Abstract
PIWI proteins, a subfamily of the ARGONAUTE/PIWI protein family, have been implicated in transcriptional and posttranscriptional gene regulation and transposon silencing mediated by small non-coding RNAs, especially piRNAs. Although these proteins are known to be required for germline development, their somatic function remains elusive. Here, we examine the maternal function of all three PIWI proteins in Drosophila; Piwi, Aubergine (Aub) and Argonaute3 (Ago3) during early embryogenesis. In syncytial embryos, Piwi displays an embryonic stage-dependent localization pattern. Piwi is localized in the cytoplasm during mitotic cycles 1-10. Between cycles 11 and 14, Piwi remains in the cytoplasm during mitosis but moves into the somatic nucleus during interphase. Beyond cycle 14, it stays in the nucleus. Aub and Ago3 are diffusely cytoplasmic from cycle 1-14. Embryos maternally depleted of any one of the three PIWI proteins display severe mitotic defects, including abnormal chromosome and nuclear morphology, cell cycle arrest, asynchronous nuclear division and aberrant nuclear migration. Furthermore, all three PIWI proteins are required for the assembly of mitotic machinery and progression through mitosis. Embryos depleted of maternal PIWI proteins also exhibit chromatin organization abnormalities. These observations indicate that maternal Piwi, Aub and Ago3 play a critical role in the maintenance of chromatin structure and cell cycle progression during early embryogenesis, with compromised chromatin integrity as a possible cause of the observed mitotic defects. Our study demonstrates the essential function of PIWI proteins in the first phase of somatic development.
Keywords: PIWI, argonaute 3, aubergine, maternal, mitosis, embryonic, chromosome
INTRODUCTION
The ARGONAUTE/PIWI (AGO/PIWI) protein family is a central player in small-RNA- mediated gene regulation. The PIWI subfamily associates with a unique class of small noncoding RNA partners called PIWI-interacting RNAs (piRNAs) and are crucial for a wide variety of developmental processes. In Drosophila Melanogaster, the PIWI protein family consists of Piwi, Aubergine (Aub) and Argonaute3 (Ago3). For clarity, in this paper, PIWI refers to the protein subfamily whereas Piwi refers to the Piwi protein itself. Mechanistic details of PIWI/piRNA complex function are not well understood but are thought to involve gene and transposon regulation at the epigenetic, post-transcriptional and translational levels (Juliano et al, 2011). The focus of PIWI/piRNA function thus far has been in germline development. All three PIWI proteins facilitate key aspects of germline development including germ cell specification (Megosh et al, 2006; Harris and MacDonald, 2001), germline stem cell function (Cox et al 1998; Cox et al 2000) and axis determination during oogenesis (Klattenhoff et al, 2007; Li et al, 2009).
In contrast to the well-known roles of PIWI in the germline, the somatic function of these proteins has yet to be clearly demonstrated. In this study, we focus on early embryogenesis to study the somatic function of PIWI for three major reasons: First, it represents the first and most important stage of somatic development. Second, in early embryos, all three proteins are expressed throughout the embryo in both germline and somatic cells. This is in contrast to their expression in adult flies where Aub and Ago3 are restricted to the germline. Thus, the early embryonic stage provides a useful setting for uncovering possible somatic functions for PIWI proteins. Third, piwi, aub and ago3 mutants are viable but infertile, as they lay eggs that do not hatch, implying their requirement for embryogenesis. This requirement could be ascribed to the oocyte patterning abnormalities seen in several piRNA pathway mutants. However, defects during oogenesis can be rescued via inactivation of DNA damage signaling whereas embryonic lethality persists, suggesting added complexity that requires further examination (Klattenhoff et al 2007 ; Khurana and Theurkauf, 2010).
Recent studies further implicate PIWI proteins in early embryogenesis. Embryos laid by piRNA pathway mutants display fragmentation of the zygotic genome after normal fertilization and deficiencies in assembly of the telomere protection complex (Khurana et al 2010). A role for Aub and Ago3 in regulating the maternal-to-zygotic transition via degradation of maternal transcripts was also recently described (Rouget et al 2010) . The PIWI/piRNA pathway thus merits careful examination for its role in embryogenesis and any understanding gained could shed light on somatic functions mediated by this important family of proteins. In this study, we systematically analyze the maternal requirement of each PIWI protein during early embryogenesis and demonstrate their shared role in mitosis and chromatin organization.
MATERIALS AND METHODS
Drosophila strains and culture
The following Drosophila melanogaster strains were used to generate maternally depleted mutant embryos: for the piwi mutant, w; piwi1, FRT40A/CyO (Cox et al,1998), yw P[hsFLP] ; P[ovoD1] 2L , FRT 40A /CyO (Chou and Perrimon, 1996), and yw P{hsFLP}12; Sco/Cyo (Bloomington); for the aub mutant, aubHN2/CyO and aubQC42/CyO (Bloomington), and both alleles are EMS induced point mutations (Schupbach and Wieschaus, 2001; Harris and Macdonald, 2001); for the ago3 mutant: bw1; st1 ago3t2/TM6B, Tb+ and bw1; st1 Ago3t3/TM6B, Tb1 (Bloomington), both of which are loss of-function alleles with premature stop codons (Li et al, 2009); and for the mnk;ago3 double mutant: w; mnk[P6]/ SM6, TM6B, Tb (from M. Brodsky). The w1118 strain was used as wild-type. All strains were grown at 22-25°C on yeast-containing molasses/agar medium.
Collection of embryos depleted of maternal Piwi, Aub or Ago3
Embryos depleted of maternal piwi were generated through the following genetic crosses: yw P[hsFLP]12; P[ovoD1]2L, FRT40A/CyO males were crossed to w; piwi1, FRT40A/CyO virgin females to produce yw P[hsFLP]12; P[ovoD1]2L, FRT40A/piwi1, FRT40A progeny. Larvae were heat shocked on days 3-6 for one hour in a 37°C incubator to induce mitotic recombination. The heat-shocked females with germline clones were crossed to w−males. Due to the presence of a copy of the ovoD1 gene in these females, only piwi− germline clones can give rise to mature eggs.
To obtain embryos depleted of maternal Aub or Ago3, the following transheterozygotic combinations were made by crossing heterozygotic males and virgin females: aubHN2/ aubQC42 and ago3t2/ago3t3. aub and ago3 null females that resulted from these crosses were then mated with heterozygotic aub and ago3 males respectively to give embryos maternally depleted of PIWI protein.
