Abstract
Surface-enhanced Raman spectroscopy (SERS) has been used in a variety of biological applications due to its high sensitivity and specificity. Here, we report a SERS-based biosensing approach for quantitative detection of biomolecules. A SERS substrate bearing gold-decorated silicon nanopillars is functionalized with aptamers for sensitive and specific detection of target molecules. In this study, TAMRA-labeled vasopressin molecules in the picomolar regime (1 pM to 1 nM) are specifically captured by aptamers on the nanostructured SERS substrate and monitored by using an automated SERS signal mapping technique. From the experimental results, we show concentration-dependent SERS responses in the picomolar range by integrating SERS signal intensities over a scanning area. It is also noted that our signal mapping approach significantly improves statistical reproducibility and accounts for spot-to-spot variation in conventional SERS quantification. Furthermore, we have developed an analytical model capable of predicting experimental intensity distributions on the substrates for reliable quantification of biomolecules. Lastly, we have calculated the minimum needed area of Raman mapping for efficient and reliable analysis of each measurement. Combining our SERS mapping analysis with an aptamer-functionalized nanopillar substrate is found to be extremely efficient for detection of low-abundance biomolecules.
Keywords: surface-enhanced Raman spectroscopy, aptamer, nanopillar array, Raman mapping, vasopressin, biosensing
Surface-enhanced Raman spectroscopy (SERS) has been widely used as an analytical tool in biosensing applications1–5 due its powerful advantages including amplifying Raman signals by factors up to 10–14 orders of magnitude, providing ultrahigh sensitivity, with the potential for single-molecule detection; and featuring the specificity of Raman spectra based on the unique molecular fingerprinting information.6–11 Nevertheless, reliable quantification of biomolecules (e.g., DNA, peptides, and proteins)5,12–17 using SERS has been hindered by poor uniformity and reproducibility of SERS-active nanostructured platforms.18–20 Furthermore, lack of specific binding and fluctuations in signal intensities21,22 impede the development of SERS-based quantitative bioassays.
To overcome the aforementioned challenges in SERS-based biomolecular quantification, we report here an approach using large area mapping of Raman signals on a SERS-active substrate, densely packed with aptamer-functionalized nanopillars. Simple and fine-tuned nanofabrication processes ensure reproducible production of substrates, which was demonstrated in our previous work.23 The high uniformity of the nanostructure distribution over a substrate minimizes the variation within a scanning area on which the Raman mapping is performed. For the specific detection of biomolecules, aptamers that are short single-stranded oligonucleotides, which specifically bind to their target molecules with high afinity, are immobilized on nanopillar surfaces. In addition, a monolayer of 6-mercapto-1-hexanol (MCH) spacer molecules is self-assembled onto the gold surface to prevent nonspecific adsorption of molecules and to improve the capturing efficiency of aptamers. Consequently, the surface functionalization helps analyte molecules to be uniformly distributed over the substrate area. Lastly, large area Raman mapping provides statistical reliability and reproducibility of signal intensity quantification by accounting for spot-to-spot intensity fluctuations typically occurring with low-abundance molecules.
To demonstrate the significance of our approach, the following issues will be examined throughout this paper: (1) the effect of surface functionalization with aptamers and spacers respectively, (2) the dependence of SERS responses to analyte concentrations at sub-nanomolar level, (3) the development of a theoretical model to predict experimental results, and (4) size determination of statistically relevant mapping area.
In this work, TAMRA (5-carboxytetramethylrhodamine)-labeled vasopressin (TVP) is used as an analyte for the purpose of biomolecular quantification. Vasopressin is an antidiuretic peptide hormone whose plasma level normally ranges from 0 to 10 pM and markedly increases up to 350 pM in late-phase hemorrhagic shock.24 Thus, accurate and rapid subnanomolar quantification of vasopressin is clinically important as a lifesaving technology.
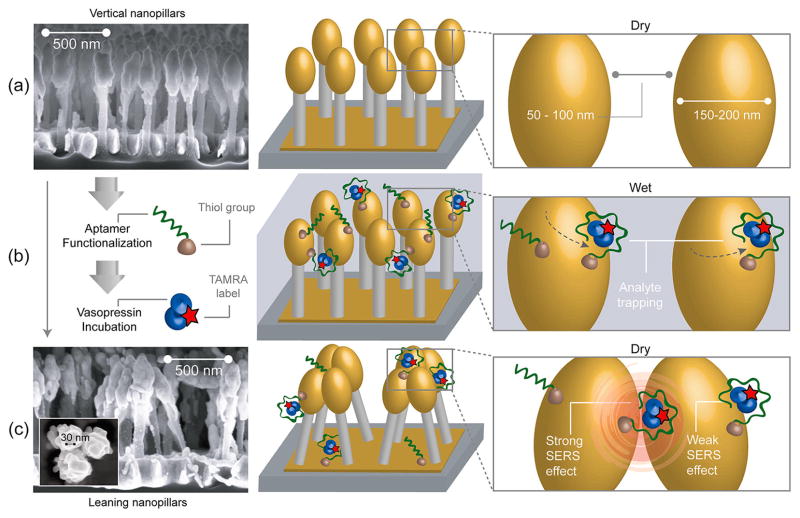
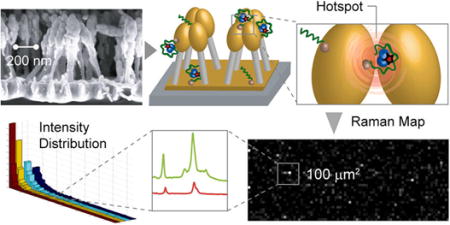
Figure 1 illustrates the basic principle of SERS generation by aptamer-functionalized nanopillars. Gold-coated silicon nanopillar structures were fabricated by maskless reactive ion etching of silicon wafers followed by electron beam evaporation for gold deposition (Figure 1a).23 Vasopressin-specific aptamers were immobilized onto the gold surfaces of the nanopillars to selectively and efficiently recognize vasopressin molecules. The aptamer-functionalized nanopillar substrate was incubated with a TVP sample solution at 37 °C, the optimal temperature for aptamer–vasopressin binding25 (Figure 1b). After sequential washing steps using buffer solution and water to remove nonspecifically bound molecules, the wetted substrate was then allowed to dry (Figure 1c). Due to the surface tension between pillars during liquid evaporation,26 the nanopillars lean toward each other, forming micro-sized clusters and inducing near-field coupling effects between electronic excitations on gold surfaces of nanopillars known as localized surface plasmons (LSPs). In small metallic junctions (less than 10 nm) between adjacent nanostructures, the coupling effect of LSPs produces even larger plasmonic fields, so-called hotspots.19,27–29 Raman scattering of the incident light from molecules located in these regions is greatly enhanced by additional high-order LSP modes. Since the interpillar distances after liquid evaporation are clearly less than 10 nm (Figure 1c), vasopressin molecules trapped within these clusters exhibit a pronounced Raman scattering signal from TAMRA tags even with a small number of molecules.
