Abstract
Protein polymers can assemble switchable nanostructures with emerging applications as biomaterials and nanomedicines. For example, above a critical micelle temperature (CMT) some elastin-like polypeptide (ELP) diblock copolymers assemble spherical nanoparticles, which may modulate cellular internalization and in vivo biodistribution. To achieve engineering-level control over their properties, this report explores a comprehensive library of ELP monoblock and diblock polymers. For the first time, we report that a surprisingly high core molecular weight is required for stable nanoparticle formation; furthermore, nanoparticle size depends on polymer molecular weight. A mathematical model was developed to characterize four ELP monoblock libraries and to predict the phase behavior of corresponding diblock copolymers. The CMT was almost entirely dependent on the hydrophobic core ELP, while the bulk phase transition temperature (Tt,bulk) depends predominantly on the hydrophilic block. Nanoparticle assembly was accompanied by a conversion in secondary structure of the hydrophobic block from random coil and beta-sheets to type-2 β turns. For the first time, this report enables the rational design of ELP protein polymer nanoparticles with physico-chemico properties that will be suitable for biological applications.
Keywords: Elastin-like polypeptide, block copolymer, self-assembly, micelles
Introduction
Protein polymers are repetitive amino acid sequences that combine the versatile properties of polymers with the ability to be genetically engineered1. As such, they are emerging as a platform for the assembly and controlled delivery of therapeutic peptides, proteins, and small molecules2. Protein polymers derived from silk3,4,5, collagen6 and elastin7,8,9 each have unique stimulus-responsive properties10, 11 and present opportunities to control assembly and biodegradation12. Recently, it was reported that protein polymers designed as block copolymers can assemble monodisperse nanostructures13-16. These nanostructures have applications in drug delivery, tissue engineering and biosensors17,18-20. To this end, recombinant DNA technology has been utilized to synthesize various protein-based block copolymers that include silk-like3,21, resilin-like22 and elastin-like polypeptides23,24,16, coiled-coil25 and leucine zipper domains26, and various peptide amphiphiles27. In comparison to synthetic polymer chemistry, genetic engineering offers exquisite control of the composition, length, and molecular weight of the block copolymer. Moreover, repetitive protein polymer genes can be constructed with high fidelity using concatamerization28,29,30, step-by-step directional approaches (seamless cloning)31 and recursive directional ligation32,33.
Elastin like polypeptides (ELPs) are protein polymers inspired from a structural motif found in human tropoelastin34. They are composed of a five amino acid repeat (Val-Pro-Gly-Xaa-Gly)l, where the identities of Xaa and l determine their phase behavior. ELPs are attractive as polymeric carriers for therapeutics because they undergo an inverse phase transition. Below a characteristic transition temperature (Tt), ELPs are highly water-soluble. When the temperature is raised above Tt, they undergo a sharp (∼1 °C) phase separation, which is known as coacervation34. Unlike many peptide aggregation phenomena, this process is fully reversible. Block copolymers with elastin-like sequences composed of blocks with different hydrophobicities have emerged as biomaterials for tissue engineering and delivery of therapeutics12, 13, 35. Hybrid block copolymers of ELPs as well as other protein polymers have also been reported including silk-elastin-like polypeptides36,37, ODN-ELP38, and cartilage-oligomeric matrix protein39. While promising, this strategy is currently limited due to a scarcity of studies designed to rationally predict the properties of novel block copolymers with the properties of the ELP monoblock.
To facilitate the rational engineering of the chemico-physico properties of ELP nanoparticles, our group has synthesized a library of ELP block copolymers of different molecular weights (MW), orientations, and amino acid sequences. Several groups have previously characterized ELP-based block copolymers7 and ELP fusion proteins40, 41 but to date, no comprehensive studies have been made that connect the behavior of ELP monoblocks with related ELP nanoparticles. This deficiency has presented challenges to their development as therapeutics. To facilitate the engineering of ELP block copolymers with targeted assembly temperatures, a quantitative mathematical model was developed to relate the CMT and Tt,bulk with the respective phase behavior of the hydrophobic and hydrophilic ELP monoblocks. For the purpose of this manuscript, the critical micelle temperature (CMT) is defined as the temperature at which micelle formation occurs. At a temperature higher than the CMT, the construct undergoes hydrophobic collapse, which is defined as the bulk transition temperature (Tt,bulk). Based on this model and dataset, this manuscript documents multiple important findings including: i) assembly of stable ELP nanoparticles requires a minimum ELP molecular weight; ii) assembly of ELP nanoparticles is accompanied by a change in secondary structure to favor type-2 β turns along the polypeptide backbone; and iii) the bulk phase transition temperature (Tt,bulk) of ELP nanoparticles can be controlled by changing the amino acid sequence of the hydrophilic block without significantly influencing the critical micelle temperature (CMT).
