Abstract
The methyltransferase like 3 (METTL3)-containing methyltransferase complex catalyzes the N6-methyladenosine (m6A) formation, a novel epitranscriptomic marker; however, the nature of this complex remains largely unknown. Here we report two new components of the human m6A methyltransferase complex, Wilms' tumor 1-associating protein (WTAP) and methyltransferase like 14 (METTL14). WTAP interacts with METTL3 and METTL14, and is required for their localization into nuclear speckles enriched with pre-mRNA processing factors and for catalytic activity of the m6A methyltransferase in vivo. The majority of RNAs bound by WTAP and METTL3 in vivo represent mRNAs containing the consensus m6A motif. In the absence of WTAP, the RNA-binding capability of METTL3 is strongly reduced, suggesting that WTAP may function to regulate recruitment of the m6A methyltransferase complex to mRNA targets. Furthermore, transcriptomic analyses in combination with photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) illustrate that WTAP and METTL3 regulate expression and alternative splicing of genes involved in transcription and RNA processing. Morpholino-mediated knockdown targeting WTAP and/or METTL3 in zebrafish embryos caused tissue differentiation defects and increased apoptosis. These findings provide strong evidence that WTAP may function as a regulatory subunit in the m6A methyltransferase complex and play a critical role in epitranscriptomic regulation of RNA metabolism.
Keywords: WTAP, m6A methyltransferase, METTL3, METTL14, mRNA
Introduction
Nitrogen or oxygen atoms in RNA are ubiquitously methylated with S-adenosylmethionine (SAM, Adomet) serving as the methyl donor1,2,3. The N6-methyladenosine (m6A) modification is one of the most frequent methylations in eukaryotic mRNA, accounting for over 80% of all RNA base methylations and, it has been observed in various species4,5,6,7,8. Recently, two independent studies combining m6A immunoprecipitation with high-throughput analysis revealed that the m6A modification tends to mainly occur in intragenic regions. The frequency is particularly high in the 3′-end of coding sequence (CDS) and the first quarter of the 3′-UTR, with an average of 1 m6A per 2 000 ribonucleotides. The m6A modification occurs in highly conserved regions with the consensus sequence, RRACH (R = G or A; H = A, C or U)9,10,11,12,13. During brain development a dynamic change in m6A levels on RNA has been observed10, suggesting the existence of RNA m6A demethylase(s). Indeed, two AlkB dioxygenase family proteins, fat mass and obesity associated gene (FTO) and ALKBH5, were recently demonstrated to catalyze the removal of the m6A mark both in vitro and in vivo14,15. These two enzymes function in various biological processes such as development, metabolism, and fertility. They regulate the expression levels of thousands of genes, suggesting a pivotal epitranscriptomic function of m6A modification in regulating RNA metabolism8,16,17,18,19.
In contrast to the discovery of RNA m6A demethylases, the nature of the methyltransferase complex responsible for catalyzing m6A formation in RNA still remains largely unknown. The mammalian m6A methyltransferase complex is generally perceived to contain at least two key subcomplexes designated MT-A (200 kD) and MT-B (800 kD)20,21. So far only a 70-kD protein, methyltransferase like 3 (METTL3) has been identified as one component of the 200-kD MT-A complex. METTL3 homologs have been identified in S. cerevisiae, Drosophila and Arabidopsis22,23,24,25,26. Although mRNA methylation occurs primarily within the RRACH consensus sequence in mammals, only a portion of the RRACH sites contain the m6A modification9,10,27. Mechanistic understanding of the selectivity, regulation and function of the m6A modification depends on a complete characterization of the methyltransferase complex.
In this study, we identify for the first time, Wilms' tumor 1 (WT1)-associating protein (WTAP) and methyltransferase like 14 (METTL14) as components of the mammalian m6A methyltransferase complex. In particular, WTAP appears to serve as a regulatory subunit essential for m6A methyltransferase activity.
Results
Identification of WTAP and METTL14 as METTL3-associating proteins
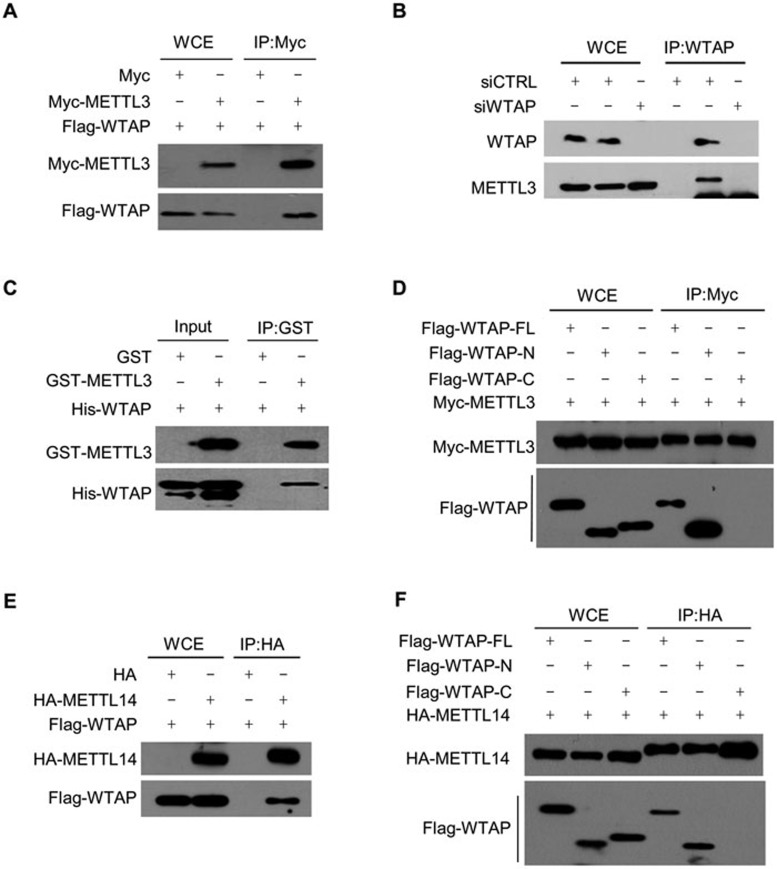
To identify mammalian METTL3-associating proteins, we utilized tandem affinity purification to pull down stably expressed SFB-tagged METTL3 from 293T cells. Subsequent mass spectrometry analysis of affinity-purified SFB-METTL3 protein complexes identified two potential METTL3-associating proteins, WTAP and METTL14 (Supplementary information, Figure S1A). Consistently, AtFIP37 and Mum2, the WTAP homologs in plant and yeast, respectively, have previously been shown to associate with METTL3 in these species26,28. METTL14 protein shares 43% identity (Supplementary information, Figure S1B) with METTL3, and a previous phylogenetic analysis has suggested that the two proteins are homologs23. Furthermore, METTL14 contains similar motifs essential for catalytic activity as METTL3 (Supplementary information, Figure S1B). The interactions among METTL3, WTAP and METT14 were confirmed by co-immunoprecipitation (Figure 1A, 1E and Supplementary information, Figure S2A and S2B). The specificity of the interaction between METTL3 and WTAP was verified by examination of the endogenous interaction between these two proteins after depletion of WTAP by small interfering RNAs (siRNA) (Figure 1B). GST pull-down using purified recombinant GST-METTL3 and 6×His-WTAP proteins demonstrated a direct interaction between METTL3 and WTAP (Figure 1C and Supplementary information, Figure S1C). By using WTAP constructs with either the C- (WTAP-N, 1-200 aa) or N-terminus (WTAP-C, 201-396 aa) truncated, we found that the interaction between WTAP and METTL3 or METTL14 depends on the N-terminus of WTAP (Figure 1D and 1F). It has been reported that RNA is not an essential component of the m6A methyltransferase complex20,29. Consistently, we observed that neither RNase A treatment nor inhibition of methylation by cycleoleucine affected the interaction between WTAP and METTL3 (Supplementary information, Figure S2C-S2E). Based on these findings, we conclude that WTAP and METTL14 are bona fide components of the m6A methyltransferase complex, and that both RNA and the m6A modification are dispensable for the interaction between WTAP and METTL3. In the remaining of this paper, we will refer to this complex as the WMM (WTAP, METTL3 and METTL14) complex.