Figure 1.
Schematics of the functionalized leaning nanopillar detection method. (a) Highly packed gold-coated nanopillars are structured on a silicon wafer. Before exposure to the analyte, pillars are vertically oriented and are spaced by 50–100 nm gaps. (b) After aptamers are immobilized on the nanopillars (via Au–thiol covalent bonding) and a TVP sample is introduced, the analyte binds to the aptamers. (c) Scanning electron microscope (SEM) image of leaning nanopillars after the evaporation of liquid (side view). The analyte trapped within the leaning region exhibits a strong Raman enhancement effect due to the highly localized plasmonic field around the contact region. When the vasopressin is trapped by an aptamer within a hotspot, as shown on the right panel, the TAMRA Raman signal is highly enhanced. The inset shows a high-magnification SEM image displaying one of the pillar clusters, revealing junction spaces less than 10 nm between pillar heads (top view).
In our previous study,23 we have demonstrated that as the pillar density increases, the SERS signal increases in an exponential fashion. Thus, in this work, we have used substrates with the highest obtainable pillar density (~20 pillars/μm2) to maximize the SERS signal. Also, an electron beam evaporation process was developed to coat the silicon pillars with gold, such that the metallization thickness (~30 nm) was optimized for a laser excitation wavelength of 633 nm. As can be seen in Figure 1a,c, the metal deposition on the pillar heads resulted in oval-shaped gold clusters, which can be approximated as an ellipsoid or sphere for the purposes of electromagnetic modeling.
A large area (101 μm × 33 μm) of the substrate was scanned with 1 μm step size to collect Raman spectra from each pixel (1 μm × 1 μm). Recorded signals over the area were then displayed as heat maps rendering intensities by degrees of color. Mapping measurements were performed three times at each concentration. Our experimental data were represented as intensity distributions to develop a theoretical model by expanding previously reported single hotspot theory30 to a large-scale SERS mapping model. The analytical model developed here has the potential to be used as a calibration curve in assay validation. Statistical tests between two heat maps (treatment versus control samples) were also employed to determine the minimum mapping area required to unambiguously (at 95% confidence level) distinguish those two samples. Once the mapping area was calculated, multimodal analysis, that is, simultaneous monitoring of different frequency peaks, was also introduced as a useful tool to further reduce the overall assay time, maintaining statistical significance in analyte quantification measurements. In summary, our large area Raman mapping approach on substrate incorporating uniformly distributed nanopillars improves statistical reliability and reproducibility of SERS intensity quantification. Furthermore, the analyte-specific recognition capability of aptamers shows the potential of this approach for bioanalytical quantifications in complex biological samples.
RESULTS AND DISCUSSION
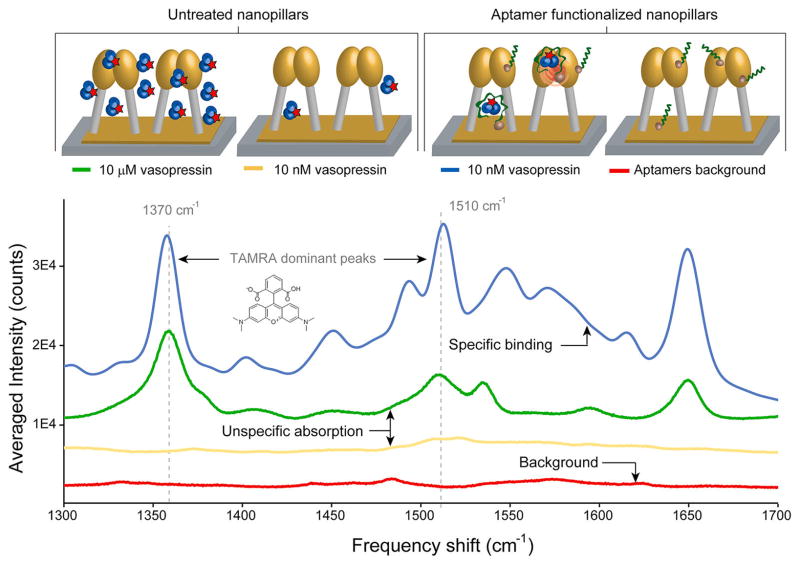
We first investigated the effect of aptamer functionalization by comparing SERS signals from substrates with untreated and aptamer-functionalized nanopillars using manual signal collection. Four substrates at various conditions were prepared (see Materials and Methods for protocol): (1) 10 μM TVP with untreated nanopillars, (2) 10 nM TVP with untreated nanopillars, (3) 10 nM TVP with aptamer-functionalized nanopillars, and (4) a blank sample with aptamer-functionalized nanopillars. As illustrated in Figure 2, the Raman spectrum from the aptamer-functionalized substrates reacted with 10 nM TVP (blue trace) displays five candidate peaks for diagnostics. Two peaks at 1450 and 1533 cm−1 can be assigned to the synergistic vibrational effects of the xanthene ring and dimethylamino group motions and the peak observed at 1651 cm−1 to the symmetric in-place C–H bend of the xanthene ring only. The two most dominant TAM-RA peaks at 1370 and 1510 cm−1, the stretching vibrations of the xanthene ring moiety of the molecule,17,30–32 were chosen to be diagnostic peaks in all further analysis because they showed the most consistent electromagnetic enhancement during the mapping experiments. The spectrum of 10 nM TVP without aptamers (yellow trace) is comparable in intensity to the aptamer background (red trace), where no significant peaks are observed. One can conclude that aptamers effectively capture TVP molecules and help position the analyte molecules close to the hotspots. Due to the SERS enhancement in the hotspot, the peak intensities for the 10 nM TVP with aptamers (blue trace) were even higher than those of the 10 μM one without aptamers (green trace). We hypothesize that at micromolar concentrations the SERS effect is hindered by a thick layer of analyte molecules formed on top of the pillars. Possibly, the measured signal at 10 μM results from bulk Raman scattering rather than SERS.