Materials and methods
Materials
Restriction enzymes and calf intestinal phosphatase (CIP) were purchased from New England Biolabs (Ipswich, MA). T4 DNA ligase was purchased from Invitrogen (Carlsbad, CA). The pET-25b+ cloning vector was obtained from Novagen Inc. (Madison, WI), and all custom oligonucleotides were synthesized by Integrated DNA Technologies Inc. (Coralville, IA). Top10™ cells were purchased from Invitrogen (Carlsbad, CA) and BLR(DE3) competent cells were purchased from Novagen (Madison, WI). All E. coli cultures were grown in TB Dry™ media purchased from MO BIO Laboratories, Inc (Carlsbad, CA). The DNA miniprep kit and the Illustra GFX Gel Band Purification Kit were purchased from Qiagen Inc. (Germantown, MD) and GE (GE Healthcare, Buckinghamshire, UK) respectively.
Recombinant synthesis of ELP and ELP diblock copolymers by plasmid reconstruction recursive directional ligation (pre-RDL)
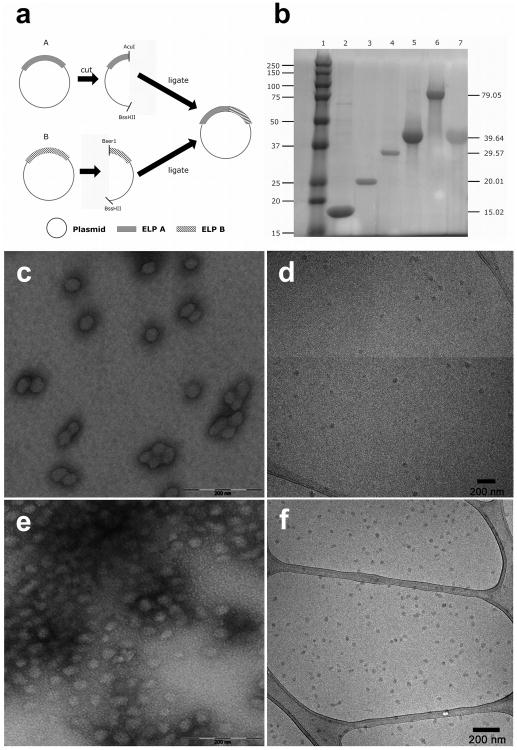
To generate ELP and ELP block copolymers of specific and pre-determined chain length the following RDL strategy was employed. Two cloning vectors containing the ELP gene were cut with two separate sets of restriction enzymes (Fig. 1a). One vector was digested using BssHII and AcuI, while BssHII and BserI cut the second vector, generating compatible sticky ends. Enzyme digestion was performed using 1μL of enzyme, each at 37°C for 3h. The two sets of cut vectors were ligated together using the T4 DNA ligase (Invitrogen, Carlsbad, CA), resulting in the extension of the pentameric repeats. Similarly for the generation of block copolymers, the N-terminal gene of one monoblock was ligated to a C-terminal ELP gene of another via pre-RDL.
Figure 1.
Biosynthesis of ELP block copolymers that assemble nanoparticles. a) ELP genes were designed using a plasmid-reconstruction recursive directional ligation method[33]. Double endonuclease digestion was used to cut plasmids at the BseR1/BssHII or AcuI/BssHII. This generates two base pair sticky ends, which enables recursive joining of ELP genes. b) Copper-staining was used to enhance an SDS PAGE gel and show a representative library of purified ELPs. The left lane contains a MW standard, which has a mass indicated to the left. Lane 1: MW marker, lane 2: I18S18, lane 3: I24S24, lane 4: I36S36, lane 5: I48S48, lane 6: I96S96 and lane 7: S96. The right column indicates the expected mass of each ELP, which was independently confirmed using MALDI (Table 1). c) Transmission electron microscopy (TEM) of I48S48 nanoparticles (stained 2% uranyl acetate), which have a particle diameter of 40 ± 5 nm. d) Cryo-TEM of I48S48 micelles, which have diameters of 29 ± 4 nm. e) TEM of I96S96 micelles (stained 2% uranyl acetate), which have diameters of 48 ± 4 nm. f) Cryo-TEM of I96S96 micelles, which have diameters of 33 ± 2 nm.
Protein purification by inverse transition cycling
pET25b(+) expression vectors containing the desired constructs were transformed into E. coli BLR (DE3) cells for protein hyperexpression and proteins were purified by inverse transition cycling42. Briefly, the overnight cultures were spun down and re-suspended in cold PBS. The proteins were liberated from inclusion bodies by sonicating the suspension for a total of 3 minutes. The sonicated product was then spun down for 15min at 4°C, 12000rpm and the supernatant transferred to another tube. PEI was added to remove any remaining nucleic acids and after 15 minutes the solution was centrifuged again. The supernatant, containing soluble ELP, was heated to 37°C and, once the solution became turbid, centrifuged at 37°C to precipitate aggregated, insoluble ELP. The pellet was then re-suspended in cold PBS and centrifuged at 4°C again. Multiple rounds of ITC were performed to ensure the overall purity of the proteins obtained (Fig. 1b). The yields observed for these ELPs ranged from 80 to 100 mg of purified ELP per liter of bacterial culture.