Figure 1.
WTAP interacts with METTL3 and METTL14. (A) 293T cells were transfected with Flag-WTAP and Myc-METTL3 constructs as indicated. Forty-eight hours later, cells were lysed and the lysates were subjected to immunoprecipitation using anti-Myc (Myc-IP) followed by immunoblotting with the anti-Flag antibodies. (B) 293T cells were treated with control siRNA (siCTRL) or siRNA targeting WTAP (siWTAP) for 48 h. Then cells were lysed and the lysates were subjected to IP using anti-WTAP. The immunoprecipitated samples were analyzed by immunoblotting with the anti-METTL3 antibodies. (C) Purified recombinant His-WTAP proteins were mixed with either GST or GST-METTL3 proteins as indicated, pulled down with GST beads, and subjected to immunoblotting with the indicated antibodies. (D) 293T cells were co-transfected with Myc-METTL3 and Flag-WTAP full-length (-FL), N-terminal (-N) or C-terminal (-C) constructs as indicated. Forty-eight hours later, cells were lysed and the lysates were subjected to Myc-IP followed by immunoblotting with the anti-Flag antibodies. (E) 293T cells were transfected with Flag-WTAP and HA-METTL14 constructs as indicated. Forty-eight hours later cells were lysed and the lysates were subjected to HA-IP followed by immunoblotting with the anti-Flag antibodies. (F) 293T cells were co-transfected with HA-METTL14 and Flag-WTAP full-length (-FL), N-terminal (-N) or C-terminal (-C) constructs as indicated. Forty-eight hours later, cells were lysed and the lysates were subjected to HA-IP followed by immunoblotting with the anti-Flag antibodies. Supportive data were included in Supplementary Information, Figures S1 and S2.
WTAP is required for m6A methyltransferase activity in vivo
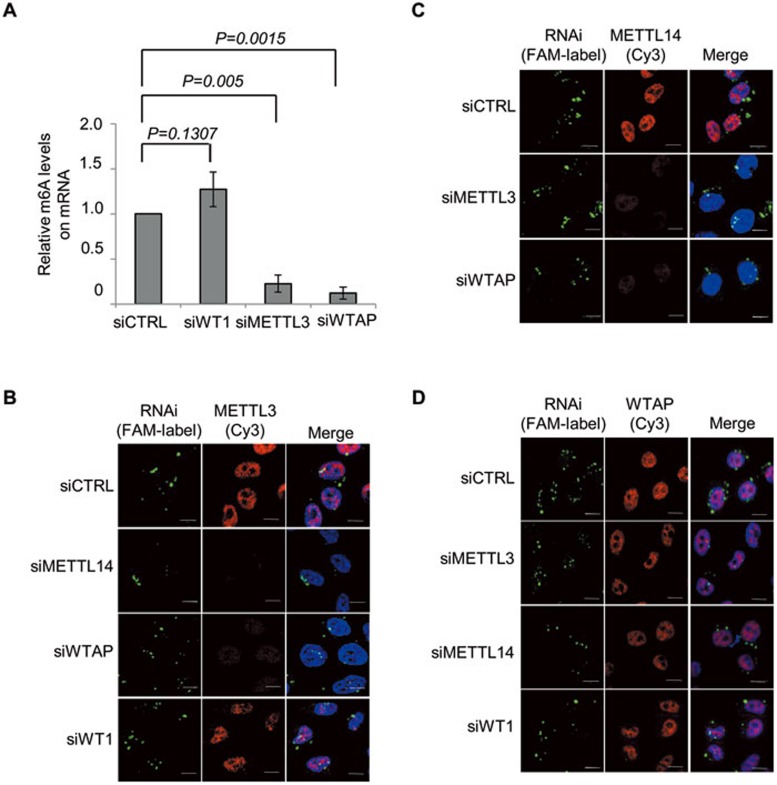
WTAP does not harbor any obvious catalytic domains, and in contrast to METTL3 and METTL14 that were recently demonstrated to form an active complex capable of catalyzing m6A formation, WTAP showed no catalytic activity itself or effect on the activity of METTL3-METTL14 complex in vitro30. Given the direct interaction between WTAP and METTL3, we speculated that WTAP may serve to regulate m6A methyltransferase activity in vivo. To test this hypothesis, we examined the m6A levels in mRNA extracted from WTAP- or METTL3-knockdown cells. Dot-blot assays showed that depletion of either of these proteins resulted in significantly lower m6A levels in mRNA (Figure 2A and Supplementary information, Figure S3A). RT-PCR and western blot analyses verified the knockdown efficiency and specificity, respectively (Supplementary information, Figure S3B and S3C). Moreover, depletion of WTAP or METTL3 did not affect the expression of the other complex components (Supplementary information, Figure S3B and S3C). WTAP was originally identified as a protein associated with WT131. We examined whether WT1 might also be required for m6A methyltransferase activity in vivo. Interestingly, we found that WT1 depletion had no influence on m6A abundance, suggesting that WTAP's role in regulating m6A modification is independent of its previously described binding partner WT1 (Figure 2A and Supplementary information, Figure S3A). Taken together, these findings suggest that WTAP affects m6A methyltransferase activity in vivo.
Figure 2.
WTAP regulates the nuclear speckle localization of METTL3 and METTL14. (A) The graph represents the quantification of three independent dot-blot experiments (raw data were included in Supplementary information, Figure S3A). The y-axis represents the relative intensity of dots relative to that of the control group. P values were calculated using a two-tailed t-test. Error bars represent SD. (B-D) After transfection (48 h) with the indicated fluorescence FAM-labeled siRNA, HeLa cells were fixed and immunostained with the indicated antibodies. DNA was visualized by DAPI. Scale bar, 10 μm. Supportive data were included in Supplementary information, Figure S3.
WTAP regulates accumulation of METTL3 and METTL14 in nuclear speckles
Previous studies have indicated that m6A modification occurs at the pre-mRNA stage within the nucleus and it thereby may affect splicing and mRNA export2,32,33,34,35. Nuclear speckles are enriched in proteins involved in RNA processing and alternative splicing. Interestingly, METTL3 and WTAP as well as the m6A demethylases FTO and ALKBH5 have previously been shown to colocalize with nuclear speckle markers14,15,31. The physical interaction between WTAP and METTL3 and METTL14 prompted us to examine the cellular distribution of METLL14. Using immunofluorescence microscopy, we found that endogenous METTL14, like METTL3 and WTAP, exhibits a diffused nucleoplasmic staining pattern with intense granule-like foci, and that it colocalizes well with the pre-mRNA splicing factor SC35 in nuclear speckles (Figure 2C and Supplementary information, Figure S3F). Interestingly, WTAP depletion decreased the accumulation of both METTL3 and METTL14 in nuclear speckles (Figure 2B, 2C and Supplementary information, Figure S3G), whereas knockdown of either METTL3 or METTL14 had no effect on the localization of WTAP (Figure 2D and Supplementary information, Figure S3G). Furthermore, METTL3 depletion affected the nuclear speckle localization of METTL14 and vice versa (Figure 2B and 2C). Consistent with our observations on m6A methyltransferase activity, WT1 depletion did not affect METTL3 or WTAP localization (Figure 2B and 2D). Based on these observations, we conclude that WTAP is required for accumulation of METTL3 and METTL14 in nuclear speckles.