Figure 2.
Raman spectra acquired on untreated substrates and aptamer-functionalized nanopillars. When nonspecific adsorption of vasopressin on the substrate surface occurs (no aptamers), signal is only obtained at very high concentrations (e.g., 10 μM TVP, green trace). At low concentrations (e.g., 10 nM TVP, yellow trace), the signal enhancement is very low, indicating that very few molecules actually reside close to a hotspot. On the other hand, when the pillars are functionalized with selective aptamers, the signal intensity greatly increases. Five possible diagnostic peaks with two of the most dominant Raman peaks (1370 and 1510 cm−1) of TAMRA were observed during experiments with different analyte concentrations.
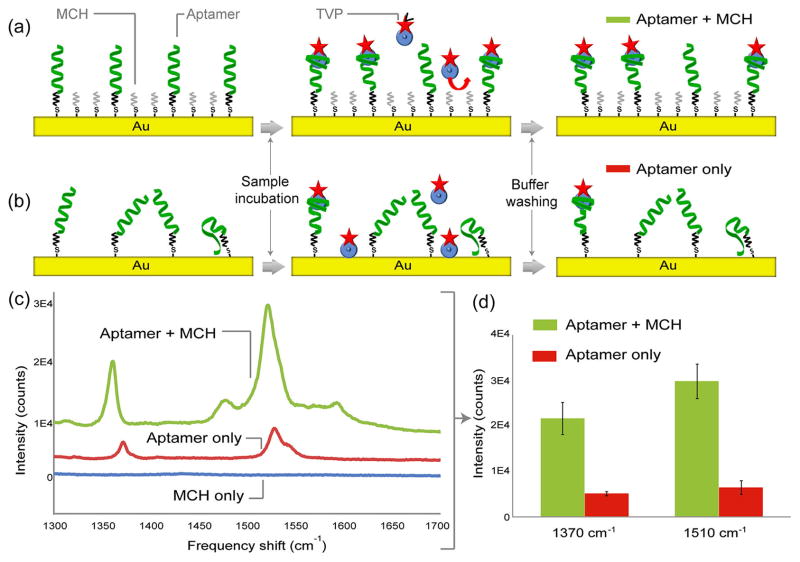
In order to optimize the aptamer–analyte binding conditions on the nanopillars, the gold surface was backfilled with MCH immediately after aptamer immobilization (see Materials and Methods for functionalization). Because MCH forms a well-organized self-assembled monolayer (SAM), it prevents nonspecific adsorption of analyte molecules by blocking access to the gold surfaces.33–35 This layer also improves the capturing efficiency of the aptamers by preventing them from collapsing and adhering to the substrate surfaces.35,36 By comparing peak intensities obtained from two substrates functionalized with and without the MCH SAM, we were able to show that MCH molecules reorient the thiolated aptamers into a vertical position favorable for aptamer–analyte binding (Figure 3). TVP samples (10 nM) were reacted with the two surfaces. The result shows that the intensities from the surfaces functionalized with MCH are roughly 2 orders of magnitude greater than those without both diagnostic peaks, thus confirming that the MCH SAM makes the majority of surface-bound aptamers more accessible for aptamer–analyte binding.
Figure 3.
Effect of 6-mercapto-1-hexanol (MCH) self-assembled monolayer (SAM) for aptamer–TVP binding. TVP molecules are bound on the aptamer-functionalized substrate (a) with and (b) without a monolayer of MCH. Due to the backfilling of MCH molecules, binding efficiency of aptamers is improved, and nonspecific adsorption of analyte is minimized. This effect is verified by experimental results shown in (c) and (d). The experiments were performed with 10 nM TVP samples on different substrates: aptamers with MCH layer (green), aptamers without MCH layer (red), and MCH only (blue).
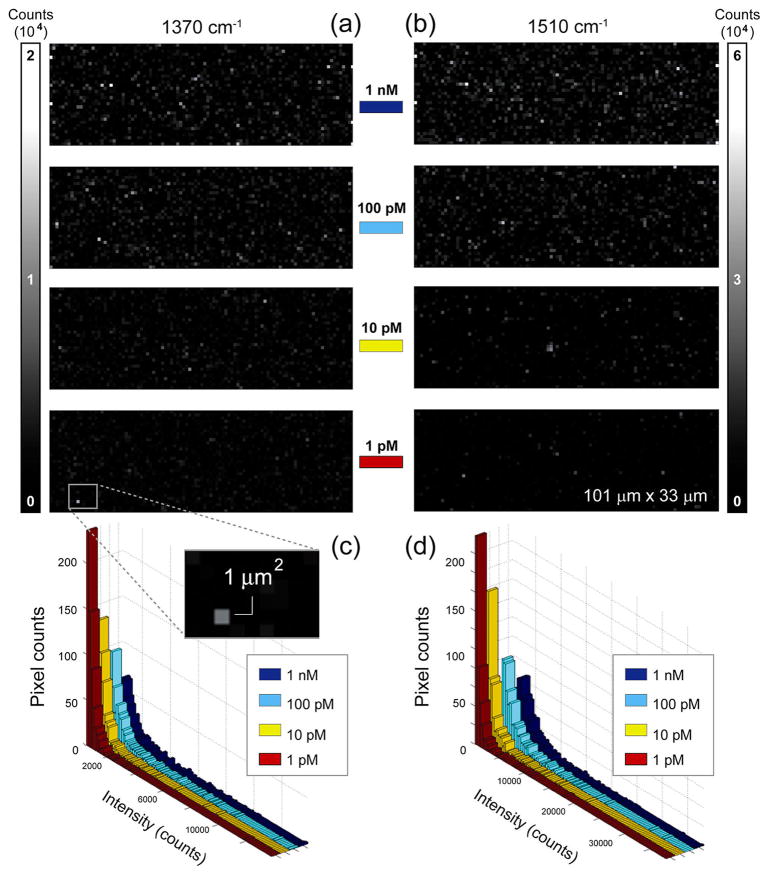
Various control experiments were performed to verify the specificity of aptamers to TVP molecules (see Supporting Information, Control Experiments). Then, several experiments were run to evaluate the ability of mapping analysis to quantify low-abundance molecules on the functionalized substrates. Around 10 000 spectra from areas of three independent substrates were collected using point-by-point Raman mapping (see Materials and Methods for instrumentation) at each concentration, recording the SERS responses from TVP molecules bound to aptamers on the gold-coated nanopillars. The intensities of the characteristic TAMRA vibrational peaks (1370 and 1510 cm−1) were extracted independently, and their spatial dependencies (Raman maps) were displayed in the format of pixel-based images at various TVP concentrations (Figure 4). For each measurement, all positive intensities were ranked and the hotspots were defined as the top 20% among such set of measurements. This percentage threshold was chosen based on the Pareto principle.37,38 Our experimental results showed that the higher the analyte concentration, the more abundant and brighter hotspots were detected for both frequency peaks, visually represented as heat maps (Figure 4a,b) and graphically as histograms (Figure 4c,d).