Transition temperature characterization of protein polymers
The transition temperature of each member of the ELP libraries was obtained by measuring solution turbidity as a function of temperature. Solutions of the polypeptide in phosphate buffered saline (PBS) were analyzed at a constant rate of 1 °C/min in a temperature controlled multicell holder of a UV visible spectrophotometer (DU800 Spectrophotometer, Beckman Coulter, CA, USA). The transition temperature (Tt) is defined at the maximum first derivative of the OD with respect to temperatures34. For diblock copolymers the critical micelle temperature (CMT) is defined as the temperature at which nanoparticles start to form as indicated where the OD first increases from baseline, and the second Tt,bulk at the maximum first derivative of OD with respect to temperature.
Circular dichroism
CD measurements were made on a Jasco CD spectrometer with a 0.1cm path length quartz cell in a 190-240 nm wavelength range. An ELP solution (25-200 μM) in deionized water was used for measurements. Data was processed using Jasco software that allowed for simultaneous processing of the recorded spectra. Each spectrum was corrected by subtracting the corresponding background spectrum recorded at the same temperature. The resulting spectra was smoothed out, and afterward converted into mean molar residue ellipticity (MRE) in mdeg cm2 dmol-1. All plots were exported using Jasco Spectral manager V2 (Easton, MD) to Excel and deconvoluted assuming that the molar ellipticity [θ] observed is a weighted linear sum of secondary structure ellipticity. The data were fit to spectra of ideal secondary structures using nonlinear regression on Microsoft Excel.
Light scattering characterization
Determination of the hydrodynamic radius of the nanoparticles was performed on a Dynapro plate reader (Wyatt Technology Inc., Santa Barbara, CA, USA). 10-25 μM of polypeptide in phosphate buffered saline (PBS) pH 7.4 was subjected to a temperature ramp between 10°C – 60 °C in 1 °C increments. Solutions were filtered through Whatman filters with a 0.02μM pore size and centrifuged at 4 °C, 1200 rpm for 10 min to remove air bubbles. Mineral oil was applied to prevent evaporation and the preparation was centrifuged again before running the samples.
Transmission electron microscopy (TEM) sample preparation
TEM samples were prepared by pipetting a small drop of heated ELP solution (37 °C) onto a carbon-coated copper grid (Electron Microscopy Sciences, Hatfield, PA). The excess solution was quickly wicked away with a small piece of filter paper. Next, approximately 10 μL of a 2 wt % aqueous uranyl acetate solution was deposited and the excess solution was carefully removed as above to leave a very thin layer. The sample grid was then allowed to dry at 37 °C prior to imaging. Bright-field TEM imaging was performed on a FEI Tecnai 12 TWIN Transmission Electron Microscope operated at an acceleration voltage of 100 kV. All TEM images were recorded by a SIS Megaview III wide-angle CCD camera or 16 bit 2K × 2K FEI Eagle bottom mount camera.
Cryogenic-transmission electron microscopy (Cryo-TEM)
Cryogenic TEM imaging was also performed on the FEI Tecnai 12 TWIN Transmission Electron Microscope, operating at 80 kV. Approximately 6μL of pre-cooled ELP solution was loaded on a lacey carbon coated TEM grid (LC325-Cu, Electron Microscopy Sciences). All the TEM grids used for cryo-TEM imaging were treated with plasma air to render the lacey carbon film hydrophilic. A thin film of the sample solution was produced using the FEI Vitrobot with a controlled humidity chamber (typically under 95% humidity). After loading of the sample solution, the lacey carbon grid was blotted using preset parameters and plunged instantly into a liquid ethane reservoir precooled by liquid nitrogen. The vitrified samples were then transferred to a Gatan 626 cryo-holder and cryo-transfer stage that was cooled by liquid nitrogen. To prevent sublimation of vitreous water and formation of other types of water crystals that may complicate the imaging process, the cryo-holder temperature was maintained below -170°C throughout the imaging process. All images were recorded at a low dose using a 16 bit 2K × 2K FEI Eagle bottom mount camera.