Nuclear speckle-associating factors can be classified as RNase-sensitive or -insensitive factors based on the nature of the association with the nuclear speckles. To characterize the nature of the association of the WMM complex with nuclear speckles, we treated cells with RNase A. Similar to the mRNA processing factor SM (smith antigen), but unlike the splicing factor SC35, RNase A treatment resulted in a dramatic decrease of WMM complex accumulation in nuclear speckles, demonstrating that the WMM complex associates with RNA, which is essential for the recruitment and/or retention of the WMM complex in nuclear speckles (Supplementary information, Figure S3D-S3F). It is worth noting that METTL3 and METTL14 staining almost disappeared upon WTAP knockdown, but the overall protein levels of METTL3 and METTL14 did not change following WTAP depletion (Supplementary information, Figure S3B and S3C). It is likely that when WTAP is depleted, METTL3 and METTL14 no longer bind to RNA in the nuclear speckles, and become diffused within the entire cell, which reduces the staining density to levels below detection. In addition, unbound METTL3 and METTL14 might easily get washed out during the permabilization and fixation procedures. A similar staining pattern in nuclear speckles has also previously been reported15.
mRNA is the major RNA species bound by WTAP and METTL3
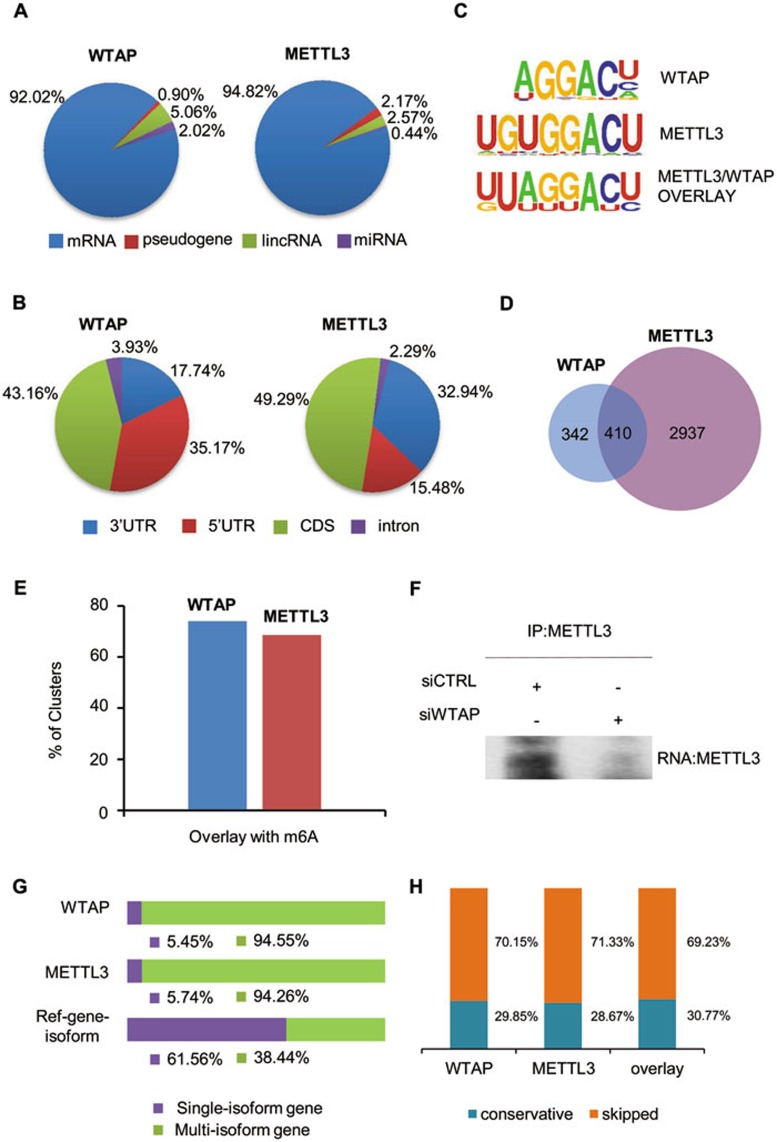
We next performed Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation (PAR-CLIP)51 to identify the targets of WTAP and METTL3 proteins. In brief, 4-thiouridine (4-SU), a photoreactive ribonucleoside analog, was incorportated into RNA transcripts, which leads to the thymidine-to-cytidine mutation in crosslinked sequences. Therefore, one unique feature of cDNA libraries prepared by PAR-CLIP is that the precise position of crosslinking can be identified by mutations residing in the sequenced cDNA. PARalyzer36,37 software was used to identify the preferred regions bound by WTAP and METTL3. 4 986 METTL3- and 922 WTAP-binding clusters were enriched and could be assigned into four RNA categories: mRNA, miRNA, large intergenic non-coding RNA (lincRNA) and pseudogene (Figure 3A). About 95% and 92% of the RNA bound by METTL3 and WTAP, respectively, represented mRNA, demonstrating that mRNA is the major substrate of the WMM complex. We further mapped the mRNA clusters into four non-overlapping transcript regions, 5′-UTR, CDS, 3′-UTR and intron, according to the human reference transcript annotation. After normalization of the number of clusters to the overall length of the different transcript regions, the majority of binding sites of WTAP and METTL3 were identified in CDS and UTR regions (Figure 3B).
Figure 3.
METTL3 and WTAP bind m6A consensus sequence in mRNA and affect gene expression and alternative splicing. (A) Percentage of various RNA species bound by WTAP and METTL3 based on PAR-CLIP analyses. The WTAP- and METTL3-binding clusters were identified by PARalyzer algorithm. Annotation of clusters was based on the human Ensembl gtf file (version 72, hg19). The majority of binding sites of WTAP and METTL3 were located in mRNA. (B) Pie chart depicting the distribution of binding clusters in mRNA based on PAR-CLIP sequence clusters for WTAP and METTL3 after normalization of the overall length of the different transcript regions. Length of these different transcript regions was extracted from Ensembl annotations and the distribution percentage of clusters in these regions were normalized by their length. (C) Enriched sequence motif analysis of binding clusters indentified by PAR-CLIP. Upper panel, WTAP-binding motif (P = 1e-14); middle panel, METTL3-binding motif (P = 1e-13); lower panel, binding motif obtained when only genes found in both WTAP- and METTL3-binding clusters were included (P = 1e-19). Binding motifs were computed by the HOMER program. (D) Venn diagram of the overlapping genes with binding clusters of WTAP and METTL3 in the PAR-CLIP samples. (E) Percentage of WTAP/METTL3 clusters in CDS and UTR regions overlapped with m6A sites. (F) HeLa cells were transfected with siCTRL or siWTAP and Myc-METTL3 for 48 h as indicated. The cell lysates were then subjected to PAR-CLIP using anti-Myc. The pulled down RNA products in the RNA-METTL3 complex were labeled by Biotin and detected by Biotin chemiluminescent nucleic acid kit. (G) Percentage of WTAP- (711 multi-isoform and 41 single-isoform) and METTLE3- (3 155 multi-isoform and 192 single-isoform) binding mRNAs derived from single-isoform or multi-isoform genes and the reference Ensembl genes of human (P = 2.2e-16, Fisher test). (H) Percentage of constitutively or alternatively spliced exons adjacent to intronic binding clusters of WTAP (left), METTL3 (middle), overlap of WTAP and METTL3 (right). Supportive data were included in Supplementary information, Figures S4 and S5.
