurs for immune range functions that are not especially steep (and hence hosts have a wide immune range) such that in fact nearby mutants gain only very small levels of infection. In addition, as the immune range function becomes steeper (and hence hosts have a narrow immune range), then evolutionary branching not only becomes more likely but also occurs more quickly, increasing the probability of diversity. Our model therefore provides evidence that partial cross-immunity may be a key explanatory factor in the high levels of parasite diversity seen in natural systems.
Models based within an evolutionary ecology framework have largely assumed that mutations lead to small phenotypic changes, for example with a parasite mutant gaining a small increase in transmission potential (and a correspondingly small increase in virulence). This is in contrast to the ‘all-or-nothing’ infection assumption of the classic gene-for-gene and matching-allele models where parasites either can or cannot infect host strains depending on their genetic configurations [1,5]. A recent direction of theoretical work has been to approximate such an ‘all-or-nothing’ function in an evolutionary ecology setting. For example, Best et al. [31] showed that if hosts coevolve the ‘range’ of parasites that they are resistant to, and parasites the ‘range’ of hosts that they can infect, then widespread static diversity, as well as coevolutionary cycles, may result. Similarly, Boots et al. [38] focused purely on host evolution and found that if infection was more common among related individuals again high levels of coexistence could emerge. Here, we have applied a similar function to parasite evolution, assuming that the host has a limited immune range that mutant parasites can partially evade. The effect of these functions is to create a considerable advantage for rare mutants over residents, leading to negative frequency dependence, and thus the potential for the creation of high levels of diversity. From an environmental feedback perspective, the effect is to make parasite fitness depend not only on the number of susceptible hosts, as in R0 maximization, but also on the number of recovered hosts. Either way, it is clear that the epidemiological feedbacks generated by the host's immune range allow for far greater diversity in parasite virulence than that predicted by classic theory.
For tractability, we have assumed a simple one-dimensional ‘antigenic space’ [34], where the degree of cross-immunity may be described solely by the difference between the virulence rates of mutant and resident parasite strains. In reality, antigenic space is likely to have multiple dimensions depending on a range of chemical factors. It is also far from clear that antigenic differences can be translated to differences in virulence and transmission in such a straightforward way. We would suppose that increasing the number of potential differences between strains could only create more potential feedbacks to parasite evolution, and therefore increase the potential level of diversity. Such a model is likely to be highly intractable for even low numbers of coexisting strains, however, but further work in this area may yield important insights.
In our Introduction, we outlined the variety of processes identified by mathematical models that can lead to coexistence of parasite strains in host populations, rather than simply the type that maximizes R0. An interesting avenue for further study would be to identify tests that would allow us to discriminate between these different processes using empirical data. Such a task would not be straightforward, because multiple processes are likely to exist in many natural and laboratory populations. However, we might hope to be able to discriminate between ecological (density dependence/predation) and epidemiological (superinfection/specialism/cross-immunity) causes of coexistence in specific systems.
The importance of the epidemiological R0 to parasite evolution is not in doubt. However, our key message is that the classic result of parasite evolution, that there is a single optimal parasite strain, does not always hold true as there are a variety of ecological and epidemiological processes that can encourage the creation and maintenance of diversity in parasite virulence. Furthermore, we have shown that, for small immune ranges (high specificity to immunity), pathogen diversity is not only possible but almost certain.
Acknowledgements
We are grateful to the organizers of and contributors to the Modelling Biological Evolution 2013 conference that inspired this Theme Issue. We particularly thank the speakers and keynotes in the ‘Evolution in host–parasite interactions and immunity’ session for an inspiring set of talks.
Appendix A. Simulation details
(1) Start with a monomorphic population, with trait value α (taken as 0.5 in all simulations).
(2) Run the ODE model for time tf. If any of the final densities are below a (low) threshold, then set them to zero. For each positive parasite population and host density, record the final density values and define them as Nres. (If there is more than one parasite strain, label them Nres1, Nres2, etc.)