Results
Generation of ELP diblock copolymers that assemble nanoparticles
To facilitate the deliberate engineering of ELP nanoparticles for specific therapeutic applications, it was first necessary to collect a comprehensive dataset of ELP monoblock and diblock copolymers. Depending on the application, it may be necessary to generate ELPs that assemble below, at, or above body temperature. The molecular weight of the ELP polymers may need to be adjusted to exceed or fall below the renal filtration cutoff. The hydrodynamic radius of resulting particles may need to be adjusted to control the route of physiological clearance43. To control these fundamental variables, this manuscript explores the relationship between the behavior of monoblock and diblock ELPs (Table 1). To accomplish this, our group genetically engineered four new gene libraries of ELPs (supplementary Table s1). Libraries with Xaa= Ser, Ala, Val, and Ile were constructed of various lengths, l, using a modification of the plasmid reconstruction-recursive directional ligation approach (pre-RDL)33. In pre-RDL, two cloning vectors that contain the desired oligomers are cut with 2 separate sets of restriction enzymes (Fig. 1a). When the two fragments of digested plasmid are ligated together, the resulting oligomeric sequence is extended and reconstituted on an intact plasmid. Each of the gene products resulting from pre-RDL was explored for expression and purification by phase separation, a technique known as inverse phase transition cycling44. ELPs long enough to be expressed and purified were characterized (supplementary Table s1). Selections from this library were then used to generate a library of ELP diblock copolymers (Table 1). Purification of the ELPs by inverse phase transition cycling yielded proteins of high purity at their expected molecular weight (>95%) (Fig. 1b). The identity of each diblock copolymer was confirmed independently using MALDI TOF (Table 1).
Table 1. Libraries of ELP diblock copolymers characterized.
| ELP diblock libraries [Xaa:Yaa] | ELP Nomenclature | *Amino acid sequence | **Calculated ELP MW (Da) | ***Observed ELP MW (Da) |
|---|---|---|---|---|
| Ile:Ser | I18S18 | G(VPGIG)18(VPGSG)18Y | 15,015 | 14,599 |
| I24S24 | G(VPGIG)24(VPGSG)24Y | 20,012 | 19,820 | |
| I36S36 | G(VPGIG)36(VPGSG)36Y | 29,572 | 29,826 | |
| I48S48 | G(VPGIG)48(VPGSG)48Y | 39,643 | 39,749 | |
| I96S96 | G(VPGIG)96(VPGSG)96Y | 79,049 | 79,216 | |
| Ser:Ile | S48I48 | G(VPGSG)48(VPGIG)48Y | 39,643 | 39,670 |
| Ile:Ala | I48A48 | G(VPGIG)48(VPGAG)48Y | 38,946 | 38,940 |
| I96A96 | G(VPGIG)96(VPGAG)96Y | 77,655 | 77,233 | |
| Ala:Ile | A48I48 | G(VPGAG)48(VPGIG)48Y | 38,946 | 38,952 |
| A96I96 | G(VPGAG)96(VPGIG)96Y | 77,655 | 77,840 |
Gene sequence confirmed by N and C terminal DNA sequencing and diagnostic digestion.
Estimated from open reading frame excluding methionine start codon
Results from matrix assisted laser desorption ion time of flight mass spectrometry
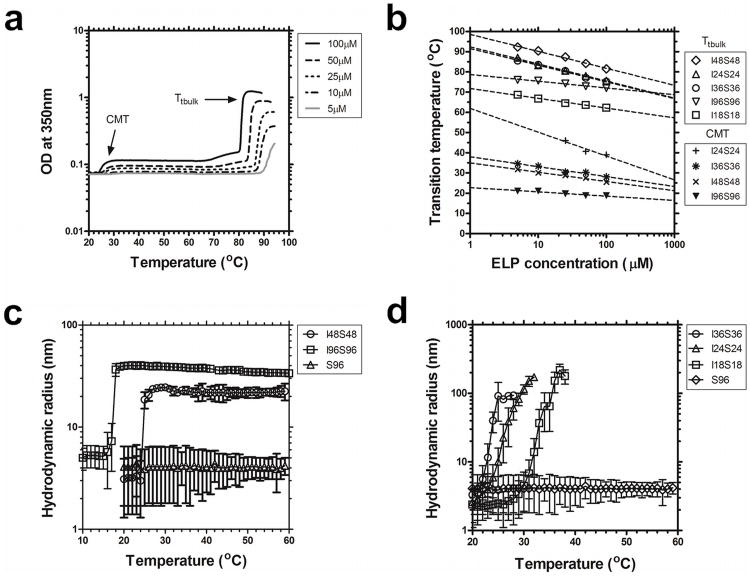
The phase diagram of each member of the monoblock library was studied by measuring optical density as a function of temperature and concentration. The temperature at which optical density changed the fastest was defined as the ELP transition temperature, Tt. ELP phase diagrams are strong functions of the hydrophobicity of Xaa and length (or MW) (Fig 2). As reported by others, ELPs followed a log-linear relationship between Tt and concentration. For all ELPs, except where Xaa=Ser, there was also a strong dependence on the polymer length. Both of the hydrophilic libraries (Xaa=Ser, Ala) phase separate above physiological temperatures, thus they were selected as the hydrophilic component of subsequent diblock copolymers. The hydrophobic library with Xaa=Val, had a Tt near physiological temperature, which may lead to unstable assembly during dilution in the body (Supplementary Fig. s1c). At constant length and concentration, the most hydrophobic ELP monoblock Xaa=Ile had the lowest Tt, close to room temperature (Supplementary Fig. s1d). To identify nanoparticles that are stable under physiological conditions, hydrophobic blocks with Xaa=Ile were selected as the core for diblock copolymer nanoparticles. This strategy was intended to generate diblock copolymers that can easily disassemble under refrigeration and assemble at physiological temperatures.
