Abstract
In a recent paper published in Cell Research, Yan Bao and colleagues characterize a new population of IFNγ-producing innate-like B cells that promotes innate immune responses and contributes to early pathogen control following intracellular bacterial infection.
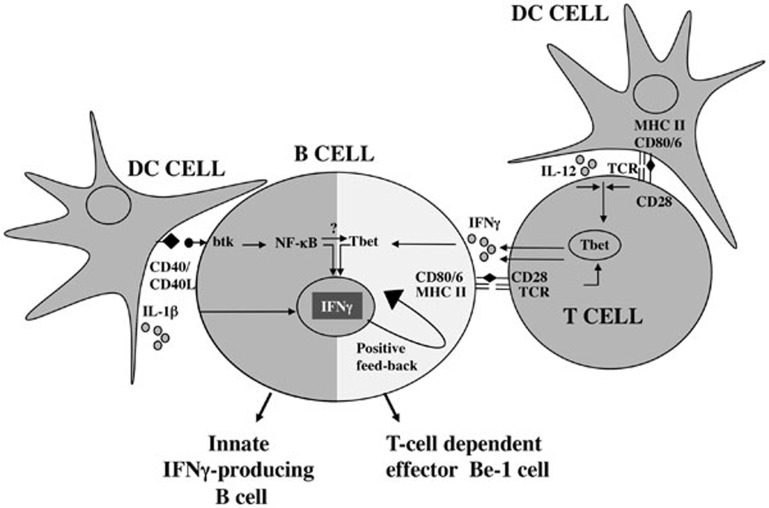
Innate immune cells provide protection against bacterial pathogens by secreting pro-inflammatory cytokines that activate the rapid innate antimicrobial response and initiate adaptive immunity1. Although myeloid-derived innate cells represent one key source of pro-inflammatory cytokines, lymphoid lineage cells, including B cells, can be an early source of cytokines following infection. Indeed, B cells activated by pathogen-associated TLR ligands rapidly produce cytokines2 (“innate” cytokine-producing B cells, Figure 1). B cells also produce cytokines later in the immune response following cognate interactions with helper T cells3 (“T-cell dependent” cytokine-producing B cells, Figure 1). Although most B lineage cells can produce IL-6, IL-10 and TNFα, some B cell subpopulations secrete additional proinflammatory antimicrobial cytokines. For example, mature follicular B cells from Toxoplasma gondii-infected mice produce IFNγ and IL-12p40 following restimulation with pathogen extracts4 while plasmablasts from Trypanosoma cruzi-infected mice produce IL-17 directly ex vivo5. Thus, the “cytokine profile” of B cells appears to be dependent on many factors including the stimuli used to activate the B cells, the cytokine microenvironment and the origin or differentiation status of the B cells.
Figure 1.
“Innate” and “adaptive” cytokine-producing B cells. Development of “Innate” and “adaptive” cytokine-producing B cells is differentially regulated by the cross-talk between B cells, innate cells and Ag-experienced T cells.
In a recent paper published in Cell Research, Yan Bao and colleagues identify a novel subset of innate-like B cells that produces IFNγ following infection with bacterial and viral pathogens6. This new CD11ahiCD16/32hi B cell subpopulation rapidly expands following infection and peaks by day 3 post-infection. These B cells, which are also induced following TLR activation, appear to represent a distinct subpopulation that expresses lower levels of MHCII, CD80 and CD86 and increased levels of CD40, CD40L, CD11a and CD16/32 compared to conventional follicular B cells. Transcriptome analysis of the CD11ahiCD16/32hi B cells also reveals important differences between these pathogen-induced innate-like B cells and the CD11aloCD16/32lo conventional B cells. In particular, cytokine genes and receptors are significantly upregulated in the innate-like B cells. In agreement with this finding, CD11ahiCD16/32hi B cells, but not the remaining CD11aloCD16/32lo B cells, secrete IFNγ following ex vivo restimulation with anti-CD40. Interestingly, the amount of IFNγ produced by the purified innate B cells is similar to that produced by an equal number of NK cells, suggesting that the innate B cells are important source of IFNγ following infection. In support of this hypothesis, CD40-activated innate CD11ahiCD16/32hi B cells can, in an IFNγ-dependent fashion, induce infected macrophages to produce TNFα and nitric oxide. Furthermore, transfer of the CD11ahiCD16/32hiB cells, but not conventional B cells, to Listeria monocytogenes-infected Btk-deficient mice, which lack CD11ahiCD16/32hi B cells and exhibit increased susceptibility to L. monocytogenes infection, significantly increased the concentration of systemic IFNγ and decreased pathogen burden in both spleen and liver. Importantly, transfer of IFNγ-deficient CD11ahiCD16/32hi B cells has no effect on pathogen load in the Listeria-infected Btk−/− mice. Based on these data, the authors conclude that the innate IFNγ-producing B cells may contribute to L. monocytogenes clearance by activating the antimicrobial properties of the Listeria-infected macrophages.
