The Hippo pathway controls organ size by regulating the expression of genes that promote cell proliferation, stemness, and cell survival, and its deregulation promotes cancer1,2,3,4. The core of the Hippo pathway contains a highly conserved kinase cascade where the MST1/2 kinases (Hippo in Drosophila) activate the LATS1/2 kinases (Warts in Drosophila), which then phosphorylate and inactivate the transcriptional co-activators YAP and TAZ (Yorkie in Drosophila). Unphosphorylated YAP and TAZ bind to the TEAD1-4 transcription factors (Scalloped in Drosophila) to drive target gene expression. Although the interactions between these components and a few associated proteins have been studied in detail, our knowledge of how the core of the Hippo pathway is regulated by upstream inputs is still incomplete.
Three recent reports now describe proteomic studies uncovering extensive protein-protein interaction networks that connect the Hippo pathway to proteins involved in other signaling pathways and processes5,6,7. All three papers used many of the known Hippo pathway components as baits to identify interacting proteins by affinity purification and mass spectrometry (AP-MS) in cultured cells. Kwon et al. from the group of Norbert Perrimon analyzed the interactome of the Drosophila Hippo pathway and identified 153 high-confidence interacting proteins5. They then tested the function of these proteins by RNAi knock-down and found that 102 of them affected the transcriptional activity of Yorkie. The two other studies by Wang et al.6 and Couzens et al.,7 from the groups of Junjie Chen and Anne-Claude Gingras, characterized the protein-protein interaction network of the human Hippo pathway and identified high-confidence networks of 343 and 445 proteins, respectively. All three studies rediscovered many of the known interactions between pathway components. In addition, they identified numerous proteins that were not previously tied to the Hippo pathway such as components associated with cell junctions, the cytoskeleton, the mitotic spindle, phosphatase complexes, and chromatin remodeling proteins. They also discovered that vesicle trafficking regulates Yorkie stability and that core components change binding partners in response to phosphorylation.
Protein phosphorylation plays a central role in the regulation of the Hippo pathway. Couzens et al.7 addressed how phosphorylation affects the Hippo pathway interaction network. They found that phosphorylation triggered remodeling of the interactomes of MST, LATS, and the MOB cofactors for LATS. For example, treatment of cells with the phosphatase inhibitor okadaic acid, which leads to increased phosphorylation of the MST and LATS kinases, promoted the formation of LATS complexes with the upstream MST kinases and the AMOT cofactors, and decreased their interaction with ubiquitin ligases, PP2A phosphatases, and cell polarity proteins. Thus, phosphorylation caused a shift in the LATS and MST interactomes from proteins that inhibit their activity to proteins that promote their activity. Phosphorylation therefore not only modulates MST/LATS kinase activity directly by phosphorylation of the kinases themselves but also affects the composition of the kinase complexes.
Several Hippo pathway components and regulators localize to adherens and tight junctions, such as the AMOT family of adaptor proteins that bind to the WW domains of YAP and promote its cytoplasmic accumulation4. Wang et al.6 identified a novel regulator of YAP, the coiled-coil domain-containing protein 85C (CCDC85C) that also localizes to tight junctions8. Similar to AMOT, CCDC85C binds to the WW domains of YAP and induces its nuclear export, thereby inhibiting YAP activity6. Thus, AMOT and CCDC85C may compete for binding to YAP and may be part of a mechanism that integrates different signaling inputs coming from cell junctions.
Endocytosis and vesicle trafficking regulate many signaling pathways and all three studies now also link them to the Hippo pathway. The interactomes of several Hippo pathway components contain proteins that are associated with endocytosis and vesicle trafficking, including Rab and Arf GTPases, coat protein complex II (COPII) components, and the endocytosis protein α-adaptin. Kwon et al.5 further characterized one of these vesicle trafficking proteins, a member of the α-arrestin family that they named Leash because it restricts Yorkie activity. Leash binds to Yorkie and suppresses its transcriptional activity by promoting its localization to endosomes and degradation through the lysosomal pathway. Binding of Leash to Yorkie requires ubiquitination by the HECT ubiquitin ligase Nedd4, and Warts activation enhances Leash-mediated degradation of Yorkie. Leash regulates the activity of Yorkie in vivo, because depletion of Leash enhanced the hyperproliferation of gut stem cells caused by Hippo RNAi, while overexpression of Leash reduced Yorkie-driven gut stem cell hyperproliferation and wing size. These studies thus identify the endocytic pathway as an important regulator of Hippo signaling activity.
Until now, discovery of Hippo pathway components has been biased towards negative regulators of YAP/TAZ and Yorkie, and only a few positive regulators were known so far. Kwon et al. now expand the repertoire of positive regulators: For example, they identified the fly homologs of NUDT21 (CPSF5) and CPSF65 that were also identified as YAP interactors by Wang and colleagues6. NUDT21 and CPSF6 are involved in the cleavage and polyadenylation of mRNAs9,10, although how they regulate YAP/Yorkie remains to be investigated. The discovery of proteins that link YAP/Yorkie with RNA processing is an interesting new twist in the regulation and function of the Hippo pathway.