Figure 2.
Particle diameter and micelle stability depend on molecular weight. ELP block copolymers were characterized for assembly using optical density and dynamic light scattering in phosphate buffered saline. a) Optical density (350 nm) for I48S48 as a function of temperature and concentration. b) Concentration-temperature phase diagrams for ELP diblock copolymers I18S18, I24S24, I36S36, I48S48, I96S96. Detectable critical micelle temperatures (CMT) and bulk inverse phase transition temperatures (Tt,bulk) are indicated. c) Longer ELP block copolymers (I48S48, I96S96) form nanoparticles of stable hydrodynamic radius at 25 μM. A monoblock ELP, S96, does not assemble and is included as a negative control. d) Shorter ELP diblock copolymers (I18S18, I24S24, I36S36) form larger nanostructures above their CMT with unstable hydrodynamic radii (25 μM).
From ELP monoblocks, a library of diblock copolymers was constructed where a gene encoding a hydrophilic ELP with high Tt (Yaa=Ser, Ala) was ligated to a gene encoding a hydrophobic ELP with a lower Tt (Xaa=Ile) (Table 1). A 1:1 ratio of hydrophobic to hydrophilic lengths was held constant to maximize the formation of stable spherical nanoparticles. By probing this library with dynamic light scattering (DLS), it was possible to identify multiple nanostructures with stability at physiologically relevant temperatures and concentrations (supplementary Fig. s2). The homogeneity and diameter of nanoparticles formed by these diblock copolymers (I48S48, I96S96) are consistent with those observed using transmission electron microscopy (Fig. 1c,e) and cryo-TEM (Fig. 1d,f).
A minimum molecular weight is necessary for nanoparticle assembly
As with ELP monoblocks, the phase diagrams of diblock copolymers were characterized by monitoring the optical density as a function of temperature, concentration, and molecular weight. Upon heating, a two-step response was observed (Fig. 2a). At low temperatures (< 24 to 32 °C), the ELP block copolymers remain in solution and maintain transparency. As the temperature rises (26 to 66 °C), the optical density increases and persists at a constant level. The lower temperature increase in optical density correlates with the phase separation of the more hydrophobic block (Xaa=Ile) and was defined as the critical micelle temperature (CMT) (Fig. 2b). Upon further heating, the hydrophilic block (Yaa=Ser) underwent phase separation, which induced the formation of larger nanostructures. This higher temperature was defined as the Tt,bulk. Both the CMT and Tt,bulk exhibited a negative log-linear relationship with concentration (Fig. 2b). The resulting phase behavior for these ELPs are consistent with the assembly of nanoparticles with a hydrophobic core (Xaa=Ile) that is stabilized by the more hydrophilic shell (Yaa=Ser).
Unexpectedly, the shortest diblock copolymer, I18S18 exhibited only a single bulk phase transition temperature by optical density (Fig. 2b). To determine if this and other diblock copolymers assembled stable nanostructures, DLS was used to verify the CMT, hydrodynamic radius, polydispersity, and thermal stability of nanoparticles. Based on diblock copolymers containing Xaa=Ile and Yaa=Ser, we identified a minimal molecular weight necessary for the assembly of stable ELP-based micelles. I48S48 and I96S96 formed large stable nanoparticles above a CMT (25, 20°C respectively), with a hydrodynamic radius proportional to their MW (Rh= ∼20, ∼40 nm respectively). Formation of these stable nanoparticles persists up to temperatures of 60 °C (Fig. 2c) and for at least two days at 37 °C (supplementary Fig s1). As a control, the hydrodynamic radius of an ELP monoblock (S96) shows no assembly. Below a minimum length (l = 48 pentamers) of the hydrophobic block, the diblock copolymers (I18S18, I24S24, I36S36) formed larger intermediate nanostructures that were unstable over the period of DLS measurement (Fig. 2d). Thus, based on a combination of optical density and DLS measurements, we determined that for hydrophobic ELPs with Xaa=Ile, at least 18 pentamers are necessary to form unstable nanoparticles and 48 pentamers are required to form stable nanoparticles.