To define the in vivo RNA recognition elements for WTAP and METTL3, the binding cluster data were analyzed by HOMER, a suite of tools for motif discovery and next-generation sequencing analysis38. In this analysis procedure, the WTAP and METTL3 clusters were set as the target sequences, and a set of background clusters was generated with the BEDTools' shuffleBed program39 to randomly shuffle regions of the same size as the clusters throughout the gene regions, with the parameter for HOMER motif length from 5 to 8. The motifs AGGACU (P = 1e-14) and UGUGGACU (P = 1e-13) were enriched in WTAP- and METTL3-binding clusters, respectively (Figure 3C). When we only included genes found in both WTAP- and METTL3-binding clusters, the highest scoring motif was UUAGGACU (P = 1e-19) (Figure 3C). In addition, AGGAC (P = 1e-12), UGGAC (P = 1e-12), and AGACUAA (P = 1e-10) were also highly enriched in WTAP, METTL3 and WTAP/METTL3 overlay clusters, respectively (Figure 3D and Supplementary information, Figure S4B). This is in accordance with the reported consensus m6A motif RRACH (R = G or A; H = A, C or U)9,10. The high degree of similarity of the mRNA binding motifs between WTAP and METTL3 are consistent with the results that WTAP and METTL3 form a complex.
We next calculated the distance between WTAP/METTL3 clusters and the m6A sites. To obtain the best coverage of m6A sites, we downloaded human m6A data released by Dominissini et al.9 and Meyer et al.10 and aligned the data to the hg19 genome by Tophat240. We then used MACS41 and Fisher test9,10 methods to determine m6A peaks. The m6A peaks identified in this analysis were combined with the previously published m6A sites, and the duplicates were removed. Then we used the BEDTools' closestBed39 to calculate the distance between WTAP/METTL3 clusters and m6A sites. We generated control clusters with BEDTools' shuffleBed39 to randomly shuffle regions of the same size as the clusters. The result demonstrated a significant spatial correlation (P = 2.2e-16) with 74% WTAP- and 69% METTL3-binding clusters in CDS and UTR regions overlapping with m6A sites (Figure 3E and Supplementary information, Figure S4C and S4D). These results strongly suggest a significant role of both WTAP and METTL3 in m6A modification.
The results that WTAP is required for nuclear speckle localization of METTL3 and METTL14 and for m6A modification prompted us to test whether the absence of WTAP might influence the binding of METTL3 to RNA. Interestingly, we found a significantly reduced amount of RNA associated with METTL3 upon WTAP depletion (Figure 3F and Supplementary information, Figure S5B), suggesting that WTAP is likely responsible for recruiting the m6A methyltransferase complex to target RNAs. Taken together, these findings support the existence of a WTAP-METTL3-containing complex that methylates RNA at specific sites in mammals.
The WMM complex regulates transcription and alternative splicing
Gene Ontology (GO) analyses of mRNA captured by METTL3 and WTAP using PAR-CLIP revealed that a large proportion of mRNA species associated with both proteins were derived from genes involved in transcription and RNA metabolism/splicing (Supplementary information, Figure S5A). To further investigate the potential roles of the WMM complex in RNA metabolism, transcriptome analyses were performed to compare the genome-wide gene expression and alternative splicing events between wild-type cells and WTAP- or METTL3-deficient cells. The transcriptome analyses revealed a significant overlap in differentially expressed genes (DEGs) between WTAP- and METTL3-deficient cells (Supplementary information, Figure S5C). Consistent with the GO analysis of the PAR-CLIP data, GO analysis of the overlapping DEGs from WTAP- and METTL3-deficient cells demonstrated that most of these genes were related to transcription and RNA processing (Supplementary information, Figure S5D), further emphasizing the potential significance of WTAP and METTL3 in RNA metabolism.
Interestingly, both METTL3 and WTAP knockdowns resulted in a pronounced variation in isoform numbers with about half of the variations (2 296) present in both WTAP- and METTL3-deficient cells (Supplementary information, Figure S5E). When comparing the 2 296 genes with changes in isoform numbers with the 410 genes bound by the two proteins (Figure 3D), 99 genes were overlapped (Supplementary information, Figure S5E). Furthermore, assigning genes with METTL3- or WTAP-binding clusters to either single- or multi-isoform genes revealed that the majority of mRNA species bound by WTAP or METT3 were derived from multi-isoform genes (Figure 3G; P = 2.2e-16, Fisher test). Refining this observation further, assignment of all intronic clusters to either constitutively or alternatively spliced exons revealed that these clusters are significantly closer to alternatively spliced exons compared to constitutively spliced exons (Figure 3H, P = 2.2e-16, Fisher test), further indicating a role of the WMM complex and mRNA m6A modification in RNA splicing as proposed previously9.