Immunostaining
Embryos were collected, dechorionated in 50% bleach, and fixed in 50% heptane, 50% fixative (3 parts fixing buffer, 1.33X PBS and 67mM EGTA :1 part 37% formaldehyde) for 10 mins. Embryos were then washed and devitellinized in methanol (MeOH) and stored at -20 degrees. Before staining, embryos were washed in a rehydration series consisting of 70%MeOH: 30%PBST, 50%MeOH: 50%PBST, 30%MeOH:70% PBST and finally 100% PBST for 5 mins each, where PBST is PBS with 0.2% Triton X Embryos were blocked in 5% normal goat serum for 1hour.
The following antisera were used for immunofluorescent staining: guinea pig Piwi generated against peptide residues 826-844 (1:200), mouse Aub (1:500, gift from H.Siomi), mouse Ago3 (1:500, gift from H.Siomi), mouse monoclonal alpha tubulin antibody (1:200, Sigma, St. Louis, MO), rabbit centrosomin antisera (1:200, gift from T. Kaufman), mouse monoclonal lamin antibody (1:200, Iowa Hybridoma Bank) ,rabbit Ser 10 Phospho-Histone H3 (1:200, Cell Signalling Technology), mouse HP1a antisera (1:200, Iowa Hybridoma Bank), rabbit methly3lysine9, (1:200, Upstate Biotechnology Co., Lake Placid, NY), rabbit ORC2 antisera (1:500, gift from S.Bell), rabbit γH2Av (1:2000, Rockland Immunochemicals). All the fluorescence-conjugated secondary antibodies were Alexa-Fluor from Invitrogen (Carlsbad, CA) and were used at 1:400 dilution. All dilutions were made in 5% normal goat serum in PBST.
Live imaging of wildtype and PIWI-depleted early embryos
Embryos depleted of maternal PIWI were produced as described above. Immediately after egg laying, embryos were dechorionated in bleach, rinsed, and suspended in halocarbon oil 27 (Sigma, St. Louis, MO) in an embryo chamber containing air-permeable Teflon on the top of the chamber and a vacuum grease sealed coverslip on the bottom. Images were collected every five minutes for six hours using a Leica ASMDW confocal microscope.
Statistical Analysis
Statistical significance for cellularization frequency in the movies was assessed via Chi-square analysis performed with one degree of freedom using the wild-type cellularization frequency as a control.
RESULTS
Localization of Piwi, Aub and Ago3 during early embryogenesis
To understand the function of maternal PIWI proteins during early embryogenesis, we first examined the localization of Piwi, Aub and Ago3 using antibodies against the endogenous protein. After fertilization and pronuclear fusion, Drosophila embryos undergo 13 rapid synchronous nuclear mitotic divisions without cytokinesis, forming a syncytial blastoderm. The founding germ cells or pole cells are the first to cellularize at cycle 9, while somatic nuclei continue to divide as a syncytium before cellularizing at cycle 14, followed by gastrulation (Campos-Ortéga and Hartenstein, 1997).
Piwi displays a developmentally dynamic pattern of localization in somatic cells as well as in germline cells in early embryos, while Aub and Ago3 constantly stay cytoplasmic. During mitotic cycles 1- 8 of early embryogenesis, all three proteins localize diffusely throughout the embryo (Fig. 1A-C’). At cycle 11, Piwi in pole cells enters the nucleus. Between cycle 11 and 14, Piwi appears to be in the somatic nucleus during interphase (Fig.1D,D’, J, J’) but moves into the cytoplasm during mitosis ( with prophase shown in Fig.1G,G’) as reported previously (Rouget et al, 2010).
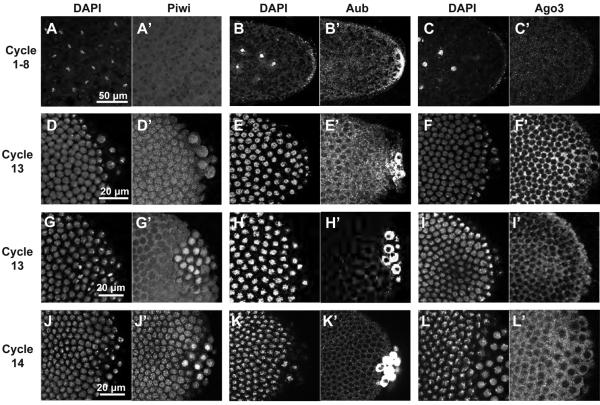
Figure 1. Piwi exhibits a dynamic localization during early embryogenesis while Aub and Ago3 are cytoplasmic.
Confocal micrographs of wildtype embryos stained for DAPI (A-L) Piwi (A’, D’, G’, J’), Aub (B’, E’, H’, K’) or Ago3 (C’, F’, I’, L’). Scale bars on the leftmost image denote scale bars for the whole row. Between cycles 1-8, embryos reveal diffuse cytoplasmic staining for Piwi, Aub and Ago3(A-C’). At cycle 13, Piwi is the nuclei in both somatic pole cells at interphase (D, D’) but is localized to the cytoplasm of somatic nuclei at prophase (G, G’). It remains nuclear in both somatic and pole cells at cycle 14 (J, J’). At cycle 13, Aub remains cytoplasmic at early prophase (E, E’) and metaphase (H,H’) in both somatic embryo and pole cells, and remains so at cycle 14 after the embryo is cellularized (K, K’). Ago3 shows similar cytpolasmic localization in both somatic and pole cell nuclei as Aub from cycle 13 (F, F’, I, I’) to cycle 14 (L, L’). Scale bars on the leftmost image denote scale bars for the whole row.
Aub is initially concentrated within the pole plasm at the posterior end of embryos (Fig 1B’) and later incorporated into pole cells, where it continues to be present in the cytoplasm. While enriched in the germline, it is also clearly present in somatic cell cytoplasm (Fig. 1E’). Ago3 appears to be equally abundant in the cytoplasm of both germline and somatic cells (Fig. 1F’). After pole cell formation (cycle 9), Aub and Ago3 also become enriched peri-nuclearly (compare: Fig. 1B’, E’ and C’, F’). Unlike Piwi, Aub and Ago3 remain in the cytoplasm throughout the cell cycle during cycle 11-13. (Fig. 1E’,F’,H’,I’). At interphase of cycle 14, Aub and Ago3 continue to be localized to the cytoplasm (Fig. 1K’, L’).