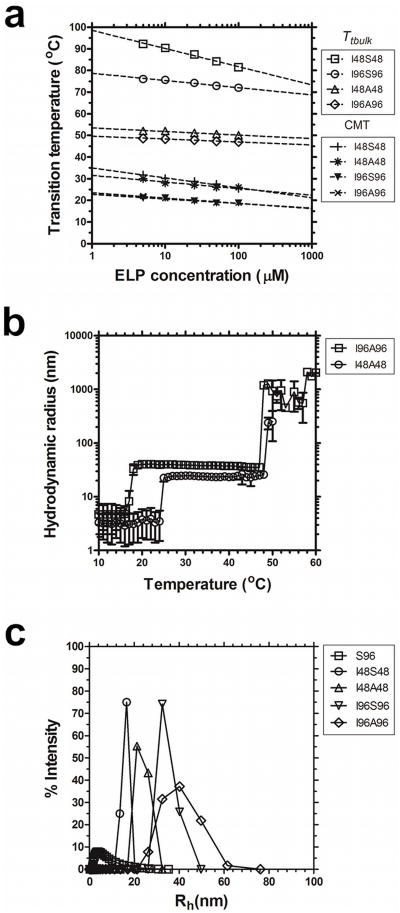
Selection of hydrophilic block determines the bulk phase transition temperature
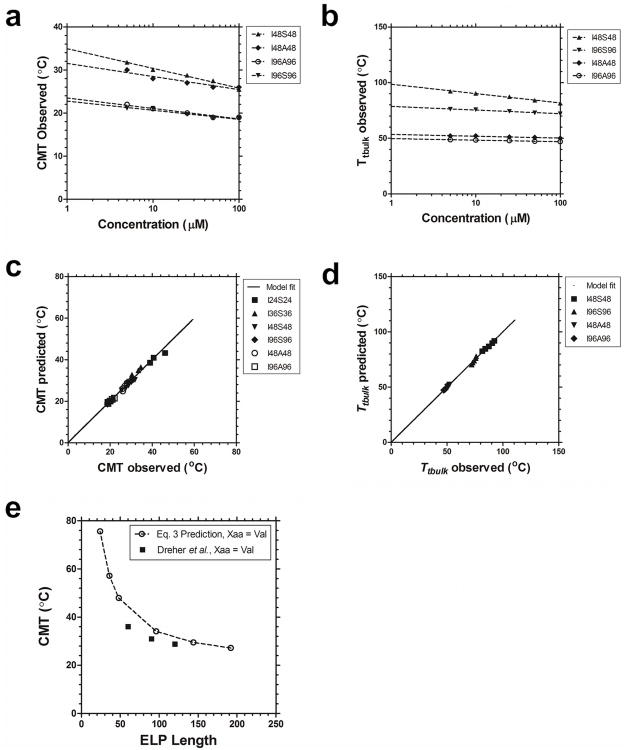
Having determined that a minimum molecular weight is required to assemble the hydrophobic core for stable nanoparticles, we further probed how changes in the hydrophilic block would influence the bulk phase separation of ELP nanoparticles. To test this, a library composed of Yaa=Ala as the hydrophilic block was constructed and characterized (Table 1, Fig. 3). When compared to the thermal behavior of the more hydrophilic Yaa=Ser library, a nearly identical profile was obtained for the onset of micelle formation (CMT) (Fig. 3a). However the less hydrophilic Yaa=Ala nanoparticles have a significantly lower Tt,bulk compared to the Yaa=Ser nanoparticles (Fig. 3a). This is consistent with the observation that serine monoblock ELPs have a higher Tt than alanine ELPs (Fig 2). Comparable CMT and hydrodynamic radii (Rh) were observed with the Yaa=Ala blocks (Fig. 3b). In addition, DLS confirmed that the Yaa=Ala nanoparticles undergo measurable bulk phase separation slightly above physiological temperatures (Fig. 3b). Further investigation of the particle size distribution at 37 °C revealed differences between the two types of block copolymers (Fig. 3c). The Yaa=Ala blocks is slightly larger and form slightly more stable structures (supplementary Fig. s2). This stability issue may have implications for in vivo applications, as it may be favorable to use a construct with increased stability at physiologically relevant temperatures.
Figure 3.
Bulk phase transition temperature for stable ELP nanoparticles depends on the hydrophilic block. ELP block copolymers with two different hydrophilic blocks (Xaa = Ser, Ala) and two lengths (l = 48, 96) were characterized for their assembly properties using optical density and DLS in phosphate buffered saline. a) Concentration-temperature phase diagrams for ELP diblock copolymers I48S48, I48A48, I96S96, I96A96. The critical micelle temperature (CMT) was minimally influenced by the sequence of the more hydrophilic block. In contrast, the bulk phase transition temperature (Tt,bulk) was highly dependent on the sequence of the hydrophilic block. b) Above their CMT, ELP diblock copolymers with Xaa = Ala (I48A48, I96A96) assemble micelles that are stable at physiological temperatures and undergo bulk phase separation ∼10 °C above body temperature (25 μM). c) Higher molecular weight ELP diblock copolymers have a larger hydrodynamic radius than smaller ELPs. The distribution of hydrodynamic radii for a soluble monoblock ELP, S96, is indicated as a negative control.