The WMM complex is required for tissue differentiation in zebrafish
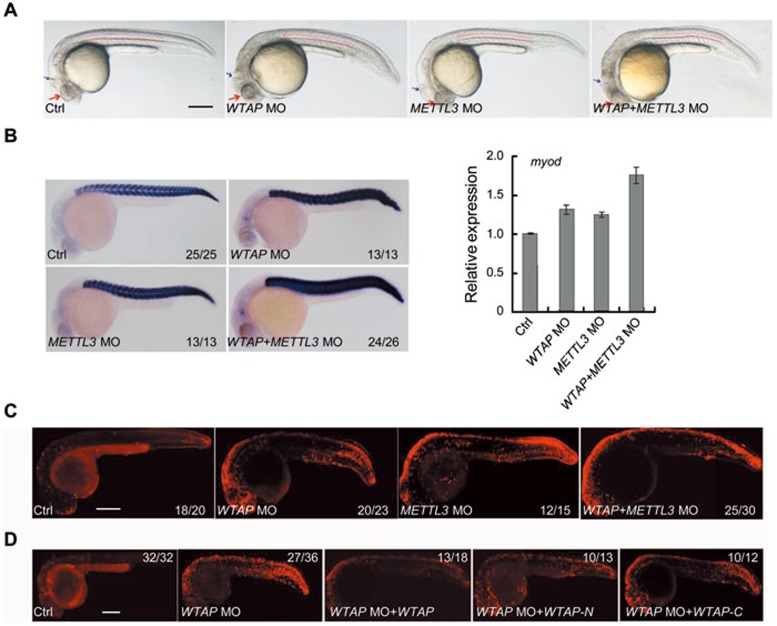
The important role of the WMM complex in the regulation of m6A modification, gene expression and mRNA processing in cultured cells prompted us to investigate its functions at the organismal level using zebrafish embryos as the model system. Human and zebrafish WTAP proteins share 81% identity with a high degree of conservation in N-terminal region (Supplementary information, Figure S5F). Whole-mount in situ hybridization (WISH) showed that WTAP and METTL3 were maternally expressed from the 4-cell stage and ubiquitously expressed through early embryogenesis, with enriched expression in the brain region at 36 hpf (hours post fertilization) (Supplementary information, Figure S6A). To determine whether WTAP and METTL3 are functionally involved in early embryonic development, WTAP and METTL3 were knocked down in zebrafish embryos using antisense morpholinos (MOs). Embryos injected with WTAP MO displayed multiple developmental defects at 24 hpf including smaller head and eyes, smaller brain ventricle, and curved notochord, while embryos injected with METTL3 MO were only modestly affected (Figure 4A). However, simultaneous knockdown of these two genes caused much more severe defects in embryos compared to parallel control and embryos injected with individual MOs (Figure 4A). GFP expression of the constructs of GFP-WTAP and GFP-METTL3 was markedly decreased by co-injection of corresponding MO, verifying efficient knockdown (Supplementary information, Figure S6B). WISH showed that the expression of neuro-ectoderm marker goosecoid, and mesoderm marker no tail at the 50% epiboly stage was not changed, suggesting normal germ layer formation in the morphants (Supplementary information, Figure S6C). However, the expression of somite marker myod was increased in WTAP-, METTL3-, and double knockdown embryos (Figure 4B), indicating that the WMM complex might play an inhibitory role in muscle development. Although the proportion of apoptotic cells detected by the TUNEL assay was slightly increased in individual morphants, double knockdown of WTAP and METTL3 led to a striking increase in apoptosis (Figure 4C), consistent with observations in human cells (Supplementary information, Figure S6D-S6F). The apoptotic cell death observed in WTAP MO-treated zebrafish embryos was rescued by either full-length or an N-terminal region of zebrafish WTAP (Figure 4D). Consistent with their role in mammalian cells, both METTL3 and WTAP are required for the m6A methyltransferase activity in zebrafish embryos (Supplementary information, Figure S6G). These data suggests that the m6A methyltransferase is required for normal tissue differentiation in zebrafish.
Figure 4.
The WMM complex plays essential roles during zebrafish embryogenesis. (A) Embryos injected with individual (WTAP-, METTL3-) or combined (METTL3+WTAP) MOs. The double knockdown showed more severe morphological defects, compared to other groups, at 24 hpf. Red arrows mark head and eyes, while blue arrows mark brain ventricle. The curve of the notochord is labeled by the double dashed lines. (B) Expression of somite marker myod in the morphants at 24 hpf. myod expression was increased in WTAP-, METTL3-, or double morphants. (C) Increased apoptosis (TUNEL assay) was observed in embryos injected with individual or combined MOs. (D) Overexpression of full-length or N-terminal but not C-terminal zebrafish WTAP mRNA prevented apoptosis in zebrafish WTAP-MO embryos. Note that there is auto-fluorescence on the yolk. Anterior to the left and dorsal is up. Scale bar, 250 μm. Supportive data were included in Supplementary information, Figures S5F and S6.
Discussion
METTL3 homologs have been found in various species including S. cerevisiae (IME4), Drosophila (Ime4) and Arabidopsis (MTA)23,24,25,26,42. The two consensus methyltransferase motifs CM I and CM II are present in all homologs. Single amino acid substitutions in the putative catalytic domain of Ime4 lead to loss of m6A modification in mRNA and severe sporulation defects42,43. In Arabidopsis, MTA is mainly expressed in dividing tissues, particularly in reproductive organs, shoot meristems and emerging lateral roots. Inactivation of MTA results in m6A modification defects and developmental failure beyond the globular stage26.
Despite the identification of a multi-subunit complex possessing m6A methyltransferase activity more than 15 years ago, METTL3 has remained the only known component of the mammalian complex22. Therefore, unveiling proteins associated with METTL3 in the methyltransferase complex is of key importance for understanding how m6A modifications in RNA might impact and regulate cellular functions. Previously, AtFIP37 and Mum2 were found to associate with plant and yeast METTL3 homologs, respectively26,28. Like MTA, disruption of AtFIP37 also results in developmental arrest at the globular stage26,44. In the present study, we have identified WTAP, the human homolog of AtFIP37/Mum2, and a novel protein METTL14 as two new components of the human m6A methyltransferase complex. Like METTL3, METTL14 contains a catalytic domain and harbors catalytic activity30; while our data suggest that WTAP likely have very important regulatory functions, including tethering the m6A methyltransferase complex to RNA and facilitating its accumulation in nuclear speckles (Figures 2B, 2C and 3F). The fact that m6A demethylases also localize in nuclear speckles strongly suggests that a dynamic regulation of m6A levels in mRNA may occur in nuclear speckles3,14. Furthermore, our studies in zebrafish revealed that METTL3 mRNA expression level changed during embryonic development (also observed in vitro in bovine embryos45), and that depletion of WTAP and METTL3 compromised tissue differentiation (Figure 4 and Supplementary information, Figure S6), strongly suggesting that m6A may play a key role in regulating organismal development.
On average, there is 1 m6A modification per 2 000 ribonucleotides2. Motif analyses of m6A peaks enriched in both human and mouse cells have revealed that m6A modification occurs in highly conserved regions with a consensus sequence RRACH (R = G or A; H= A, C or U)9,10,11,12,13. The m6A modification occurs in intragenic regions particularly in the 3′-end of CDS and the first quarter of the 3′-UTR9,10. Our PAR-CLIP data demonstrate that the conserved WTAP- and METTL3-binding motifs both contain the consensus m6A sequence (Figure 3C), and their binding clusters in CDS and UTR regions significantly overlap with m6A sites (Figure 3E), strongly suggesting a direct connection between the WMM complex and m6A modification.
Interestingly, without normalization of the overall length of the various transcript regions, the majority of WTAP- or METTL3-binding clusters were located in introns (Supplementary information, Figure S4A), which suggests that WMM-dependent m6A modification may be coupled to gene transcription as previously proposed1,2,3. For the relationship of m6A and mRNA splicing, Dominissini and colleagues calculated the average number of m6A peaks in multi-isoform and single-isoform genes, and assigned all m6A peaks to either constitutively or alternatively spliced exons based on the list of splicing events of all coding Ensembl genes. Their study revealed that m6A modification is significantly over-represented in multi-isoform genes and alternatively spliced exons, proposing that m6A may affect RNA splicing9. Consistent with this observation, we found an enrichment of WTAP/METTL3-binding clusters in genes with multi-isoforms and alternatively spliced exon junctions based on our PAR-CLIP+RNA-seq dataset (Figure 3G and 3H). Taken together, these results suggest that WTAP/METTL3 and thereby the m6A methyltransferase may influence RNA splicing. However, a thorough understanding of the relationship between m6A and transcription and/or splicing awaits future investigations.