Embryos examined for Piwi localization were also co-stained for heterochromatin protein 1a (HP1a), a non-histone protein primarily associated with heterochromatin. Localization patterns were compared since HP1a interacts with Piwi (Brower-Toland et al. 2007), exhibits dynamic localization during the mitotic cell cycle in Drosophila and has an essential role during embryogenesis (Kellum and Alberts, 1995) . HP1a is diffusely cytoplasmic till Cycle 7 (Supp Fig. 1C), after which it enters the nucleus but is apparently excluded from the bulk of chromosomes (Supp Fig.1G). From Cycle 11 onwards, it appears to coincide with chromatin within the nucleus (Supp Fig 1 K and O). The difference between the nuclear localization of Piwi and and that of HP1a is quite striking from cycle 8-13, where PIWI is excluded from the nucleus during mitosis while HP1a is enriched in the nucleus (compare: Supp Fig 1F,G and 1J,K).
Embryos depleted of maternal Piwi, Aub or Ago3 display various mitotic defects
Mutations in all three PIWI proteins lead to embryonic lethality, suggesting an essential function in embryogenesis (Cox et al.1998; Schüpbach and Wieschaus, 1991; Li et al. 2009). Keeping in mind the widespread and dynamic localization of all three proteins in somatic cells of the embryo, we analyzed embryos laid by PIWI protein-null females, herein called PIWI-deficient embryos for simplicity (where PIWI will be specified as Piwi, Aub, or Ago3 when appropriate), to investigate a possible requirement of PIWI proteins for normal cell cycle progression and embryonic development distinct from pole cell formation. Our analysis shows that PIWI-deficient embryos have more than one defect as assayed by DAPI staining, and these defects span most of early embryogenesis, so that the majority of the embryos arrest before gastrulation.
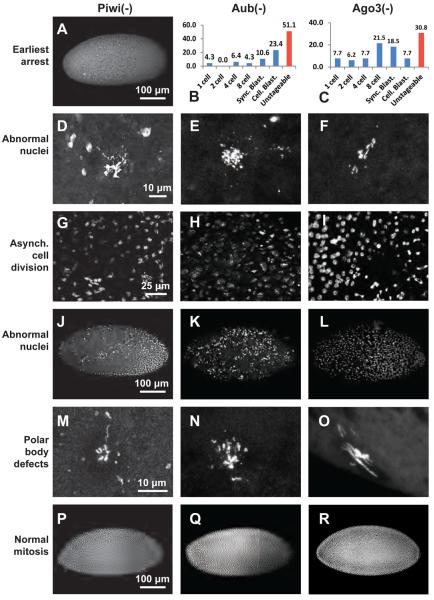
PIWI-deficient embryos were collected at 1-hour intervals and were allowed to age for 3 hours before analysis to ensure that the phenotypes seen were true arrest and not developmental delays. The earliest and most severe defect in all three PIWI-deficient embryos is complete mitotic arrest at cycle one, which occurred in 53% of the Piwi-deficient embryos (n=200) but only in 8% (n=65) and 4% (n = 47) of Ago3- and Aub-deficient embryos, respectively (Fig. 2A-C). The early arrest of Piwi-deficient embryos was confirmed by live imaging. These embryos fail to cellularize at a significantly higher rate than wildtype embryos (p<0.001, and see Sup. Movie A, B). Many of the embryos fail to cellularize in seven hours, indicating that these embryos are truly arrested, and not developmentally delayed. A more gradual arrest phenotype was seen for Aub- and Ago3-deficient embryos that show variable developmental stopping points spanning mitotic cycles 1 through 14 (Figs. 2B and 2C). 51% and 31% of the Aub- and Ago3-deficient embryos, respectively, are unstageable, owing to severe defects in nuclear morphology as discussed below.
Figure 2. Maternally PIWI-depleted embryos display various mitotic defects.
Confocal micrographs of DAPI-stained Piwi-depleted (A,D,G,J,M,P), Aub-depleted(E,H,K,N,Q) and Ago3-depleted (F,I,L,O,R) embryos displaying: (A) Arrest at one cell stage (B,C) Stage of arrest distribution (D-F) Abnormal nuclear morphology, (G-I) Asynchronous nuclear division, (J-L) Abnormal nuclear migration, (M-O) Polar body defects and (P-R) No mitotic defects. Scale bars on the leftmost image denote scale bars for the whole row.
In PIWI-deficient embryos that do not arrest at cycle 1 but enter mitosis, dividing nuclei exhibit a wide range of defects. Abnormal nuclear morphology was observed in 20% of piwi-deficient embryos (Fig. 2D). Strikingly, 85% of Aub-deficient (Fig 2E) and 92% of Ago3-deficient (Fig. 2F) embryos show nuclear abnormalities, including aberrantly condensed chromosomes, abnormal ploidy, and severe fragmentation. For zoomed in images of these defects, see Figure 4. Furthermore, while wildtype embryos undergo synchronized nuclear divisions in the syncytial embryo, 16% of the Piwi-deficient (Fig. 2G), 11% of the Aub-deficient (Fig. 2H and 10% of the Ago3-deficient (Fig.2I) embryos exhibit asynchronous nuclear divisions, where nuclei in different stages of mitosis can be seen in the same embryo. Additionally, 24% of the Piwi-deficient embryos (Fig. 2J), 28% of Aub-deficient (Fig.2K) and 22% of Ago3-deficient (Fig. 2L) embryos have abnormally spaced nuclei, either due to migration defects or nuclear fallout. Finally, defects in polar body morphology that could indicate meiotic failures during oogenesis are seen in 2%, 11%, and 15% of Piwi-, Aub-, and Ago3-deficient embryos, respectively (Fig 2M-O). The remaining 17% of Piwi-deficient (Fig. 2P), 15% of Aub-deficient (Fig.2Q) and 7% of the Ago3-deficient (Fig. 2R) embryos exhibit no obvious mitotic defects. These defects seen in the majority of embryos laid by PIWI-null mothers indicate that all three maternal PIWI proteins are required for normal mitosis during early embryogenesis.
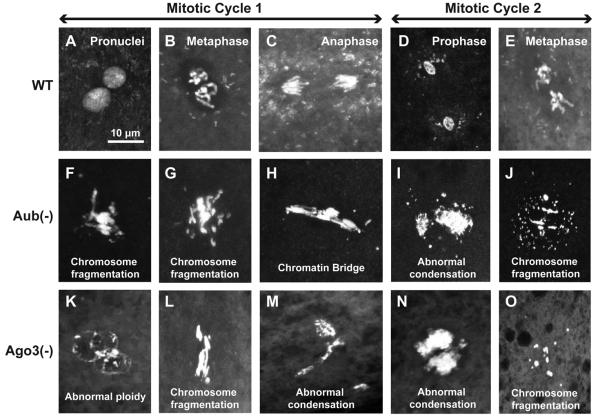
Figure 4. Mitotic defects are present during very early nuclear divisions in Aub- and Ago3- deficient embryos.