As diblocks with a hydrophobic domain at either the N or C terminus may have advantages, a library of related ELP diblocks were similarly prepared with the opposite orientation (supplementary Fig. s3). Here the Xaa=Ile blocks are reversed to give Yaa=Ile. The ability to switch the block's orientation exposes different functional groups e.g COO- or NH3+ at the coronal surface for bioconjugation. The physico-chemico properties of the reverse orientation diblocks surprisingly yielded nearly superimposable CMT values to each other and to some extent the Tt,bulk (supplementary Fig. s3a). The Rh of the reverse orientation blocks also does not differ significantly (supplementary Fig. s3b).
Nanoparticle assembly correlates with shift in secondary structure
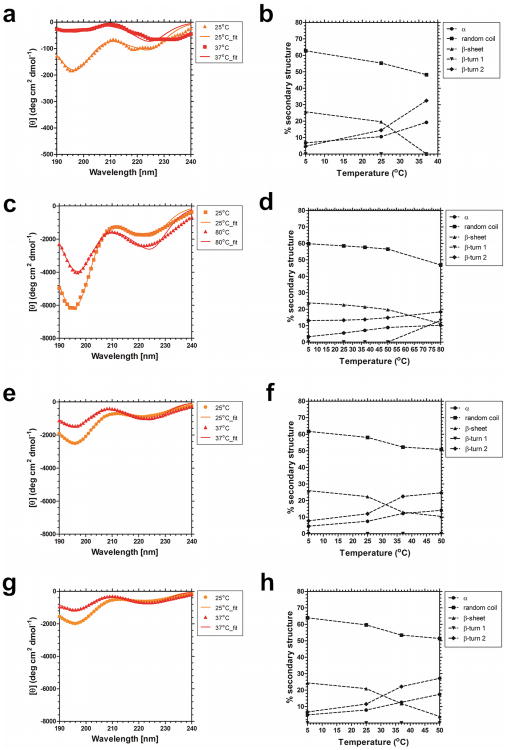
To determine if ELP-mediated nanoparticle assembly is associated with a shift in the secondary structure of the peptide backbone, far UV CD measurements were obtained. The CD spectra of S96, I48, I48S48 and I48A48 taken at various temperatures are depicted (Fig. 4). At low temperatures (< CMT), the spectra of S96, I48, I48S48 and I48A48 (Figs. 6a,c,e,g) consist of a large negative band at ∼195nm and a smaller trough at ∼220nm, which are characteristic for random coil and β-turn structures respectively. The monoblock I48 was studied (Fig. 4a) because it provides the hydrophobic core to the ELP diblock copolymers I48S48 and I48A48. The monoblock S96 was evaluated (Fig. 4c) to determine if the hydrophilic block adopts any structure over the same temperature range. With increasing temperature, the random coil signal becomes less pronounced, while the β-turn increases, indicating a transition toward a more ordered secondary structur45,46. β-turns play an important role in protein folding and stability47; furthermore, they have been previously been associated with the onset of ELP-mediated phase separation48. In a β-turn, a tight loop is formed when the carbonyl oxygen of one residue forms a hydrogen bond with the amide proton of an amino acid three residues down the chain. This hydrogen bond stabilizes the β-turn structure. This β-turn structure confers folded proteins globularity by allowing proteins to reverse direction of the peptide chain49.
Figure 4.
Nanoparticle assembly is accompanied by formation of secondary structure on the hydrophobic block. Circular dichroism was used to probe ELPs in water below, near, and above the Tt or CMT. a) The monoblock ELP I48, Tt = 31.0°C. b) Changes in percent secondary structure of ELP monoblock I48. c) The monoblock ELP S96, Tt = 55.5 °C. d) Changes in percent secondary structure of ELP monoblock S96. e) The diblock copolymer I48S48, CMT = 27.5°C. f) Changes in percent secondary structure of the ELP diblock copolymer I48S48. g) The diblock copolymer I48A48, CMT = 26°C. h) Changes in percent secondary structure of the ELP diblock copolymer I48A48.
Deconvolution, via non-linear regression, of each spectra yielded a detailed breakdown of secondary structure during nanoparticle assembly (Figs. 6b,d,f,h). The general trend shows a decrease in random coil and β-sheet content, and an increase in the β-turn 2 content. This suggests that the peptide backbone adopts a more ordered state upon phase separation. No significant change in the β-turn 1 content was observed for the polypeptides studied except for S96 at the highest temperature observed. For reference, β-turn 1 and β-turn 2 are diametrically opposed with the essential difference being the orientation of the peptide bond residues at (i+1) and (i+2).