Figure 4.
Spectroscopic Raman mapping of SERS intensity distribution. (a,b) Intensity maps of diagnostics TAMRA peaks 1370 cm−1 (left) and 1510 cm−1 (right) in a series of mapping experiments for quantification of intensities with 1 nM, 100 pM, 10 pM, and 1 pM TVP. A substrate area of about 101 μm × 33 μm is shown in each case, along with the two intensity scale bars labeled with the maximum intensity values (in arbitrary units) for each diagnostic peak. The inset highlights a 1 μm2 area, which corresponds to a typical cluster of leaning nanopillars forming hotspots. The Raman map pixel size is 1 μm × 1 μm. (c,d) Histogram representation of SERS intensity distributions of hotspots at different concentrations of TVP. Four histograms are displayed in overlaid configuration for both TAMRA dominant peaks, (a) 1370 cm−1 and (b) 1510 cm−1, corresponding to various analyte concentrations.
In order to determine the consistency and/or repeatability of SERS mapping measurements for individual substrates with the same functionalization/treatment conditions, the Kolmogorov–Smirnov (KS)39–41 significance test was used. The KS test is a nonparametric test that makes no assumptions regarding the distribution of the collected data. It determines if two random variables are sampled from identical distributions. Substrates bearing vasopressin-specific aptamer-functionalized nanopillars were reacted with 1 nM TVP samples. During mapping experiments, three individual substrate areas were measured, collecting 3333 spectra (101 μm × 33 μm) for each measurement. The KS significance test verified that the measurements from the different substrates were not statistically different (see Supporting Information, Table S-I), confirming the reproducibility of the experimental mapping technique.
To further understand the statistical distribution of hotspots, we plotted the intensity histogram in which the smallest and largest value on the x axis corresponds to the minimum and maximum hotspot intensity value of the Raman maps. Such histograms were constructed for both diagnostic peaks and different TVP concentrations (Figure 4c,d). It was observed that these histograms follow a highly skewed or long-tailed truncated Pareto distribution (TPD), with coefficient of determination values greater than 0.95 for both major TAMRA peaks at the four difierent concentrations (see Supporting Information, Table S-II). TPDs have previously been discovered to describe the intensity distribution characteristics of closely spaced SERS hostpots.22 Since our experimentally obtained hot-spot intensity distribution fits the predicted long-tailed TPD well, we can adapt an analytical framework for single hotspot theory30 into our large-scale SERS mapping model (see Supporting Information, Hotspot Distribution Theory). Now, by arbitrarily defining α = 1 + k, we can rewrite the intensity distribution function (see Supporting Information, equation (III)) for a given analyte concentration (C) as
| (1) |
where parameters α and A can depend on the analyte concentration. In particular, α determines how fast the enhancement decreases when moving away from the hotspot, while A is an indicator of how probable it is for a molecule to be located close to the hotspot. Parameter A is generally assumed to be an intrinsic characteristic of the SERS substrate.30 We observed that for both diagnostic peaks there is a strong power relationship between the experimentally obtained α and A parameters and the analyte concentration (see Supporting Information, Figures S2 and S3). Now, since both α and A are a power function of the analyte concentration, we have general forms
| (2) |
where b, d, e, and f are constants whose values are determined by the power fitting shown in Figures S2 and S3 (see also Supporting Information, Tables S-III and S-IV). Subsequently, we can express the hotspot intensity distribution analytically as a function of concentration by substituting (2) into (1). Then, the integral of the intensity distribution between the minimum (Imin) and maximum (Imax) intensity values gives the sum of all hotspot intensities (Isum) in a given mapping experiment. This integral can be expressed as a function of analyte concentration (C) such that
| (3) |
By substituting (2) into (3), we obtain an analytical expression for the total intensity of hotspots as a function of TVP concentration:
| (4) |
This expression is believed to properly describe the collection of SERS signals measured on the uniform nanopillar substrates for different analyte concentrations. Figure 5 shows the experimentally obtained intensity integrals of hotspots for both diagnostic peaks along with their respected power fit (black lines). The high coefficient of determination (R2) value (>0.93) demonstrates a strong power relationship between TVP concentration and SERS intensity integrals. It is also observed that the experimentally fitted power curves reasonably agree with the theoretical ones (red lines) calculated using the analytical expression shown in (4) with low percentage error values (see Supporting Information, Table S-V). The theoretical model is found to be more accurate for the 1370 cm−1 peak, with the R2 value of 0.98. The fitting for the 1510 cm−1 peak is believed to be less accurate due to the broader shape of the band associated with this vibrational frequency, compared to the sharper 1370 cm−1 peak. The result shows that the analytical model developed here agrees with the experimental data, particularly well in the low concentration regimes. Thus, it can potentially be utilized to predict experimental hotspot intensity distributions on the substrate for quantitative measurements of low-abundance biomolecules.
Figure 5.
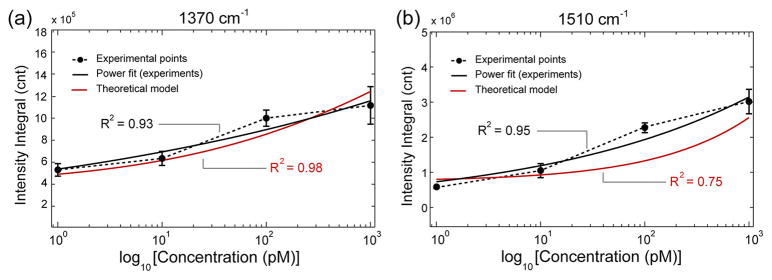
Experimental hotspot intensity integral plots for the diagnostic TVP peaks, (a) 1370 cm−1 and (b) 1510 cm−1, as a function of analyte concentration on a semilog scale (dotted curves). The error bars represent 2 standard deviations on three different samples for the same concentration. Both curves fit power functions (solid black lines). The red curves are calculated from the hotspot intensity distribution of mapping experiments using the statistical model of eq 4.