Confocal micrographs of DAPI-stained embryos 30 mins after egg laying (A-E) Wildtype embryos in each stage of mitotic cycle 1 and 2 are easily discernable (F-J) Early Aub-deficient embryos display severe defects including chromosome fragmentation, chromatin bridge formation and condensation abnormalities (K-O) Nuclei in very early Ago3-depleted embryos show segregation defects, chromosome fragmentation and inappropriate condensation. The scale bar in A represents 10μm for A-O.
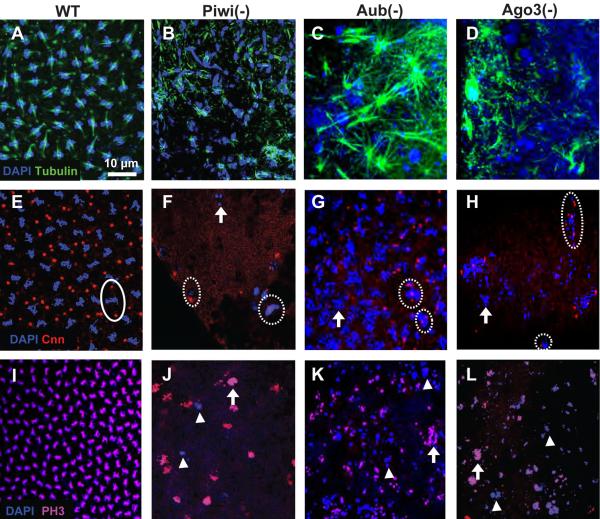
To explore these mitotic defects in more detail, we examined the assembly of the mitotic machinery by assaying spindle formation. Wildtype embryos stained with tubulin and treated with 2uM Taxol (which stabilizes microtubules) contain well-ordered microtubule arrays emanating from chromosomes aligned on the metaphase plate (Fig. 3A). Microtubule organization in Piwi-, Aub-, and Ago3-deficient embryos appears disrupted, often with microtubules present in many asters that do not fuse to form spindles (Fig. 3B-D). In addition, the distribution, morphology, and size of the asters are highly variable both between and within the embryos. Consistent with this finding, Centrosomin (Cnn), a microtubule organizing center component, also exhibits defects. Instead of finding two centrosomes per nucleus as in the mitotic nuclei of wildtype embryos (Fig. 3E, closed circles represent one nucleus bound by two centrosomes), we saw nuclei with abnormal numbers of centrosomes, i.e. nuclei with one, or more than two centrosomes (Fig. 3F-H, dashed circles) as well as nuclei with no Cnn signal (Fig.3F-H, arrows). This further indicates a mitotic defect in the PIWI-depleted embryos.
Figure 3. Maternally PIWI-depleted embryos exhibit mitotic abnormalities.
Confocal micrographs of wildtype (A, E, I), Piwi- (B, F, J), Aub- (C, G, K) and Ago3- deficient (D, H, L) embryos. (A) Wildtype embryos show normal organization of the spindle apparatus. DAPI is in blue and alpha tubulin is in green. (B-D) Alpha tubulin (green) is stretched and spindle morphology is abnormal in mutants. (E) Two centrosomes per nucleus (closed circle) is seen in wildtype embryos. Centrosomes are highlighted by Centrosomin (Cnn), in red. (F-H) Abnormal Cnn (red) expression is seen in mutant embryos. Dashed circles represent nuclei with too few or too many Cnn positive signals and arrows highlight nuclei with no Cnn signal. (I) All nuclei in wildtype embryos concurrently stain positively for phosphorylated histone 3 (red) (J-L) Nuclei in mutant embryos show variability in PH3 staining. Some nuclei are positive for PH3 while neighboring nuclei are negative (arrowheads). Very fragmented nuclei are also able to enter mitosis (arrows). The scale bar in A represents 10μm for A-L.
To assay for mitotic synchrony, we looked at Ser10 phosphorylation on histone H3 (PH3), a commonly used marker for mitotic entry. The first 14 mitotic divisions in wild type embryos are synchronous. PH3 staining is thus detected simultaneously throughout the embryo in all nuclei (Fig. 3I). In contrast, Piwi-, Aub-, and Ago3-deficient embryos show asynchronies in mitosis as seen by variable PH3 staining; while some nuclei are in M phase of mitosis, several neighboring nuclei are not PH3-positive (arrowheads, Fig. 3J-L). Several nuclei that do enter mitosis contain fragmented and morphologically abnormal chromosomes (arrows, Fig 3J-L), suggesting that mitotic entry is not prohibited by obvious defects in chromosome structure in these mutants. To further explore cell cycle regulation in PIWI-deficient embryos, we looked at Cyclin levels. Early embryonic mitotic cycles in Drosophila are non-canonical and characterized by rapid S-to-M phase transitions with little gap between them (Garcia et al, 2007). A key regulator of these early cell divisions is Cyclin dose; appropriate Cyclin A and B levels appear to be especially important for mitotic entry, progression through the cell cycle and mitotic exit (Lee and Orr-Weaver, 2003). Aub and Ago3 mutants show marked overexpression of both Cyclin A and B at the protein level (Supp Figs. 2A and B) indicating possible cell cycle regulation defects and substantiating the mitotic entry asynchronies seen with PH3 staining. All mitotic defects found were highly penetrant; majority of the embryos analyzed displayed the abnormalities described.
Mitotic defects during the 1st nuclear division in Aub- and Ago3- deficient embryos
Considering the variable arrest phenotype and severe nuclear morphology abnormalities seen in Aub- and Ago3-deficient embryos, we examined newly laid embryos to pinpoint when these defects start occurring. 30 minutes after egg laying, wild type embryos in each stage of mitotic cycle 1 and 2 can be easily discerned based on number and morphology of nuclei (Fig. 4A-E).
84% (n=25) of early Aub-deficient embryos display severe defects within the first two mitotic divisions. These early nuclei show excessive chromosome fragmentation (Fig. 4F, G and J) and chromatin bridge formation at an anaphase-like stage (Fig. 4H). Abnormal condensation is frequently seen with certain regions being more highly condensed than normal (Fig. 4F and G) as well as highly decondensed nuclei (Fig. 4I). A large fraction of Ago3- deficient embryos (93%, n=30) also display early abnormalities, with nuclei most frequently being severely fragmented (Fig. 4L and O). Segregation abnormalities, as scored by uneven numbers of DAPI positive signal, are additionally noticed (Fig 4K). Similar to Aub-deficient embryos, a subset of the nuclei also exhibits chromosome condensation aberrations (Fig. 4M and N).