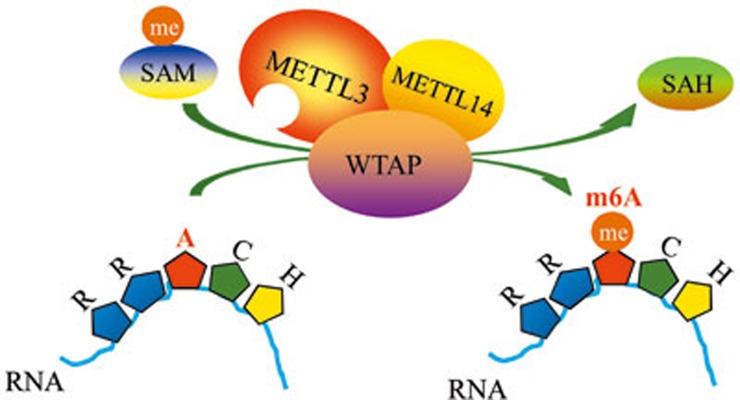
Drosophilia female-lethal (2)D (FL (2)D, WTAP homolog) also interacts with the female-specific RNA-binding protein sex-lethal (SXL)46,47, VIR46, Snf, U2AF, U2A50 and U2AF3847. This together with the results showing that there is a difference in the numbers of genes containing METTL3- or WTAP-binding clusters might suggest that METTL3 is the core component of the m6A methyltransferase, whereas WTAP serves as its regulatory subunit. Based on the findings that WTAP interacts directly with METTL3 (Figure 1) and mediates nuclear speckle localization of both METTL3 and METTL14, we propose a model, in which METTL3 and METTL14 are anchored to mRNA by WTAP, allowing subsequent methylation of target adenosine residues (Figure 5).
Figure 5.
A schematic model of the WMM complex function. The WMM complex mediates methylation of internal adenosine residues in eukaryotic mRNA, forming N6-methyladenosine. WTAP binds to the m6A consensus RRACH motif of mRNA and recruits catalytic subunits METTL3 and METTL14. Then the METTL3-METTL14 complex carries out m6A methyltransferase activity in the m6A motif.
In summary, our findings identify WTAP as a regulatory subunit required for formation of a functional m6A methyltransferase complex including METTL3 and METTL14, which play an important role in the regulation of gene expression and alternative splicing. Thus, the discovery and characterization of the RNA m6A methyltransferase complex, together with the recent advances in the understanding of the demethylases FTO and ALKBH5, is an important step towards a more thorough understanding of the biological significance of the epitranscriptomic marker m6A.
Materials and Methods
Cell culture, plasmids and antibodies
Human cells were cultured in standard cell culture Dulbecco's modified Eagle's medium at 37 °C in a humidified incubator with 5% CO2(v/v). The human METTL3 gene was cloned into pCMV-Myc (Invitrogen), S-protein/FLAG/SBP (streptavidin-binding protein) triple-tagged destination vector48 or pGEX-5×-2 (GE healthcare). The human WTAP gene was cloned into p3XFLAG-CMV-14 (Sigma) or pProEX-HTb (Invitrogen). The human METTL14 gene was cloned into pcDNA3-HA (Invitrogen). The following antibodies were used: rabbit-anti-METTL3 (Abcam), mouse-anti-METTL3 (Abnova), mouse-anti-WTAP (Santa Cruz), rabbit-anti-METTL14 (Atlas), rabbit-anti-m6A (Synaptic Systems) and other antibodies included in Supplementary information, Data S1.
Tandem affinity purification and mass spectrometry
293T cells transiently transfected with triply-tagged METTTL3 plasmid (SFB-METTL3) were used for tandem affinity purification. Cell lysates were incubated with streptavidin-Sepharose beads (Amersham Biosciences), and eluted with 1 mg/ml biotin (Sigma). The eluates were further incubated with S-protein-agarose (Novagen). The proteins bound to S-protein-agarose beads were subjected to SDS-PAGE and visualized by coomassie blue staining. The identities of eluted proteins were revealed by mass spectrometry analysis. The Q Exactive mass spectrometry data (Thermo Fisher Scientific) were searched against SwissProt human database using 15 ppm peptide mass tolerance and 20mmu fragment mass tolerance.
Immunoprecipitation
293T cells transfected with the indicated siRNAs and/or DNA constructs were lysed in buffer (100 mM NaCl, 20 mM Tris-HCl (pH 7.4), 0.5% NP-40, 1 mM PMSF, 1 mM Na3VO4, 1 mM β-glycerophosphate, 1 mM NaF and 1× Cocktail), and subjected to immunoprecipitation (IP) followed by immunoblotting with the indicated antibodies.
The following antibodies were used in the study: mouse-anti-Flag (Sigma), rabbit-anti-Myc (Abcam), rabbit-anti-HA(Clontech), rabbit-anti-WTAP (Atlas), Anti-Rabbit IgG-HRP (Dakocytomation), Anti-Mouse IgG-HRP (Dakocytomation).
GST pull-down assay
The human METTL3 gene was subcloned into pGEX-5X-2 expression plasmid with GST-tag and the human WTAP gene was subcloned into pProEX-HTb expression plasmid with His tag. Recombinant GST-METTL3 and His-WTAP proteins were induced to express in E. coli strain BL21(DE3) and purified by FPLC using Bio-Scale Mini Profinity GST cartridge (Biorad) and Bio-Scale Mini Profinity IMAC cartridge (Biorad) as described in the commercial instruction manual. The quality of proteins was tested by western blot and coomassie brilliant blue staining. His-WTAP proteins were incubated with glutathione sepharose (GE Healthcare) to be pre-cleared in NETN buffer (100 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl pH 7.4, 0.5% NP-40). Then pre-cleared His-WTAP was mixed with GST or GST-METTL3 protein and incubated overnight at 4 °C with equal amount of glutathione sepharose beads. After washing the beads five times with NETN buffer, proteins bound to the beads were analyzed by 8% SDS-PAGE followed by immunoblotting with HRP-conjugated anti-His and anti-GST antibodies (Abcam).
Immunofluorescence staining
HeLa cells grown on coverslips were transfected with siRNA (50 nM) or DNA constructs as indicated for about 4 h, and 24 h later fixed with 4% paraformaldehyde on ice followed by permeabilization with 0.1% Triton X-100 and 0.05% NP-40 on ice. After pre-blocking with 5% non-fat dried milk in TBST, coverslips were first incubated with primary antibody for 1 h and then with fluorescent dye-conjugated secondary antibody for 0.5 h and mounted with DAPI-containing mounting medium (Vector Laboratories, Burlingame, CA, USA) and visualized under immunofluorescence confocal microscope Leica TCS SP5 (Leica). The method used for immunofluorescence staining following RNase A treatment was adapted from Mayer et al.49 and Sytnikova et al.50. In brief, cells grown on coverslips were permeabilized with 0.05% Triton X-100 in 20 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 0.5 mM EDTA, and 25% glycerol; washed twice with PBS; and treated with RNase A (1 mg/ml) (diluted in PBS) in 37 °C for 7 min. After washing, cells were fixed with cold methanol (15 min) and stained with the indicated antibodies following the blocking procedure. The immunofluorescence images were digitally recorded by using confocal microscope. The following antibodies were used in immunofluorescence staining: mouse-anti-Flag (Sigma), rabbit-anti-Myc (Abcam), rabbit-anti-HA (Clontech), mouse-anti-METTL3 (Abnova), rabbit-anti-METTL14 (Atlas), mouse-anti-WTAP (Santa Cruz), mouse-anti-SC35 (Sigma), anti-mouse IgG-Cy3 (Sigma), anti-rabbit IgG-Cy3 (Sigma), anti-mouse IgG-FITC (Sigma), anti-rabbit IgG-FITC (Sigma).