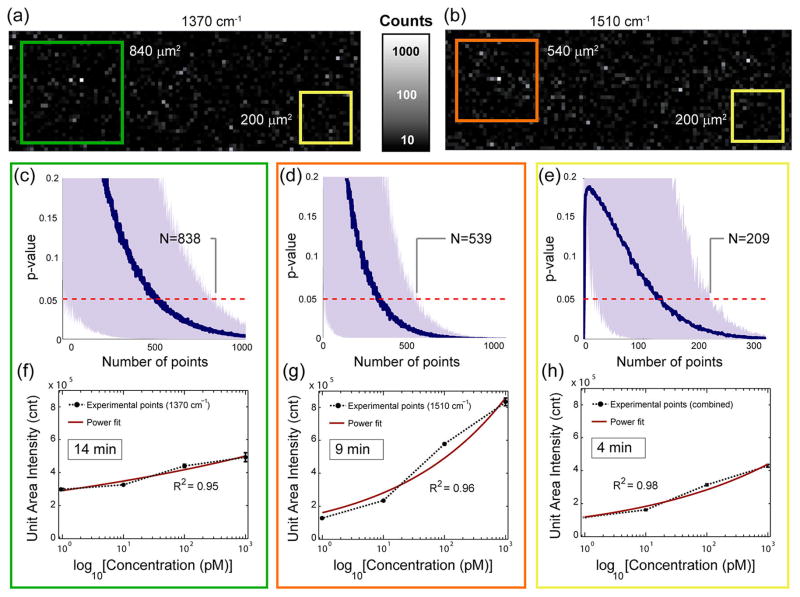
Next, we aim to determine the statistically significant minimum mapping area required to unambiguously distinguish between treatment and control samples. The p values from KS tests were calculated comparing a collection of SERS signals measured on two substrates treated with 1 nM TVP samples, each functionalized with vasopressin-specific aptamers and with immuno-globulin E (IgE)-specific aptamers (as described in Materials and Methods). Figure 6 indicates that when using the one-dimensional KS test (1d-KS), in which each peak is independently investigated, a total of 838 and 539 pixels are required to reach the p value threshold (p = 0.05) for the 1370 and 1510 cm−1 peaks, respectively (Figure 6c,d). Furthermore the two-dimensional KS test41 (2d-KS) was utilized to evaluate the effect of the combined multimodal analysis. Using the 2d-KS test in which the two distinct frequency peaks (1370 and 1510 cm−1) were evaluated simultaneously, only 209 pixels were needed to be 95% confident that the treatment is distinguishable from the control (Figure 6e). These results suggest that the combined multimodal sensory analysis outperforms the analysis where just one frequency shift is used (see Supporting Information, Table S-VI). Using 1 s exposure time during acquisition, the mapping time can be reduced from 10 to 15 min (539–838 pixels) to less than 4 min (209 pixels) by applying the multimodal analysis. Thus, this SERS mapping technique has great potential to be integrated into a rapid bioassay. Figure 6 also shows the intensities of hotspots obtained for various analyte concentrations per unit area, where the unit area is defined as the statistically significant minimum mapping area (Figure 6f–h). The intensities per unit area averaged over 100 randomly selected unit areas are presented for both individual peak analysis (Figure 6f, g) and the combined multimodal analysis (Figure 6h) along with their respected power fit (solid red lines). Both analyses show concentration-dependent SERS intensity integrals over the unit area in the subnanomolar range. The error bars are less than 5% of the measured data for all analyses (Supporting Information, Table S-VII). Similarly to the theoretically derived eq 4 to fit the experimental data, the high experimental R2 values (>0.95) indicate a strong power relationship between the unit area intensity and the TVP concentration, as shown in the insets.
Figure 6.
Advantages of combined sensory readings using the two-sample 2d Kolmogorov–Smirnov test. The p values of the KS test as a function of μm2 area (number of pixels) collected from heat maps based on the comparison of IgE-specific aptamer (control) and vasopressin-specific aptamer (treatment) data sets at 1 nM concentration. Hotspot intensity maps obtained for diagnostic TVP peaks, (a) 1370 cm−1 and (b) 1510 cm−1, in the treatment experiments. The insets show a green (1370 cm−1), orange (1510 cm−1), and yellow (combined) rectangular area, which correspond to unit areas determined by the KS tests; (c) 1d-KS test for diagnostic frequency of 1370 cm−1, (d) 1510 cm−1; and (e) 2d-KS test for the combined diagnostic TAMRA frequencies (1370 and 1510 cm−1). Unit area intensity plots corresponding to individual peaks (f) 1370 cm−1, (g) 1510 cm−1; and (h) combined multimodal analysis as a function of analyte concentration on a semilog scale. The error bars represent 2 standard deviations on 100 different unit areas for the same concentration and are all less than 5% of measured data values. All curves fit power functions (solid red line).
These findings demonstrate that one can employ multimodal analysis without any observable change in final SERS result. The multimodal analysis allows one to significantly reduce time to perform a reliable and statistically significant analyte quantification experiment.
CONCLUSIONS
In this paper, we have demonstrated a SERS-based quantitative biosensing on aptamer-functionalized nanopillars utilizing large area Raman mapping technique. Ultrasensitive and specific detection of TAMRA-labeled vasopressin molecules was achieved at picomolar levels by collecting highly enhanced SERS signals emanating from nanojunctions formed between the leaning nanopillars and functionalizing surfaces with target-specific aptamers. Reliability and statistical reproducibility of quantitative SERS analysis were improved by a large area Raman mapping on the highly uniform substrates. On the basis of our experimental mapping results, we successfully developed a theoretical model to be utilized as an analytical tool for biomolecular quantification on the aptamer-functionalized nanopillar substrates. Furthermore, for the assay optimization, we determined the minimum size of mapping area providing statistical reliability by applying a KS test between treatment and control data. Finally, by employing a multimodal analysis combining two frequency readings, we showed that one can significantly reduce the assay time. In conclusion, we believe that the approach of functionalizing uniform SERS substrates and subsequently performing large area multimodal SERS mapping is a promising technique for specific detection of picomolar concentrations of biomolecules. For our method to be implemented, it is crucial that the substrate is reproducible and uniform. Otherwise, large background variations will occur.