Although most of Aub and Ago3- deficient embryos display severe defects within the first two mitotic divisions, not all of them arrest within the first two mitotic divisions. A small fraction of them can divide further, albeit aberrantly, up to the cellular blastoderm stage before arrest, which contributes to the distribution of Aub- and Ago3-deficient embryos with diverse defects seen in later mitotic cycles (Fig. 2). This, combined with phosphohistone-3 staining of fragmented nuclei (Fig. 3J-L) adds support to the observation that mitotic cycling is not prohibited by chromosomal defects in these early embryos. No correlation exists between embryos that display early nuclear abnormalities and dorsal appendage number, suggesting that the patterning defects seen in these PIWI-deficient embryos are separable from these additional mitotic defects. Live imaging was not possible immediately after fertilization since these nuclear divisions mostly occur in utero and deep inside the embryo. This excluded the possibility of following these early abnormal embryos to see if they could proceed further in development. However the fact there was no clear 2-4 cell arrest phenotype seen suggests that most of these abnormal nuclei continue to divide (abnormally) a variable number of times.
PIWI-deficient embryos exhibit defects in nuclear and chromatin organization
Because all three types of PIWI-deficient embryos show distinct nuclear morphology abnormalities, including fragmentation and condensation defects, we were interested in whether the loss of a PIWI protein affects nuclear and chromatin organization in embryos aged 3-4 hours. To evaluate nuclear organization, we stained PIWI-depleted embryos with lamin, a major component of the nuclear lamina. Wildtype embryos display even, circular localization of lamin that closely surround the chromosomes within (Fig. 5A). In contrast, Piwi-, Aub-, or Ago3- deficient embryos display lamin localization that is highly spatially distorted. Notably, nuclear lamina appears stretched, and not in proportion to the area occupied by the chromosomes (Fig. 5B-D). The nuclear lamina has been implicated in anchoring heterochromatin during interphase, as well as in DNA replication (Dechat et al, 2008). The observed laminal defects could potentially link the observed mitotic defects (Figure 3) to a chromatin-organizational defect.
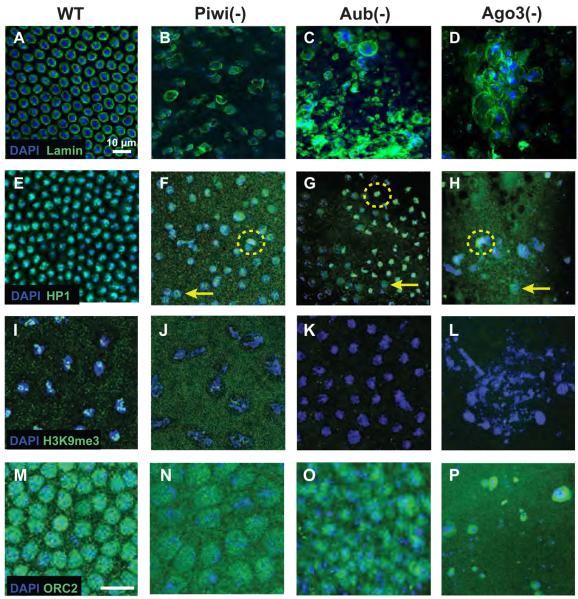
Figure 5. Maternally PIWI-depleted embryos exhibit defects in nuclear and chromatin organization.
Confocal micrographs of wildtype (A,E,I,M) , Piwi-depleted (B,F,J,N), Aub-depleted (C,G,K,O) and Ago3-depleted (D,H,J,P) embryos. (A) Lamin expression in wildtype embryos is even and circular (B-D) Lamin staining in mutant nuclei is highly abnormal and indicates nuclear morphology disruption (E) Even HP1a expression is seen on chromosomes (F-H) Abnormally high HP1a expression (dashed circles) and highly diffuse signal (arrows) are both present in the nuclei examined. (I) Wild type expression of histone 3 Me3K9, a chromatin modification involved in HP1a localization (J) Piwi mutants show a reduction in histone 3 Me3K9 expression (K,L) Histone 3 Me3K9 is below the threshold of detection in Aub and Ago3 mutants (M) Staining against ORC2 in wildtype embryos shows a pattern similar to HP1a (N-P) ORC2 staining in PIWI-deficient embryos is diffuse and excluded from chromatin. The scale bar in A represents 10μm for A-L. The scale bar in (M) represents 10μm in M-P.
We next analyzed chromatin organization by studying the localization of HP1a, other known chromatin-binding proteins and chromatin-modifying markers. In Piwi-, Aub- or Ago3- deficient embryos, HP1a appears to be nuclearly localized, but not tightly bound to chromatin (compare Fig. 5E to 5F-H). HP1a is more diffuse as compared to wildtype in certain regions (Fig.5F-H, arrows) while regions of abnormally high HP1a localization are seen also seen (Fig 5F-H, dashed circles). Intriguingly, these polarized differences in HP1a localization do not correlate to DAPI intensity, which can be used as a rough estimation of chromosome condensation. We next looked at histone3 lysine9 methylation (H3K9me3), a histone modification required for HP1a localization (Lachner et al, 2001). Compared to wildtype embryos (Fig 5I), there appears to be a reduction and diffusion of the H3K9me3 signal in Piwi- deficient embryos (Fig 5J), and a complete loss in Aub- or Ago3-deficient embryos (Fig 5K, L). Another candidate for HP1a localization to chromatin is the origin recognition complex (ORC) that is thought to provide a DNA-targeting mechanism for the initiation of heterochromatin formation (Pak et al, 1997; Huang et al, 1998). ORC2 localization in wildtype embryos is nuclear and coincided with HP1a localization (Fig. 5M). In Piwi-, Aub-, or Ago3-deficient embryos, ORC staining was present and in the vicinity of the nucleus but appears diffuse and excluded from chromatin (Fig. 5N-P). This suggests that the defects in HP1a localization could also result from a failure to properly initiate heterochromatin formation via ORC2-mediated mechanism. This would be expected to cause problems in maintenance of heterochromatin, abnormalities in gene expression, and defects in organizing mitotic chromosomes.