(3) Label the existing strain as αres. (If there is more than one strain, label them αres1, αres2, etc.)
(4) When multiple parasite strains are present, choose which parasite strain gives rise to a mutation based on relative population sizes of hosts infected with each parasite (i.e. the probability that parasite i gives rise to a mutation is ‘total number of hosts infected with parasite i/total number of infected hosts’).
(5) Create a mutant strain by drawing a number at random from a normal distribution with mean αres and standard deviation σmut (taken as 0.01 in all simulations). Label this value as αmut. If there is more than one parasite strain present, randomly select from which strain the mutation will occur (with appropriate bias for population size—i.e. the larger the population, the more likely it is the mutation will come from that population).
(6) Add equations for this mutant parasite type to the existing equations, creating an (n + 1)-strain model (where n is the number of trait values, i.e. 1 mutant and n resident values). For the initial densities, we take αres to have densities Nres and αmut to have ‘low’ density.
(7) Repeat from step 2.
Appendix B. Mutual invasibility result in two-strain system
Let us set X = S + σ(αi, αj)Rj. The fitness gradient of a strain i can then be written as
| B1 |
which for a (co)-singular point requires
| B2 |
When evaluated at the singular point, 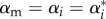 , the MI condition will be
, the MI condition will be
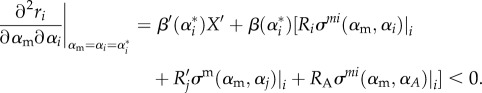 |
B3 |
As σ(αm, αA) = min{σ(αm, α1), σ(αm, α2)}, then locally we can write this as
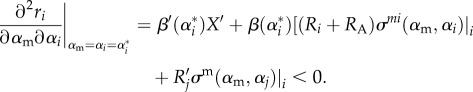 |
By considering equations (2.21) and (2.22), we find that
| B4 |
giving the equilibrium condition
| B5 |
and, therefore,
| B6 |
Substituting the singular point condition of (B 2) into (B 6) gives
| B7 |
Thus, the MI condition becomes
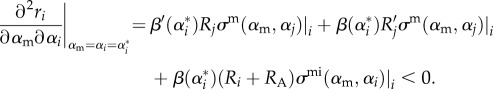 |
B8 |
References
- 1.Flor HH. 1956. The complementary genic systems in flax and flax rust. Adv. Genet. 8, 29–54. ( 10.1016/S0065-2660(08)60498-8) [DOI] [Google Scholar]
- 2.Jayakar SD. 1970. A mathematical model for interaction of gene frequencies in a parasite and its host. Theor. Popul. Biol. 1, 140–164. ( 10.1016/0040-5809(70)90032-8) [DOI] [PubMed] [Google Scholar]
- 3.Thompson JN, Burdon JJ. 1992. Gene-for-gene coevolution between plants and parasites. Nature 360, 121–125. ( 10.1038/360121a0) [DOI] [Google Scholar]
- 4.Sasaki A. 2000. Host–parasite coevolution in a multilocus gene-for-gene system. Proc. R. Soc. Lond. B 267, 2183–2188. ( 10.1098/rspb.2000.1267) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agrawal A, Lively CM. 2002. Infection genetics: gene-for-gene versus matching-allele models and all points in between. Evol. Ecol. Res. 4, 79–90. [Google Scholar]
- 6.Anderson RM, May RM. 1981. The population dynamics of microparasites and their invertebrate hosts. Phil. Trans. R. Soc. Lond. B 291, 452–524. ( 10.1098/rstb.1981.0005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kermack WO, McKendrick AG. 1927. Contributions to the mathematical theory of epidemics. Proc. R. Soc. Lond. A 115, 700–721. ( 10.1098/rspa.1927.0118) [DOI] [Google Scholar]