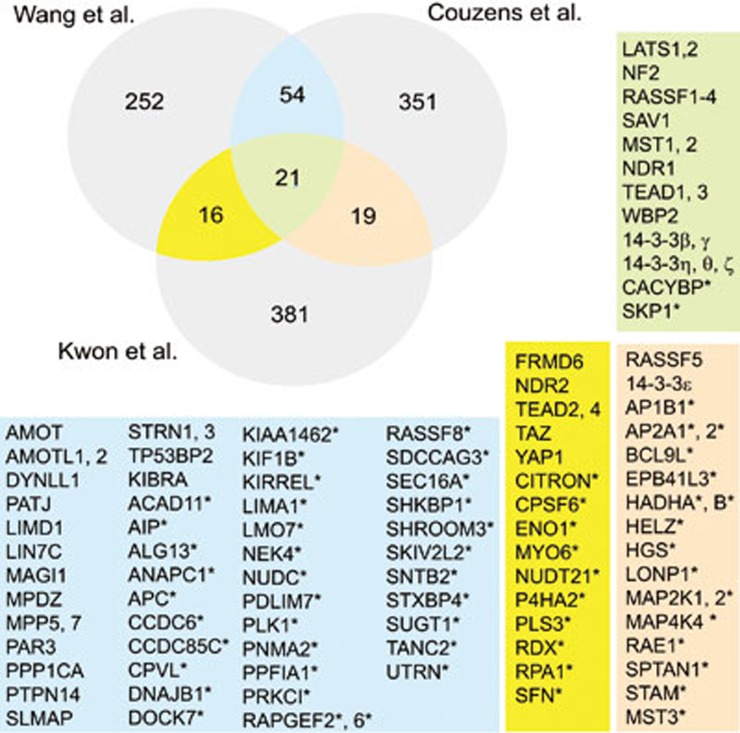
What else is new and how much do the different interactomes overlap? First comparing the two human interactomes, we found that they share 75 proteins, which is about 20% of either interactome (Figure 1). Of these 75 proteins, over 70% are novel Hippo pathway interacting proteins and many of them are associated with the cytoskeleton, cell-cell junctions or vesicle trafficking. For YAP interactors, however, three of the 20 common proteins are novel interactors: the RA-domain factor RASSF8, RAPGEF2, and the above discussed cell junction protein CCDC85C. Comparing all three interactomes revealed 21 shared proteins, most of which are known core members and only two novel interactors: the E3 ubiquitin ligase associated CACYBP (SIP1)11 and SKP112, which may identify novel mechanisms of pathway component turnover. However, the number of novel and potentially conserved interactors is greatly expanded to nearly half of the overlapping proteins when comparing the fly interactome (human homologs) to either human interactome individually; then 30 proteins are novel shared interactors, which are associated with the cytoskeleton, RNA processing, vesicle trafficking, and other processes (Figure 1). In conclusion, the three proteomic studies identified hundreds of potential new components and modulators of the Hippo pathway, many of which are shared between the different interactomes. New interactors may act as regulators of the Hippo pathway as many affect Yorkie transcriptional activity, or they may constitute novel downstream effectors of the Hippo pathway that act independently of Yorkie/YAP. The function for most of these interactions is not yet known, thus they promise great potential to discover novel aspects of the biology of the Hippo pathway.
Figure 1.
Comparison between the two human and Drosophila Hippo pathway protein-protein interacting networks. Venn diagram indicating the number of shared Hippo pathway interacting proteins reported with high confidence in the three proteomic studies5,6,7. The background colors in the lists correspond to the colors of the overlapping areas of the Venn diagram. Asterisks indicate novel Hippo pathway interacting proteins reported in these studies. Known Hippo pathway components are listed before novel interactors.
References
- Halder G, Johnson RL. Development. 2011. pp. 9–22. [DOI] [PMC free article] [PubMed]
- Pan D. Dev Cell. 2010. pp. 491–505. [DOI] [PMC free article] [PubMed]
- Harvey KF, Zhang X, Thomas DM. Nat Rev Cancer. 2013. pp. 246–257. [DOI] [PubMed]
- Yu FX, Guan KL. Genes Dev. 2013. pp. 355–371. [DOI] [PMC free article] [PubMed]
- Kwon Y, Vinayagam A, Sun X, et al. Science. 2013. pp. 737–740. [DOI] [PMC free article] [PubMed]
- Wang W, Li X, Huang J, et al. Mol Cell Proteomics. 2014. pp. 119–131. [DOI] [PMC free article] [PubMed]
- Couzens AL, Knight JD, Kean MJ, et al. Sci Signal. 2013. p. rs15. [DOI] [PubMed]
- Mori N, Kuwamura M, Tanaka N, et al. Am J Pathol. 2012. pp. 314–327. [DOI] [PubMed]
- de Vries H, Ruegsegger U, Hubner W, et al. EMBO J. 2000. pp. 5895–5904. [DOI] [PMC free article] [PubMed]
- Ruegsegger U, Blank D, Keller W. Mol Cell. 1998. pp. 243–253. [DOI] [PubMed]
- Matsuzawa SI, Reed JC. Mol Cell. 2001. pp. 915–926. [DOI] [PubMed]
- Lisztwan J, Marti A, Sutterluty H, et al. EMBO J. 1998. pp. 368–383. [DOI] [PMC free article] [PubMed]