Embryonic defects in PIWI-deficient embryos are independent of activation of the DNA damage signaling pathway
Patterning and egg-laying defects in several PIWI/piRNA pathway mutants can be rescued by inactivation of DNA damage checkpoints, suggesting that these defects are a consequence of activation of the DNA damage signaling pathway (Klattenhoff et al, 2007). To evaluate if the defects seen in embryos depleted of maternal PIWI are similarly a consequence of DNA damage checkpoint activation, we first looked for the presence of DNA damage via phosphorylated H2Av staining (γH2Av) in very early embryos. Embryos depleted of Aub and Ago3 both show a significant upregulation in γH2Av, suggesting the presence of unrepaired double-stranded breaks (Fig. 6A-D).
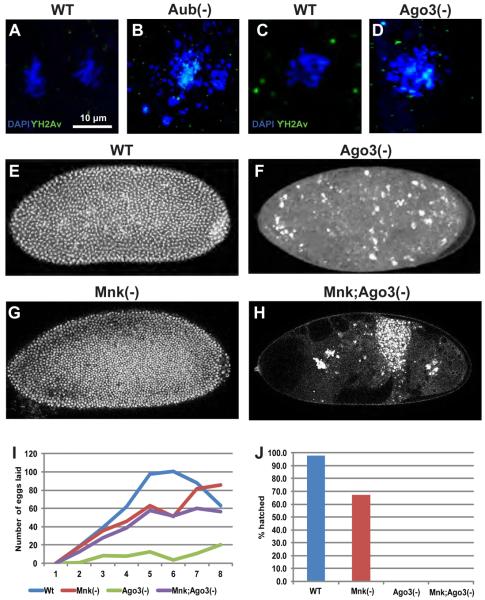
Figure 6. Defects in PIWI-deficient embryos are independent the DNA damage pathway.
(A,C) Nuclei in early wildtype embryos show no phosphorylated H2Av (γH2Av) signal (green) (B,D) Upregulation of γH2Av signal is seen in fragmented nuclei of very early Aub- and Ago3- deficient embryos respectively. The scale bar in (A) represents 10μm for A-D. (E) Confocal image of DAPI stained wildtype embryo after 2-3 hours of development. (F) DAPI stained image of age-matched Ago3-depleted embryo shows highly abnormal nuclei. (G) Mnk(-) mutant embryos show normal early development. (H) Embryos laid by Mnk;Ago3 double mutants are phenotypically similar to those laid by Ago3(−) females. (I) The graph shows the number of eggs laid (y axis) as a function of time (x axis) by females of the indicated genotypes. (J) The graph shows percentage of eggs laid that hatch (y axis) for each indicated genotype (x axis)
Klattenhoff and colleagues reported that suppressing DNA damage signaling in aub mutants could not rescue egg hatching defects (Klattenhoff et al, 2007). We similarly made double mutants of mnk, the Drosophila homologue of Chk2, and ago3 to examine the role of DNA damage signaling in Ago3-depleted embryo defects. While wild type and embryos laid by mnk mutants show normal development (Fig. 6E and G), inactivation of DNA damage signaling could not rescue embryonic defects in ago3 mutants. Embryos laid by mnk;ago3 double mutants appeared phenotypically similar to embryos laid by ago3 mutants (compare Fig. 6F to 6H). Additionally, while egg laying was rescued in embryos depleted of both Mnk and Ago3 (Fig. 6I), egg hatching remained defective (Fig.6J), suggesting that embryonic defects in PIWI-deficient embryos are independent of activation of the DNA damage signaling pathway.
piwi mutants could lay only a few eggs, which precluded similar analysis. Despite this, these results suggest that unlike the axis determination defects seen in PIWI mutants, embryonic defects appear independent of DNA damage pathway activation.
DISCUSSION
In this study, we report an essential somatic function of PIWI proteins in early embryonic mitosis. Although PIWI proteins are commonly regarded as essential only for germline development, exploration of somatic functions for the PIWI/piRNA pathway is becoming increasingly relevant in Drosophila and other organisms. Expression of PIWI proteins and piRNAs is not limited to the germline, but is present in a variety of adult stem cells, tissue types and cancer models (Thomson and Lin, 2009; Juliano et al, 2011; Yan et al, 2011). Functionally, the initially reported involvement of Piwi in germline stem cell self-renewal is driven by expression in somatic niche cells (Cox et al, 1998). Recently, a prominent role for PIWI proteins in the nervous system was described (Lee et al, 2011; Rajasethupathy et al, 2012). Indeed, a molecular mechanism for this family of proteins in epigenetic regulation has been studied mainly in somatic cells (Brower-Toland et al, 2007). Our study contributes significantly to the understanding of essential somatic functions for PIWI proteinsoutside the germline and adds on to an increasing base of evidence for important PIWI/piRNA pathway functions in the soma.
The depletion of maternal PIWI proteins results in a variety of mitotic and chromatin abnormalities that arrests embryo development prior to gastrulation. The severity and pleiotropic nature of defects makes the primary role of the PIWI/piRNA pathway during embryogenesis difficult to determine. The most prominent defects are the one-cell stage arrest of Piwi-deficient embryos and the morphological abnormalities seen in the nuclei of Aub/Ago3-deficient embryos (Figure 2 and 4). Piwi-deficient embryos that manage to escape arrest show similar nuclear abnormalities. Complete arrest at the one-cell stage in most Piwi-deficient embryos hints at a more pronounced role for this protein as compared to Aub and Ago3, where PIWI protein depletion allows most embryos to escape initial arrest.
All these prominent defects as well as the known function of PIWI proteins in epigenetic regulation and in protecting genome integrity (Yin and Lin, 2007; Klattenhoff et al, 2007; Huang et al, 2013) point to a likely defect in chromatin organization as the root of other defects in mitosis. Abnormal staining patterns for lamin and chromatin markers in all three PIWI-deficient embryos (Figure 5) indicate a requirement for maternal PIWI proteins and associated piRNAs in appropriate nuclear and chromatin organization. Indeed, the loss of HP1a and inappropriate histone modification are increasingly being associated with defects during mitotic and meiotic progression due to kinetochore depletion and centromere abnormalities, leading to segregation defects and telomere shortening, all of which could compromise chromosome stability and result in abnormal cell division (Dialynas et al, 2009; Heit et al, 2009; Xu et al, 2009). Chromatin organization mediated by Piwi, Aub and Ago3 could therefore be critical in maintaining chromosome structure and stability during early embryonic mitosis. While it is certainly possible that the primary role of PIWI proteins is in cell cycle progression and chromatin deficiencies are secondary, we favor the first alternative due to two major reasons: 1) Defects in nuclear morphology are seen at the very first nuclear division. A primary role in the mechanics of cell division would result in a more gradual and cell-cycle stage-specific appearance of abnormalities on depletion of PIWI proteins. 2). Cell cycle-specific defects usually result in the activation of cell cycle checkpoints and uniform cell-cycle arrest of all nuclei in the same embryo (Lee and Orr-Weaver, 2003) which does not adequately encapsulate the variety of defects seen in PIWI-deficient embryos. Chromatin organization abnormalities could sufficiently impact early mitotic divisions at the structural level, easily causing abnormal chromosome segregation, defects in spindle assembly, uneven nuclear distribution, and their asynchrony within the same cytoplasm.