This is not the first demonstration that B cells make IFNγ or that IFNγ made by B cells can influence immune responses to pathogens. Indeed, immature bone marrow B cells constitutively express low levels of IFNγ7, and mature follicular and marginal zone B cells can be induced to secrete IFNγ following stimulation with pathogens, TLR ligands or Th1 cells2,3. Furthermore, prior reports indicate that IFNγ-producing B cells can induce differentiation of Th1 cells in vitro4 and can regulate the Th1 response to Salmonella enterica infection8. Thus, IFNγ-producing B cells appear to modulate both innate and adaptive immunity. However, it is not yet clear whether the same subpopulation of B cells is responsible for regulating the activation and differentiation of both macrophages and CD4 T cells.
To assess the origin of the IFNγ-producing innate B cells and identify the signals that control the development of these cells, the authors established an in vitro model system. Using this system they demonstrate that co-incubation of follicular B cells, but not marginal zone or B1 B cells, with DCs and heat-killed L. monocytogenes induces the CD11ahiCD16/32hi B cell subset in culture. The authors also show that Btk, IL-1β and CD40/CD40L-dependent signals are important for the development of the innate B cells in vitro and in vivo. Interestingly, Btk is also required for the development of innate-like IL-17-producing plasmablasts following infection with T. cruzi5. However, unlike the Listeria-induced IFNγ-producing B cell subpopulation, the T. cruzi-induced IL-17-producing B cells can develop in the absence of TLR and CD40 signaling5, demonstrating clear heterogeneity among innate-like Btk-dependent B cell subpopulations.
CD11ahiCD16/32hi B cell differentiation also differs in regard to the requirement of IFNγR. Previous work from our lab has demonstrated that differentiation of IFNγ-producing B-effector 1 (Be-1) cells requires IFNγR and T-bet9. However, in IFNγR−/− mice, the CD11ahiCD16/32hi B cell population expands normally following L. monocytogenes infection. Therefore, it seems likely that multiple types of IFNγ-producing B cell “subsets” can be induced following infection and that the signals used to induce these cells are context-specific and will vary depending on whether the B cell is activated in the presence of different types of “innate” cells or T cells (Figure 1).
Collectively, the data presented by Bao and colleagues suggest a functional role for IFNγ-producing B cells following infection. Since NK cells, and not B cells, make IFNγ at the very early time points post-infection, future studies are required to delineate the individual contributions of NK cells and the innate-like IFNγ-producing B cells to pathogen control. Likewise, in the future it will be important to assess whether the IFNγ-producing B cells are dependent on NK cells for their development and whether the B cells from NK-deficient mice are still able to decrease bacterial burden when adoptively transferred to B cell-deficient hosts. If CD11ahiCD16/32hi IFNγ-producing B cells can decrease L. monocytogenes burden independent of NK cells then it will be interesting to assess whether these cells can also influence the development of the adaptive immune response, particularly the differentiation of Th1 cells. If so, then the IFNγ-producing B cells described in the current report may represent a new B cell population that contributes to protection from pathogens and spans the gap between the early innate and later adaptive immune responses.
References
- Kumar H, Kawai T, Akira S. Int Rev Immunol. 2011. pp. 16–34. [DOI] [PubMed]
- Gray D, Gray M, Barr T. Eur J Immunol. 2007. pp. 3304–3310. [DOI] [PubMed]
- Lund FE, Randall TD. Nat Rev Immunol. 2010. pp. 236–247. [DOI] [PMC free article] [PubMed]
- Harris DP, Haynes L, Sayles PC, et al. Nat Immunol. 2000. pp. 475–482. [DOI] [PubMed]
- Bermejo DA, Jackson SW, Gorosito-Serran M, et al. Nat Immunol. 2013. pp. 514–522. [DOI] [PMC free article] [PubMed]
- Bao Y, Liu X, Han C, et al. Cell Res. 2014. pp. 161–176. [DOI] [PMC free article] [PubMed]
- Hart G, Flaishon L, Becker-Herman S, et al. J Immunol. 2005. pp. 5034–5042. [DOI] [PubMed]
- Barr TA, Brown S, Mastroeni P, et al. J Immunol. 2010. pp. 2783–2789. [DOI] [PMC free article] [PubMed]
- Harris DP, Goodrich S, Gerth AJ, et al. J Immunol. 2005. pp. 6781–6790. [DOI] [PubMed]
- Sun JC, Lanier LL. Nat Rev Immunol. 2011. pp. 645–657. [DOI] [PMC free article] [PubMed]