MATERIALS AND METHODS
Materials
5-TAMRA-labeled synthetic arginine vasopressin (TVP) was used as analyte: 5-TAMRA-CYFQNCPRG-NH2. The peptide hormone contains an N-terminus conjugated 5-TAMRA fluorophore obtained from American Peptide Company (Sunnyvale, CA). Vasopressin- and IgE-specific aptamers modified with thiol linkers (HS) were acquired from ChemGenes Corporation (Wilmington, MA) and Integrated DNA Technologies, Inc. (Coralville, IA), respectively (vasopressin aptamer: 5′-GGG GUA GGG CUU GGA UGG GUA GUA CAC GUG UGC GUG GU-HS-3′; IgE-aptamer: 5′-HS-TTT TTT TTG GGG CAC GTT TAT CCG TCC CTC CTA GTG GCG TGC CCC-3′). Other chemicals used in buffer preparation and surface functionalization are listed as follows: TCEP (tris(2-carboxyethyl)phosphine hydrochloride), MCH (6-mercapto-1-hexanol), Tris-HCl, NaCl, KCl, CaCl2, MgCl2, urea, EDTA (ethylenediaminetetraacetic acid), and PBS (phosphate buffered saline, 1×) were all obtained from Sigma-Aldrich (St. Louis, MO).
Substrate Fabrication
The nanostructured pattern was created on an undoped polished 〈100〉 silicon wafer surface with free-standing vertical silicon pillars of 50–80 nm width; subsequently, the wafer was diced into about 5 mm × 5 mm squares to form the active sensor surface. High-density nanopillars were formed using a reactive ion etching (RIE) method without any lithographic steps as described in a previous publication.23 After the RIE step, the nanopillars were exposed to argon plasma for 1 min to completely remove the etchant gas residuals and the decomposition products. Finally, ~30 nm gold was deposited on the heads of the nanopillars by electron beam evaporation to obtain the plasmonic property needed for SERS enhancement.
Surface Functionalization
The substrate was repeatedly cleaned with ethanol and deionized water rinsing. Aptamer solution (5 μM) was mixed with a stable reducing agent, TCEP (500 μM), for 30 min at pH 4 to expose the HS groups at the terminal of the aptamer sequences. PBS solution was added to the solution right before the substrate incubation to adjust the pH to 7. Then the substrate was incubated in the prepared aptamer solution for 1 h followed by immersion in MCH solution (2 mM) for 2 h. After the functionalization, the substrate was rinsed with the mixture of 4 M urea and 15 mM EDTA and water sequentially.
Experimental Protocol
TVP samples were prepared in vasopressin buffer solution (pH 7.4) consisting of deionized water, Tris-HCl (20 mM), NaCl (150 mM), KCl (5 mM), CaCl2 (1 mM), and MgCl2 (1 mM). Standard samples were prepared in the concentration range of 1 pM to 1 nM by serial dilution. Functionalized substrates were incubated with the TVP samples for 1 h at 37 °C in a thermal-cycler (Eppendorf AG, Germany). Then, the substrates were transferred into vasopressin buffer solutions and kept at 37 °C for 15 min to remove unbound molecules. Lastly, the substrates were quickly rinsed with water to prevent salt aggregation on the dried substrates.
Raman Mapping Measurement
SERS measurements were performed under the 632.8 nm excitation line of a He–Ne laser on an in Via Raman microscope (Renishaw, UK) in a standard backscattering configuration. The Raman signal was collected through a confocal pinhole of 25 μm diameter by using a 0.75 NA dry objective of 50× magnification (Leica Microsystems, Germany). A computer-controlled XY translation stage was used to acquire a total of 3333 SERS spectra within the scan area of 101 μm × 33 μm divided into an acquisition grid by 1 μm step size in both dimensions. All spectra in this work were obtained with an exposure time of 1 s and at 0.3 mW laser power before the objective. The SERS intensity maps of each major TAMRA vibration mode (1370 and 1510 cm−1) were derived using the signal to baseline map generation algorithm of WiRE 3.2 software (Renishaw, UK) for spectral ranges of 1350–1390 and 1490–1530 cm−1, respectively.
Signal Combination
The two-sample Kolmogorov–Smirnov (KS) test was used to examine if the independent signal readings of the diagnostic frequencies (1370 and 1510 cm−1) should be combined to improve sensor sensitivity. Each mapping measurement consists of 3333 spectra, and for each spectrum, the SERS signal intensity was extracted at the two aforementioned frequencies. These intensity readings were considered to be random variables. So, from each measurement, we had 3333 realizations of a random variable. These random variables followed some distribution, and the KS test determined if they are from identical distributions. Formally, the hypothesis test is H0: F(X) = G(X) and H1: F(X) ≠ G(X), where F(X) is the empirical distribution from one measurement and G(X) is the empirical distribution from another measurement. The two-sample two-dimensional Kolmogorov–Smirnov test had a similar hypothesis, but in this case, the random variable X was two-dimensional. The calculated p values are listed in Supporting Information (Table S-VI).
Determination of Raman Mapping Area
The two-sample Kolmogorov–Smirnov (KS) was utilized to determine the minimum substrate area required to unambiguously distinguish between treatment and control samples. SERS intensity measurements were considered as independent measurements from both treatment and control samples. Based on these measurements, two sets of pixels (corresponding to treatment and control) were sampled randomly at a time, starting from 1 pixel up to 300 pixels with 1 pixel increments. According to this sampling algorithm, we calculated the p values of a two-sample KS test for different sizes of substrates. This sampling was repeated 1000 times. Finally, the mean p value and the 90% highest density interval were plotted. This essentially means that the top and bottom 5% of p values were not displayed.
Supplementary Material
Acknowledgments
We gratefully acknowledge financial support from the National Science Foundation (Award No. CBET-0854030) and the National Institutes of Health (Award Nos. RR025816-02 and CA147925-01). The authors thank J.J. Russo for helpful discussions and reviewing the manuscript. We also thank M.N. Stojanovic, J.A. Oliver, and D.W. Landry for inspiring us to test with vasopressin.
Footnotes
Conflict of Interest: The authors declare no competing financial interest.