It is interesting to note that there is an upregulation of γH2Av signal in very early embryos, indicating the presence of DNA damage. However, embryonic defects are not rescued when DNA damage signaling is abrogated, suggesting that PIWI function in early embryogenesis may be independent of DNA damage checkpoint activation. It is certainly possible that defects during oogenesis could result in the drastic nuclear abnormalities seen in PIWI depleted embryos. Indeed, a characteristic abnormality displayed by piRNA pathway mutants is in compaction of the oocyte nucleus during oogenesis. Mutants show a variety of defective DNA structures instead of a compact karyosome (Gonzalez-Reyes, 1999). However, inactivation of DNA damage signaling appears to restore karyosome defects, but not embryonic nuclear morphology (Abdu et al, 2002; Klattenhoff et al, 2007). Further study of karyosome defects during oocyte formation in PIWI mutants, and examination of the progression of oocyte meiosis is required for further assessment of separable PIWI function during oogenesis and embryogenesis.
Regardless of which possibility is the actual case, PIWI proteins likely achieve their function via a small RNA pathway. Keeping in mind the dynamic subcellular localization pattern seen for all three PIWI proteins and especially Piwi, it is possible that different small RNA targets at different developmental stages allow PIWI proteins to play roles in more than one cellular process. While chromatin organization from Cycles 1-10 is probably indirect and achieved possibly via piRNA target regulation, from cycle 11 on, Piwi, now in the nucleus, might be directly involved in chromatin regulation via HP1a. This possibility is supported by work from our lab showing that Piwi interacts with HP1a and can recruit HP1a to chromatin in an RNA dependent manner (Brower-Toland et al, 2007;Huang et al, 2013).
Supplementary Material
A PIWI-depleted embryo videoed 0.5-7 hours after egg exhibits developmental and cell cycle arrest. Resumption of development is not evidenced during this time period, suggesting that the embryo is arrested and not developmentally delayed.
A wildtype embryo videoed 0.5-7 hours after egg lay develops through the cellular blastoderm stage and gastrulates.
Figure S1. HP1a and Piwi enter the nucleus at different stages during embryogenesis
Confocal micrographs of wildtype embryos stained for DAPI (A,E,I,M) Piwi (B,F,J,N) and HP1a (C,G,K,O). Merged images are represented in D,H,L and P. (A-D) Cycle 7 pro-metaphase embryo reveals diffuse cytoplasmic staining for both HP1a and Piwi. (E-H) Cycle 8 anaphase embryo reveals nuclear staining of HP1a that is excluded from the chromatin as well as diffuse cytoplasmic Piwi localization. (I-L) Cycle 11 embryo in which HP1a is nuclear and Piwi is excluded from the nucleus in somatic cells (M-P) Cycle 14 interphase embryo in which both HP1a and Piwi exhibit nuclear localization in the soma and the germline.
Figure S2. PIWI-deficient embryos contain abnormal levels of cyclin proteins
(A) Cyclin A and B protein levels are highly upregulated in Aub-depleted embryos (B) Cyclin A and B protein levels are highly upregulated in Ago3-depleted embryos. GAPDH is used as a loading control.
ACKNOWLEDGEMENTS
We thank Dr. P. Zamore for ago3 mutants, Dr. M. Brodsky for mnk mutants and Bloomington Drosophila Stock Center for aub mutants. Drs. H.Siomi, T.Kaufman , S.Bell, and Iowa Hybridoma Bank for the antibodies against Ago3, centrosomin, ORC2, and lamin and HP1a, respectively. This work was supported by NIH (DP1CA174416) and the Mathers Foundation.
REFERENCES
- Abdu U, Brodsky M, Schüpbach T. Activation of a Meiotic Checkpoint during Drosophila Oogenesis Regulates the Translation of Gurken through Chk2/Mnk. Current Biology. 200212:1645–1651. doi: 10.1016/s0960-9822(02)01165-x. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Current Opinion in Cell Biology. 200719:238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SCR, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 200721:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TB, Perrimon N. The Autosomal Flp-Dfs Technique for Generating Germline Mosaics in Drosophila Melanogaster. Genetics. 1996144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 199812:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 200822:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dialynas GK, Vitalini MW, Wallrath LL. Linking Heterochromatin Protein 1 (HP1) to cancer progression. Mutat Res. 2008647:13–20. doi: 10.1016/j.mrfmmm.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development. 2003130:859–871. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- Garcia K, Duncan T, Su TT. Analysis of the cell division cycle in Drosophila. Methods. 200741:198–205. doi: 10.1016/j.ymeth.2006.08.013. [DOI] [PubMed] [Google Scholar]
- González-Reyes A. DNA repair and pattern formation come together. Nature Cell Biology. 1999;1:E150–E152. doi: 10.1038/14103. [DOI] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM. aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Heit R, Rattner JB, Chan GKT, Hendzel MJ. G2 histone methylation is required for the proper segregation of chromosomes. J Cell Sci. 2009122:2957–2968. doi: 10.1242/jcs.045351. [DOI] [PubMed] [Google Scholar]
- Huang DW, Fanti L, Pak DTS, Botchan MR, Pimpinelli S, Kellum R. Distinct Cytoplasmic and Nuclear Fractions of Drosophila Heterochromatin Protein 1: Their Phosphorylation Levels and Associations with Origin Recognition Complex Proteins. J Cell Biol. 1998142:307–318. doi: 10.1083/jcb.142.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Yin H, Sweeney S, Raha D, Snyder M, Lin H. A Major Epigenetic Programming Mechanism Guided by piRNAs. Developmental Cell. 201324:502–516. doi: 10.1016/j.devcel.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano C, Wang J, Lin H. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu. Rev. Genet. 201145:447–469. doi: 10.1146/annurev-genet-110410-132541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R, Alberts BM. Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J Cell Sci. 1995108:1419–1431. doi: 10.1242/jcs.108.4.1419. [DOI] [PubMed] [Google Scholar]