PAR-CLIP and RNA-Seq
293T cells expressing Myc-METTL3 or Flag-WTAP were cultured in medium supplemented with 4-SU (Sigma), irradiated with 365 nm UV light to induce crosslinking as described previously51. Immunoprecipitated protein-RNA complexes were separated by SDS-PAGE and then transferred to PVDF membrane; RNA-protein complexes were cut out from the membrane corresponding to the size of Myc-METTL3 or Flag-WTAP. Purified RNAs from RNA-Protein complex were subjected to the small RNA library construction and then sequenced with HiSeq 2000 system (Illumina Inc.). Bowtie 0.12.7 software52 was applied to map sequencing reads against human (hg19) genome with up to two mismatches allowed. To multi-mapped reads, mapped locations were only reported for those with the minimum number of observed mismatches. PARalyzer software36 was used to define METTL3- and WTAP-binding sites. Sequence motifs enriched in clusters were identified by HOMER, a suite of tools for motif discovery and next-generation sequencing analysis38.
Total RNA was extracted from 293T cells transfected with siRNA using RNAzol (Molecular Research Center, Inc., USA). cDNA library was constructed using TruSeq RNA Sample Prep Kit and then sequenced with HiSeq 2000 system (Illumina Inc.). RNA-seq reads were aligned against the Ensembl genome (hg19) using TopHat240,53. The number of reads mapped to each of the Ensembl genes (version 72) was counted using the HTSeq python package54. DEGs were determined using the R-package DEGseq with the method MARS (MA-plot-based method with random sampling model), P value cutoff = 0.00155. GO analyses of DEGs and PAR-CLIP data were performed using DAVID56,57. Enrichment map of DEGs was constructed by Cytoscape 2.8.3 software57. The FPKM (Fragments Per Kilobase of exon per Million mapped reads) value for each transcript was calculated by using Cufflinks version 2.0.0 and the differentially expressed transcripts were identified by Cuffdiff program in the Cufflinks software package58. Scripture, a method for transcriptome reconstruction that relies solely on RNA-Seq reads and an assembled genome to build the transcriptome59, was applied to calculate the isoform number for each gene. Variation in the isoform number between METTL3/WTAP-depleted and control samples indicated an alternative splicing event.
Furthermore, a Bowtie library of non-redundant set of sequences consisting of all possible junctions between the exons of each Refseq gene of human was generated and all the RNA-seq reads were aligned against this library with Bowtie 0.12.7 software52. To the exon-exon junctions (EEJs) with read mapping, according to their continuous or discrete exon number, they can be divided into constitutively or alternatively spliced exons. All the WTAP/METTL3 clusters were assigned to these exons.
MO-mediated gene knockdown in zebrafish embryos
The antisense MO oligonucleotides targeting individual members of the WMM complex were synthesized (Gene Tools, LLC.). The MOs (2.5-8 ng) were injected into embryos from one-cell to four-cell stage. Zebrafish WTAP and METTL3 were amplified from zebrafish cDNA library by PCR and sequentially subcloned into the pGEM-T vector and pEGFP-N1 (Clontech) to generate pWTAP-GFP or pMETTL3-GFP constructs for checking MO efficiency
Statistical analysis
Two-tailed t-tests in grouped analyses of Prism5 software (GraphPad Software Inc., USA) were applied. P-values < 0.05 were considered significant.
Accession numbers
Mass spectrometry data have been uploaded to Peptide Atlas under accession number PASS00296. PAR-CLIP and RNA-Seq data have been uploaded to GEO database and can be accessed via accession number GSE50583.
Acknowledgments
We thank Dr Chuan He for sharing his data before publishing, Drs Dang-Sheng Li and Guo-Liang Xu for their critical reading and suggestion. This work was supported by the National Basic Research Program of China (973 program; 2011CB510103 and 2012CB518302 to YGY, 2010CB945300 and 2011CB943900 to FL), the National Natural Science Foundation of China (31370796 and 91319308 to YGY, 30971678 to FL), 100 Talents Program of Chinese Academy of Sciences (CAS) (YGY), and CAS Strategic Priority Research Program (XDA01010110 to FL), SKLEH-Pilot Research Grant to YGY, and Danish Medical Research Council (JMRD).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
(Related to Figure 1) Identification of WTAP and METTL14 as METTL3 associating proteins.
(Related to Figure 1) Validation of WTAP and METTL14 as METTL3 associating proteins.
(Related to Figure 2) WTAP is essential for in vivo methyltransferase activity; localization of METTL3, METTL14 and WTAP in nuclear speckles requires RNA: METTL3/WTAP/METTL14 knockdown efficiency detected by Western blot and RT-PCR.
(related to Figure 3) (A) Distribution of mRNA binding regions based on PAR-CLIP sequence clusters for WTAP and METTL3 without normalization of the overall length of the different transcript regions.
(Related to Figure 3) METTL3 and WTAP influence gene expression and alternative splicing (A) The 410 genes identified in both METTL3 and WTAP PAR-CLIPs were analyzed by GO analysis and an enrichment map was constructed by Cytoscape (parameters: p < 0.05, overlap cutoff > 0.5).
(Related to Figure 4). Expression pattern and functional assay of WTAP and METTL3 in zebrafish embryos (A) Whole-mount in situ hybridization (WISH) shows ubiquitous expression of WTAP and METTL3 during embryogenesis, respectively.