Supporting Information Available: Theoretical derivations to correlate distributions of SERS enhancement factor and intensity; experimental procedures for manual instrumentation and data analysis; control experiments (and figures) evaluating surface functionalization and analyte dependency; figures illustrating power fitting parameters of SERS intensity distribution as a function of analyte concentration; correlation data of SERS intensity integral versus analyte concentration; tables listing p values of the Kolmogorov–Smirnov test evaluating mapping consistency and aptamer specificity, coefficient of determination of Pareto fits, power fitting constants for the SERS intensity distributions of diagnostic TAMRA peaks, comparison of experimentally and theoretically obtained intensity integral values; and additional references. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES AND NOTES
- 1.Wang G, Park HY, Lipert RJ, Porter MD. Mixed Monolayers on Gold Nanoparticle Labels for Multiplexed Surface-Enhanced Raman Scattering Based Immunoassays. Anal Chem. 2009;81:9643–9650. doi: 10.1021/ac901711f. [DOI] [PubMed] [Google Scholar]
- 2.Kim JH, Kim JS, Choi H, Lee SM, Jun BH, Yu KN, Kuk E, Kim YK, Jeong DH, Cho MH, et al. Nanoparticle Probes with Surface Enhanced Raman Spectroscopic Tags for Cellular Cancer Targeting. Anal Chem. 2006;78:6967–6973. doi: 10.1021/ac0607663. [DOI] [PubMed] [Google Scholar]
- 3.Wu HY, Choi CJ, Cunningham BT. Plasmonic Nanogap-Enhanced Raman Scattering Using a Resonant Nanodome Array. Small. 2012;8:2878–2885. doi: 10.1002/smll.201200712. [DOI] [PubMed] [Google Scholar]
- 4.Lo HC, Hsiung HI, Chattopadhyay S, Han HC, Chen CF, Leu JP, Chen KH, Chen LC. Label Free Sub-Picomole Level DNA Detection with Ag Nanoparticle Decorated Au Nanotip Arrays as Surface Enhanced Raman Spectroscopy Platform. Biosens Bioelectron. 2011;26:2413–2418. doi: 10.1016/j.bios.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 5.Huh YS, Erickson D. Aptamer Based Surface Enhanced Raman Scattering Detection of Vasopressin Using Multilayer Nanotube Arrays. Biosens Bioelectron. 2010;25:1240–1243. doi: 10.1016/j.bios.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kneipp K, Wang Y, Kneipp H, Perelman LT, Itzkan I, Dasari RR, Feld MS. Single Molecule Detection Using Surface-Enhanced Raman Scattering (SERS) Phys Rev Lett. 1997;78:1667–1670. [Google Scholar]
- 7.Le Ru EC, Blackie E, Meyer M, Etchegoin PG. Surface Enhanced Raman Scattering Enhancement Factors: A Comprehensive Study. J Phys Chem C. 2007;111:13794–13803. [Google Scholar]
- 8.Cao YC, Jin R, Mirkin CA. Nanoparticles with Raman Spectroscopic Fingerprints for DNA and RNA Detection. Science. 2002;297:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 9.Nie S, Emory SR. Probing Single Molecules and Single Nanoparticles by Surface-Enhanced Raman Scattering. Science. 1997;275:1102–1106. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- 10.Kim NH, Lee SJ, Moskovits M. Aptamer-Mediated Surface-Enhanced Raman Spectroscopy Intensity Amplification. Nano Lett. 2010;10:4181–4185. doi: 10.1021/nl102495j. [DOI] [PubMed] [Google Scholar]
- 11.Chen G, Wang Y, Yang M, Xu J, Goh SJ, Pan M, Chen H. Measuring Ensemble-Averaged Surface-Enhanced Raman Scattering in the Hotspots of Colloidal Nanoparticle Dimers and Trimers. J Am Chem Soc. 2010;132:3644–3645. doi: 10.1021/ja9090885. [DOI] [PubMed] [Google Scholar]
- 12.Faulds K, Smith WE, Graham D. Evaluation of Surface-Enhanced Resonance Raman Scattering for Quantitative DNA Analysis. Anal Chem. 2004;76:412–417. doi: 10.1021/ac035060c. [DOI] [PubMed] [Google Scholar]
- 13.Ochsenkuhn MA, Campbell CJ. Probing Biomolecular Interactions Using Surface Enhanced Raman Spectroscopy: Label-Free Protein Detection Using a G-Quadruplex DNA Aptamer. Chem Commun. 2010;46:2799–2801. doi: 10.1039/b920941g. [DOI] [PubMed] [Google Scholar]
- 14.Lim DK, Jeon KS, Hwang JH, Kim H, Kwon S, Suh YD, Nam JM. Highly Uniform and Reproducible Surface-Enhanced Raman Scattering from DNA-Tailorable Nanoparticles with 1-nm Interior Gap. Nat Nanotechnol. 2011;6:452–460. doi: 10.1038/nnano.2011.79. [DOI] [PubMed] [Google Scholar]
- 15.Graham D, Thompson DG, Smith WE, Faulds K. Control of Enhanced Raman Scattering Using a DNA-Based Assembly Process of Dye-Coded Nanoparticles. Nat Nanotechnol. 2008;3:548–551. doi: 10.1038/nnano.2008.189. [DOI] [PubMed] [Google Scholar]
- 16.Papadopoulou E, Bell SEJ. Label-Free Detection of Single-Base Mismatches in DNA by Surface-Enhanced Raman Spectroscopy. Angew Chem, Int Ed. 2011;50:9058–9061. doi: 10.1002/anie.201102776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang T, Yoo SM, Yoon I, Lee SY, Kim B. Patterned Multiplex Pathogen DNA Detection by Au Particle-on-Wire SERS Sensor. Nano Lett. 2010;10:1189–1193. doi: 10.1021/nl1000086. [DOI] [PubMed] [Google Scholar]
- 18.Im H, Bantz KC, Lindquist NC, Haynes CL, Oh SH. Vertically Oriented Sub-10-nm Plasmonic Nanogap Arrays. Nano Lett. 2010;10:2231–2236. doi: 10.1021/nl1012085. [DOI] [PubMed] [Google Scholar]