- Khurana JS, Theurkauf W. piRNAs, transposon silencing, and Drosophila germline development. J Cell Biol. 2010191:905–913. doi: 10.1083/jcb.201006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana JS, Xu J, Weng Z, Theurkauf WE. Distinct Functions for the Drosophila piRNA Pathway in Genome Maintenance and Telomere Protection. PLoS Genet. 2010;6:e1001246. doi: 10.1371/journal.pgen.1001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell. 200712:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lau NC, Ohsumi T, Borowsky M, Kingston RE, Blower MD. Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi. EMBO J. 200928:2945–2958. doi: 10.1038/emboj.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Banerjee S, Zhou H, Jammalamadaka A, Arcila M, Manjunath BS, Kosik KS. Identification of piRNAs in the central nervous system. RNA. 201117:1090–1099. doi: 10.1261/rna.2565011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LA, Orr-Weaver TL. REGULATION OF CELL CYCLES IN DROSOPHILA DEVELOPMENT: Intrinsic and Extrinsic Cues. Annual Review of Genetics. 200337:545–578. doi: 10.1146/annurev.genet.37.110801.143149. [DOI] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Yin H. A Novel Epigenetic Mechanism in Drosophila Somatic Cells Mediated by PIWI and piRNAs. Cold Spring Harb Symp Quant Biol. 200873:273–281. doi: 10.1101/sqb.2008.73.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Liu N, Mani S, Lin H. The Diverse Functions of PAPI, a Tudor-Domain-Containing Interactor of PIWI Proteins, in Oogenesis and Embryogenesis. (unpublished) [Google Scholar]
- Mahowald AP. Assembly of the Drosophila germ plasm. Int. Rev. Cytol. 2001203:187–213. doi: 10.1016/s0074-7696(01)03007-8. [DOI] [PubMed] [Google Scholar]
- Megosh HB, Cox DN, Campbell C, Lin H. The Role of PIWI and the miRNA Machinery in Drosophila Germline Determination. Current Biology. 200616:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- Lee LA, Orr-Weaver TL. Regulation of Cell Cycles in Drosophila Development: Intrinsic and Extrinsic Cues. Annual Review of Genetics. 200337:545–578. doi: 10.1146/annurev.genet.37.110801.143149. [DOI] [PubMed] [Google Scholar]
- Pak DT, Pflumm M, Chesnokov I, Huang DW, Kellum R, Marr J, Romanowski P, Botchan MR. Association of the Origin Recognition Complex with Heterochromatin and HP1 in Higher Eukaryotes. Cell. 199791:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SCR. Heterochromatic Silencing and HP1 Localization in Drosophila Are Dependent on the RNAi Machinery. Science. 2004303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- Pek JW, Kai T. A Role for Vasa in Regulating Mitotic Chromosome Condensation in Drosophila. Current Biology. 201121:39–44. doi: 10.1016/j.cub.2010.11.051. [DOI] [PubMed] [Google Scholar]
- Pek JW, Kai T. DEAD-box RNA helicase Belle/DDX3 and the RNA interference pathway promote mitotic chromosome segregation. PNAS. 2011108:12007–12012. doi: 10.1073/pnas.1106245108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, Kandel ER. A Role for Neuronal piRNAs in the Epigenetic Control of Memory-Related Synaptic Plasticity. Cell. 2012149:693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AJ, Seipel SA, Hamill DR, Romancino DP, DI Carlo M, Suprenant KA, Bonder EM. Seawi--a sea urchin piwi/argonaute family member is a component of MT-RNP complexes. RNA. 200511:646–656. doi: 10.1261/rna.7198205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouget C, Papin C, Boureux A, Meunier A-C, Franco B, Robine N, Lai EC, Pelisson A, Simonelig M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010467:1128–1132. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma AK, Nelson MC, Brandt JE, Wessman M, Mahmud N, Weller KP, Hoffman R. Human CD34(+) stem cells express the hiwi gene, a human homologue of the Drosophila gene piwi. Blood. 200197:426–434. doi: 10.1182/blood.v97.2.426. [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes TR, Westwood JT, Smibert CA, Lipshitz HD. SMAUG Is a Major Regulator of Maternal mRNA Destabilization in Drosophila and Its Translation Is Activated by the PAN GU Kinase. Developmental Cell. 200712:143–155. doi: 10.1016/j.devcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Takada S, Kelkar A, Theurkauf WE. Drosophila Checkpoint Kinase 2 Couples Centrosome Function and Spindle Assembly to Genomic Integrity. Cell. 2003113:87–99. doi: 10.1016/s0092-8674(03)00202-2. [DOI] [PubMed] [Google Scholar]
- Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu. Rev. Cell Dev. Biol. 200925:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Bai J, Duan Q, Costa M, Dai W. Covalent modifications of histones during mitosis and meiosis. Cell Cycle. 20098:3688–3694. doi: 10.4161/cc.8.22.9908. [DOI] [PubMed] [Google Scholar]
- Yan Z, Hu HY, Jiang X, Maierhofer V, Neb E, He L, Hu Y, Hu H, Li N, Chen W, et al. Widespread expression of piRNA-like molecules in somatic tissues. Nucl. Acids Res. 201139:6596–6607. doi: 10.1093/nar/gkr298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A PIWI-depleted embryo videoed 0.5-7 hours after egg exhibits developmental and cell cycle arrest. Resumption of development is not evidenced during this time period, suggesting that the embryo is arrested and not developmentally delayed.
A wildtype embryo videoed 0.5-7 hours after egg lay develops through the cellular blastoderm stage and gastrulates.
Figure S1. HP1a and Piwi enter the nucleus at different stages during embryogenesis
Confocal micrographs of wildtype embryos stained for DAPI (A,E,I,M) Piwi (B,F,J,N) and HP1a (C,G,K,O). Merged images are represented in D,H,L and P. (A-D) Cycle 7 pro-metaphase embryo reveals diffuse cytoplasmic staining for both HP1a and Piwi. (E-H) Cycle 8 anaphase embryo reveals nuclear staining of HP1a that is excluded from the chromatin as well as diffuse cytoplasmic Piwi localization. (I-L) Cycle 11 embryo in which HP1a is nuclear and Piwi is excluded from the nucleus in somatic cells (M-P) Cycle 14 interphase embryo in which both HP1a and Piwi exhibit nuclear localization in the soma and the germline.
Figure S2. PIWI-deficient embryos contain abnormal levels of cyclin proteins
(A) Cyclin A and B protein levels are highly upregulated in Aub-depleted embryos (B) Cyclin A and B protein levels are highly upregulated in Ago3-depleted embryos. GAPDH is used as a loading control.