- 8.Alizon S, Hurford A, Mideo N, van Baalen M. 2009. Virulence evolution and the trade-off hypothesis: history, current state of affairs and the future. J. Evol. Biol. 22, 245–259. ( 10.1111/j.1420-9101.2008.01658.x) [DOI] [PubMed] [Google Scholar]
- 9.Bremermann HJ, Pickering J. 1983. A game-theoretical model of parasite virulence. J. Theor. Biol. 100, 411–426. ( 10.1016/0022-5193(83)90438-1) [DOI] [PubMed] [Google Scholar]
- 10.Bremermann HJ, Thieme H. 1989. A competitive exclusion principle for pathogen virulence. J. Math. Biol. 27, 179–190. ( 10.1007/BF00276102) [DOI] [PubMed] [Google Scholar]
- 11.Minayev P, Ferguson N. 2009. Improving the realism of deterministic multi-strain models: implications for modelling influenza A. J. R. Soc. Interface 6, 509–518. ( 10.1098/rsif.2008.0333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Restif O, Grenfell BT. 2006. Integrating life history and cross-immunity into the evolutionary dynamics of pathogens. Proc. R. Soc. B 273, 409–416. ( 10.1098/rspb.2005.3335) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams B, Holmes EC, Zhang C, Mammen MP, Nimmannitya S, Kalayanarooj S, Boots M. 2006. Cross-protective immunity can account for the alternating epidemic pattern of dengue virus serotypes circulating in Bangkok. Proc. Natl Acad. Sci. USA 103, 14 234–14 239. ( 10.1073/pnas.0602768103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metz JAJ, Geritz SAH, Meszena G, Jacobs FJA, Van Heerwaarden JS. 1996. Adaptive dynamics: a geometrical study of the consequences of nearly faithful reproduction. In Stochastic and spatial structures of dynamical systems (eds van Strein SJ, Verduyn Lunel SM.), pp. 183–231. North-Holland, The Netherlands: Elsevier. [Google Scholar]
- 15.Dieckmann U, Law R. 1996. The dynamical theory of coevolution: a derivation from stochastic ecological processes. J. Math. Biol. 34, 579–612. ( 10.1007/BF02409751) [DOI] [PubMed] [Google Scholar]
- 16.Marrow P, Dieckmann U, Law R. 1996. Evolutionary dynamics of predator–prey systems: an ecological perspective. J. Math. Biol. 34, 556–578. ( 10.1007/BF02409750) [DOI] [PubMed] [Google Scholar]
- 17.Geritz SAH, Kisdi E, Meszena G, Metz JAJ. 1998. Evolutionarily singular strategies and the adaptive growth and branching of the evolutionary tree. Evol. Ecol. 12, 35–57. ( 10.1023/A:1006554906681) [DOI] [Google Scholar]
- 18.Rueffler C, Van Dooren TJM, Metz JAJ. 2006. The evolution of resource specialization through frequency-dependent and frequency-independent mechanisms. Am. Nat. 167, 81–93. ( 10.1086/498275) [DOI] [PubMed] [Google Scholar]
- 19.Hoyle A, Bowers RG. 2008. Can possible evolutionary outcomes be determined directly from the population dynamics? Theor. Popul. Biol. 74, 311–323. ( 10.1016/j.tpb.2008.09.002) [DOI] [PubMed] [Google Scholar]
- 20.Boots M, Haraguchi Y. 1999. The evolution of costly resistance in host–parasite systems. Am. Nat. 153, 359–370. ( 10.1086/303181) [DOI] [PubMed] [Google Scholar]
- 21.Boots M, Bowers RG. 1999. Three mechanisms of host resistance to microparasites—avoidance, recovery and tolerance—show different evolutionary dynamics. J. Theor. Biol. 201, 13–23. ( 10.1006/jtbi.1999.1009) [DOI] [PubMed] [Google Scholar]