Materials and Methods
References
- Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 2013;29:108–115. doi: 10.1016/j.tig.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Zhao X, Wu YS, Li MM, Wang XJ, Yang YG. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteomics Bioinformatics. 2013;11:8–17. doi: 10.1016/j.gpb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, et al. Sprouts of RNA epigenetics: The discovery of mammalian RNA demethylases. RNA Biol. 2013;10:915–918. doi: 10.4161/rna.24711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Kiang S, Nevins JR, Darnell JE., Jr N-6-methyl-adenosine in adenovirus type 2 nuclear RNA is conserved in the formation of messenger RNA. J Mol Biol. 1979;135:733–752. doi: 10.1016/0022-2836(79)90174-8. [DOI] [PubMed] [Google Scholar]
- Desrosiers RC, Friderici KH, Rottman FM. Characterization of Novikoff hepatoma mRNA methylation and heterogeneity in the methylated 5′ terminus. Biochemistry. 1975;14:4367–4374. doi: 10.1021/bi00691a004. [DOI] [PubMed] [Google Scholar]
- Levis R, Penman S. 5′-terminal structures of poly(A)+ cytoplasmic messenger RNA and of poly(A)+ and poly(A)- heterogeneous nuclear RNA of cells of the dipteran Drosophila melanogaster. J Mol Biol. 1978;120:487–515. doi: 10.1016/0022-2836(78)90350-9. [DOI] [PubMed] [Google Scholar]
- Narayan P, Ayers DF, Rottman FM, Maroney PA, Nilsen TW. Unequal distribution of N6-methyladenosine in influenza virus mRNAs. Mol Cell Biol. 1987;7:1572–1575. doi: 10.1128/mcb.7.4.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T. N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem Sci. 2013;38:204–209. doi: 10.1016/j.tibs.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m(6)A RNA methylomes revealed by m(6)A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimock K, Stoltzfus CM. Sequence specificity of internal methylation in B77 avian sarcoma virus RNA subunits. Biochemistry. 1977;16:471–478. doi: 10.1021/bi00622a021. [DOI] [PubMed] [Google Scholar]
- Schibler U, Kelley DE, Perry RP. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J Mol Biol. 1977;115:695–714. doi: 10.1016/0022-2836(77)90110-3. [DOI] [PubMed] [Google Scholar]
- Wei CM, Moss B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry. 1977;16:1672–1676. doi: 10.1021/bi00627a023. [DOI] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C. Grand challenge commentary: RNA epigenetics. Nat Chem Biol. 2010;6:863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Agarwala SD, Mumbach MR, et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013;155:1409–1421. doi: 10.1016/j.cell.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbritt T, Patel HR, Preiss T. Mapping and significance of the mRNA methylome. Wiley Interdiscip Rev RNA. 2013;4:397–422. doi: 10.1002/wrna.1166. [DOI] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability Nature 2013. Nov 27. doi: 10.1038/nature12730 [DOI] [PMC free article] [PubMed]
- Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem. 1994;269:17697–17704. [PubMed] [Google Scholar]
- Tuck MT. Partial purification of a 6-methyladenine mRNA methyltransferase which modifies internal adenine residues. Biochem J. 1992;288:233–240. doi: 10.1042/bj2880233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Bujnicki JM, Feder M, Radlinska M, Blumenthal RM. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA: m(6)A methyltransferase. J Mol Evol. 2002;55:431–444. doi: 10.1007/s00239-002-2339-8. [DOI] [PubMed] [Google Scholar]
- Hongay CF, Orr-Weaver TL. Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis. Proc Natl Acad Sci USA. 2011;108:14855–14860. doi: 10.1073/pnas.1111577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah JC, Clancy MJ. IME4, a gene that mediates MAT and nutritional control of meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1078–1086. doi: 10.1128/mcb.12.3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Li H, Bodi Z, et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20:1278–1288. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane SE, Beemon K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Mol Cell Biol. 1985;5:2298–2306. doi: 10.1128/mcb.5.9.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwala SD, Blitzblau HG, Hochwagen A, Fink GR. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genet. 2012;8:e1002732. doi: 10.1371/journal.pgen.1002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottman FM, Bokar JA, Narayan P, Shambaugh ME, Ludwiczak R. N6-adenosine methylation in mRNA: substrate specificity and enzyme complexity. Biochimie. 1994;76:1109–1114. doi: 10.1016/0300-9084(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, et al. A hMETTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation Nat Chem Biol 2013. Dec 6. doi: 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed]
- Little NA, Hastie ND, Davies RC. Identification of WTAP, a novel Wilms' tumour 1-associating protein. Hum Mol Genet. 2000;9:2231–2239. doi: 10.1093/oxfordjournals.hmg.a018914. [DOI] [PubMed] [Google Scholar]
- Carroll SM, Narayan P, Rottman FM. N6-methyladenosine residues in an intron-specific region of prolactin pre-mRNA. Mol Cell Biol. 1990;10:4456–4465. doi: 10.1128/mcb.10.9.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel D, Groner Y. Methylations of adenosine residues (m6A) in pre-mRNA are important for formation of late simian virus 40 mRNAs. Virology. 1983;131:409–425. doi: 10.1016/0042-6822(83)90508-1. [DOI] [PubMed] [Google Scholar]
- Shimba S, Bokar JA, Rottman F, Reddy R. Accurate and efficient N-6-adenosine methylation in spliceosomal U6 small nuclear RNA by HeLa cell extract in vitro. Nucleic Acids Res. 1995;23:2421–2426. doi: 10.1093/nar/23.13.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus CM, Dane RW. Accumulation of spliced avian retrovirus mRNA is inhibited in S-adenosylmethionine-depleted chicken embryo fibroblasts. J Virol. 1982;42:918–931. doi: 10.1128/jvi.42.3.918-931.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran DL, Georgiev S, Mukherjee N, et al. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011;12:R79. doi: 10.1186/gb-2011-12-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee N, Corcoran DL, Nusbaum JD, et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, et al. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy MJ, Shambaugh ME, Timpte CS, Bokar JA. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002;30:4509–4518. doi: 10.1093/nar/gkf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodi Z, Button JD, Grierson D, Fray RG. Yeast targets for mRNA methylation. Nucleic Acids Res. 2010;38:5327–5335. doi: 10.1093/nar/gkq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vespa L, Vachon G, Berger F, Perazza D, Faure JD, Herzog M. The immunophilin-interacting protein AtFIP37 from Arabidopsis is essential for plant development and is involved in trichome endoreduplication. Plant Physiol. 2004;134:1283–1292. doi: 10.1104/pp.103.028050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw S, Vigneault C, Sirard MA. Temporal expression of factors involved in chromatin remodeling and in gene regulation during early bovine in vitro embryo development. Reproduction. 2007;133:597–608. doi: 10.1530/REP-06-0251. [DOI] [PubMed] [Google Scholar]
- Ortega A, Niksic M, Bachi A, et al. Biochemical function of female-lethal (2)D/Wilms' tumor suppressor-1-associated proteins in alternative pre-mRNA splicing. J Biol Chem. 2003;278:3040–3047. doi: 10.1074/jbc.M210737200. [DOI] [PubMed] [Google Scholar]
- Penn JK, Graham P, Deshpande G, et al. Functioning of the Drosophila Wilms'-tumor-1-associated protein homolog, Fl(2)d, in sex-lethal-dependent alternative splicing. Genetics. 2008;178:737–748. doi: 10.1534/genetics.107.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddika S, Sy SM, Chen J. Functional interaction between Chfr and Kif22 controls genomic stability. J Biol Chem. 2009;284:12998–13003. doi: 10.1074/jbc.M900333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Schmitz KM, Li J, Grummt I, Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell. 2006;22:351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Sytnikova YA, Kubarenko AV, Schäfer A, Weber AN, Niehrs C. Gadd45a is an RNA binding protein and is localized in nuclear speckles. PLoS One. 2011;6:e14500. doi: 10.1371/journal.pone.0014500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer SA, Trapnell C, Aviran S, Pachter L, Lucks JB. SHAPE-Seq: High-Throughput RNA Structure Analysis. Curr Protoc Chem Biol. 2012;4:275–297. doi: 10.1002/9780470559277.ch120019. [DOI] [PubMed] [Google Scholar]
- Anders S.HTSeq: analysing high-throughput sequencing data with Python. . http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html . 2010 [DOI] [PMC free article] [PubMed]
- Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(Related to Figure 1) Identification of WTAP and METTL14 as METTL3 associating proteins.
(Related to Figure 1) Validation of WTAP and METTL14 as METTL3 associating proteins.
(Related to Figure 2) WTAP is essential for in vivo methyltransferase activity; localization of METTL3, METTL14 and WTAP in nuclear speckles requires RNA: METTL3/WTAP/METTL14 knockdown efficiency detected by Western blot and RT-PCR.
(related to Figure 3) (A) Distribution of mRNA binding regions based on PAR-CLIP sequence clusters for WTAP and METTL3 without normalization of the overall length of the different transcript regions.
(Related to Figure 3) METTL3 and WTAP influence gene expression and alternative splicing (A) The 410 genes identified in both METTL3 and WTAP PAR-CLIPs were analyzed by GO analysis and an enrichment map was constructed by Cytoscape (parameters: p < 0.05, overlap cutoff > 0.5).
(Related to Figure 4). Expression pattern and functional assay of WTAP and METTL3 in zebrafish embryos (A) Whole-mount in situ hybridization (WISH) shows ubiquitous expression of WTAP and METTL3 during embryogenesis, respectively.
Materials and Methods