- 19.Li W, Camargo PHC, Au L, Zhang Q, Rycenga M, Xia Y. Etching and Dimerization: A Simple and Versatile Route to Dimers of Silver Nanospheres with a Range of Sizes. Angew Chem, Int Ed. 2010;49:164–168. doi: 10.1002/anie.200905245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laurence TA, Braun G, Talley C, Schwartzberg A, Moskovits M, Reich N, Huser T. Rapid, Solution-Based Characterization of Optimized SERS Nanoparticle Substrates. J Am Chem Soc. 2008;131:162–169. doi: 10.1021/ja806236k. [DOI] [PubMed] [Google Scholar]
- 21.Le Ru EC, Meyer M, Etchegoin PG. Proof of Single-Molecule Sensitivity in Surface Enhanced Raman Scattering (SERS) by Means of a Two-Analyte Technique. J Phys Chem B. 2006;110:1944–1948. doi: 10.1021/jp054732v. [DOI] [PubMed] [Google Scholar]
- 22.Etchegoin PG, Meyer M, Blackie E, Le Ru EC. Statistics of Single-Molecule Surface Enhanced Raman Scattering Signals: Fluctuation Analysis with Multiple Analyte Techniques. Anal Chem. 2007;79:8411–8415. doi: 10.1021/ac071231s. [DOI] [PubMed] [Google Scholar]
- 23.Morales D, Madigan J, Cullinane S, Chen J, Heath M, Oz M, Oliver JA, Landry DW. Reversal by Vasopressin of Intractable Hypotension in the Late Phase of Hemorrhagic Shock. Circulation. 1999;100:226–229. doi: 10.1161/01.cir.100.3.226. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt MS, Hubner J, Boisen A. Large Area Fabrication of Leaning Silicon Nanopillars for Surface Enhanced Raman Spectroscopy. Adv Mater. 2012;24:OP11–OP18. doi: 10.1002/adma.201103496. [DOI] [PubMed] [Google Scholar]
- 25.Purschke WG, Eulberg D, Buchner K, Vonhoff S, Klussmann S. An L-RNA-Based Aquaretic Agent That Inhibits Vasopressinin Vivo. Proc Natl Acad Sci US A. 2006;103:5173–5178. doi: 10.1073/pnas.0509663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim A, Ou FS, Ohlberg DAA, Hu M, Williams RS, Li Z. Study of Molecular Trapping inside Gold Nanofinger Arrays on Surface-Enhanced Raman Substrates. J Am Chem Soc. 2011;133:8234–8239. doi: 10.1021/ja200247x. [DOI] [PubMed] [Google Scholar]
- 27.Moskovits M. Imaging Spot the Hotspot. Nature. 2011;469:307–308. doi: 10.1038/469307a. [DOI] [PubMed] [Google Scholar]
- 28.Cang H, Labno A, Lu CG, Yin XB, Liu M, Gladden C, Liu YM, Zhang X. Probing the Electromagnetic Field of a 15-Nanometre Hotspot by Single Molecule Imaging. Nature. 2011;469:385–388. doi: 10.1038/nature09698. [DOI] [PubMed] [Google Scholar]
- 29.Chen T, Wang H, Chen G, Wang Y, Feng YH, Teo WS, Wu T, Chen HY. Hotspot-Induced Transformation of Surface-Enhanced Raman Scattering Fingerprints. ACS Nano. 2010;4:3087–3094. doi: 10.1021/nn100269v. [DOI] [PubMed] [Google Scholar]
- 30.Le Ru EC, Etchegoin PG, Meyer M. Enhancement Factor Distribution around a Single Surface-Enhanced Raman Scattering Hot Spot and Its Relation to Single Molecule Detection. J Chem Phys. 2006;125:204701. doi: 10.1063/1.2390694. [DOI] [PubMed] [Google Scholar]
- 31.Sarkar J, Chowdhury J, Pal P, Talapatra GB. Ab Initio, DFT Vibrational Calculations and SERRS Study of Rhodamine 123 Adsorbed on Colloidal Silver Particles. Vib Spectrosc. 2006;41:90–96. [Google Scholar]
- 32.Lowe AJ, Huh YS, Strickland AD, Erickson D, Batt CA. Multiplex Single Nucleotide Polymorphism Genotyping Utilizing Ligase Detection Reaction Coupled Surface Enhanced Raman Spectroscopy. Anal Chem. 2010;82:5810–5814. doi: 10.1021/ac100921b. [DOI] [PubMed] [Google Scholar]
- 33.Stewart S, Fredericks PM. Surface-Enhanced Raman Spectroscopy of Amino Acids Adsorbed on an Electrochemically Prepared Silver Surface. Spectrochim Acta, Part A. 1999;55:1641–1660. [Google Scholar]
- 34.Herne TM, Tarlov MJ. Characterization of DNA Probes Immobilized on Gold Surfaces. J Am Chem Soc. 1997;119:8916–8920. [Google Scholar]
- 35.Tang L, Zeng G, Shen G, Li Y, Liu C, Li Z, Luo J, Fan C, Yang C. Sensitive Detection of Lip Genes by Electrochemical DNA Sensor and Its Application in Polymerase Chain Reaction Amplicons from Phanerochaete Chrysosporium. Biosens Bioelectron. 2009;24:1474–1479. doi: 10.1016/j.bios.2008.09.031. [DOI] [PubMed] [Google Scholar]
- 36.Huh YS, Chung AJ, Cordovez B, Erickson D. Enhanced On-Chip SERS Based Biomolecular Detection Using Electrokinetically Active Microwells. Lab Chip. 2009;9:433–439. doi: 10.1039/b809702j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newman MEJ. Power Laws, Pareto Distributions and Zipf’s Law. Contemp Phys. 2005;46:323–351. [Google Scholar]
- 38.Fang Y, Seong NH, Dlott DD. Measurement of the Distribution of Site Enhancements in Surface-Enhanced Raman Scattering. Science. 2008;321:388–392. doi: 10.1126/science.1159499. [DOI] [PubMed] [Google Scholar]
- 39.Smirnov N. Table for Estimating the Goodness of Fit of Empirical Distributions. Ann Math Stat. 1948;19:279–281. [Google Scholar]
- 40.Kolmogorov AN. Sulla Determinazione Empirica Di Una Legge Di Distribuzione. G Ist Ital Attuari. 1933;4:83–91. [Google Scholar]
- 41.Peacock JA. Two-Dimensional Goodness-of-Fit Testing in Astronomy. Mon Not R Astron Soc. 1983;202:615–627. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.