- 22.Miller MR, White A, Boots M. 2005. The evolution of host resistance: tolerance and control as distinct strategies. J. Theor. Biol. 236, 198–207. ( 10.1016/j.jtbi.2005.03.005) [DOI] [PubMed] [Google Scholar]
- 23.Hoyle A, Best A, Bowers RG. 2012. Evolution of host resistance towards pathogen exclusion: the role of predators. Evol. Ecol. Res. 14, 125–146. [Google Scholar]
- 24.Levin S, Pimental D. 1981. Selection of intermediate rates of increase in parasite–host systems. Am. Nat. 117, 308–315. ( 10.1086/283708) [DOI] [Google Scholar]
- 25.Boldin B, Geritz SAH, Kisdi E. 2009. Super-infection and adaptive dynamics of pathogen virulence revisited: a critical function analysis. Evol. Ecol. Res. 11, 153–175. [Google Scholar]
- 26.Pugliese A. 2002. On the evolutionary coexistence of parasite strains. Math. Biosci. 177, 355–375. ( 10.1016/S0025-5564(02)00083-4) [DOI] [PubMed] [Google Scholar]
- 27.Svennungsen TO, Kisdi E. 2009. Evolutionary branching of virulence in a single-infection model. J. Theor. Biol. 257, 408–418. ( 10.1016/j.jtbi.2008.11.014) [DOI] [PubMed] [Google Scholar]
- 28.Best A, White A, Boots M. 2009. The implications of co-evolutionary dynamics to host–parasite interactions. Am. Nat. 173, 779–791. ( 10.1086/598494) [DOI] [PubMed] [Google Scholar]
- 29.Donnelly R, Best A, White A, Boots M. 2013. Seasonality selects for more acutely virulent parasites when virulence is density dependent. Proc. R. Soc. B 280, 20122464 ( 10.1098/rspb.2012.2464) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gudelj I, Fitt BDL, van den Bosch F. 2004. Evolution of sibling fungal plant pathogens in relation to host specialization. Anal. Theor. Plant Pathol. 94, 789–795. [DOI] [PubMed] [Google Scholar]
- 31.Best A, White A, Kisdi E, Antonovics J, Brockhurst MA, Boots M. 2010. The evolution of host–parasite range. Am. Nat. 176, 63–71. ( 10.1086/653002) [DOI] [PubMed] [Google Scholar]
- 32.Morozov A, Best A. 2012. Predation on infected host promotes evolutionary branching of virulence and pathogens’ biodiversity. J. Theor. Biol. 307, 29–36. ( 10.1016/j.jtbi.2012.04.023) [DOI] [PubMed] [Google Scholar]
- 33.Andreasen V, Sasaki A. 2006. Shaping the phylogenetic tree of influenza by cross-immunity. Theor. Popul. Biol. 70, 164–173. ( 10.1016/j.tpb.2006.04.004) [DOI] [PubMed] [Google Scholar]
- 34.Adams B, Sasaki A. 2007. Cross-immunity, invasion and coexistence of pathogen strains in epidemiological models with one-dimensional antigenic space. Math. Biosci. 210, 680–699. ( 10.1016/j.mbs.2007.08.001) [DOI] [PubMed] [Google Scholar]
- 35.Bowers RG, Hoyle A, White A, Boots M. 2005. The geometric theory of adaptive evolution: trade-off and invasion plots. J. Theor. Biol. 233, 363–377. ( 10.1016/j.jtbi.2004.10.017) [DOI] [PubMed] [Google Scholar]
- 36.Kisdi E. 2006. Trade-off geometries and the adaptive dynamics of two co-evolving species. Evol. Ecol. Res. 8, 959–973. [Google Scholar]
- 37.Adler FR, Mosquera J. 2000. Is space necessary? Interference competition and limits to biodiversity. Ecology 81, 3226–3232. ( 10.1890/0012-9658(2000)081[3226:ISNICA]2.0.CO;2) [DOI] [Google Scholar]
- 38.Boots M, White A, Best A, Bowers R. 2012. The importance of who infects whom: the evolution of diversity in host resistance to infectious disease. Ecol. Lett. 15, 1104–1111. ( 10.1111/j.1461-0248.2012.01832.x) [DOI] [PubMed] [Google Scholar]