The changes in spectra of the diblock copolymers I48S48 (Fig. 4f) and I48A48 (Fig. 4h) are of particular note because, at the examined concentration, they have CMTs that are similar to Tt of the monoblock ELP I48 (Fig. 4b). The spectra of the two diblocks are remarkably similar at the first post-assembly temperature analyzed of 37 °C (Fig. 4e, g), and the two show minimal differences in the extent of conformation change. Between 37 and 50 °C the random coil structure of I48S48 (Fig. 4f) and I48A48 (Fig. 4h) remains similar, with a decrease from 53.4% content to 51.4% in I48A48, and a decrease from 52.3% to 50.8% in I48S48. The loss of random coil along the peptide backbone is further associated with an increase in the β-turn 2 content, with an increase from 22.2% to 27.2% in I48A48, and from 22.5% to 24.6% in I48S48. The greatest change is evident in the β sheet content, which decreases from 11.8% to 4.0% in I48A48, while decreasing from 12.9% to 10.4% in I48S48. Overall, the CD data is consistent with the interpretation that adoption of secondary structure in the hydrophobic Xaa=Ile block (Fig. 4b) is responsible for the gain of structure in the diblock copolymers based upon this ELP (Fig. 4f, h). In turn, it appears that the hydrophilic blocks, Yaa=Ser, Ala do not adopt structure during nanoparticle formation. Thus, the assembly of ELP diblock copolymers provides spatial control over secondary structure, with the hydrophobic core becoming enriched in β turns, while the corona remains a mixture of random coil and beta sheets.
Quantitative model of the critical micelle temperature and bulk phase transition temperature
Having observed qualitatively that assembly of ELP nanoparticles depends on the phase separation of the hydrophobic domain, a quantitative framework was developed to predict the assembly properties (CMT, Tt,bulk) of ELP nanoparticles based on their monoblock phase behavior. To achieve this, the following model of ELP phase behavior was adopted:
| Eq. 1 |
Where Tc is a critical transition temperature, m is the dependence on the natural logarithm of concentration, n is the dependence on the inverse of the ELP length l, and k is an interaction term between length and concentration. Eq. 1 is a general form of the model validated by Meyer and Chilkoti50 (Eq. 2), where m = 0 and Cc= e-(n/k).
| Eq. 2 |
An improvement on this expression, Eq. 1 was developed because, unlike n, the critical concentration, Cc, is an extrapolated parameter that cannot be robustly estimated using multivariate curve fitting. In addition, Eq. 2 cannot fit the data for ELP libraries that are independent of length, which was observed for monoblock ELPs with Xaa=Ser (supplementary Fig. s1a).
Having characterized the phase diagrams for four ELP monoblock libraries (supplementary Fig. s1), their datasets were fit (Table 2). Parameter estimates for ELPs with Xaa = Val were previously reported by fitting to Eq. 2, which yielded Tc= 20.8 °C; k = 129 °C pentamers/ln(μM), Cc= 25,000 μM50. By fitting our library with Xaa=Val, nearly identical parameters were obtained (Table 2), which validates the use of Eq. 1 to describe ELP phase behavior. The fit results show that, with the exception of the Xaa=Ser block, the transition temperature of the ELP monoblock can be described by n, and the interaction between the ELP length and concentration. The Xaa=Ser monoblock transition temperature has no significant dependence on length, as evident in supplementary Fig. s1a, and so its monoblock behavior is described by the slope m. The fit parameters in Table 2 provide a simple way to estimate the aqueous miscibility of ELP monoblocks at any concentration, length, or temperature; furthermore, this is the first comprehensive dataset for ELPs with Xaa=Ser, Ile, or Ala. As such, these parameters enable the design of ELPs that phase separate over a broad range of target concentrations, lengths, and temperatures.
Table 2. Multivariate fit parameters describing the *phase diagram of ELP monoblock copolymers.
| Monoblock libraries [Xaa] | ELPs fit | Tc [°C] | m [°C/ln(μM)] | n [°C pentamers ] | k [°C pentamers/ln(μM)] | **Cc [μM] | data points | R2 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ser | S72, S96, S144, S192 | 63.6 | ±1.1 | -1.86 | ±0.31 | 0 | 0 | n.a. | 20 | 0.664 | ||
| Ala | A96, A144, A192 | 40.5 | ±1.6 | 0 | 7009 | ±345 | -850.1 | ±51.5 | 3,808 | 14 | 0.974 | |
| Val | V24, V36, V48, V96, V144, V192 | 21.2 | ±0.9 | 0 | 1607 | ±90 | -163.1 | ±23.3 | 19,012 | 29 | 0.965 | |
| Ile | I18, I24, I48, I96 | 13.0 | ±0.6 | 0 | 760 | ±30 | -81.2 | ±7.9 | 11,609 | 20 | 0.982 | |
Multiple linear regression was used to fit datasets with Xaa=Ser, Ala, Val, and Ile (supplementary Fig. s1) to Eq. 1. Non-predictive parameters are indicated with a value of 0. Values indicated are the mean ± standard error
For comparison with prior studies, the critical concentration, Cc, was estimated[55]. n.a. not applicable.
Using the above model, we tested the hypothesis that the assembly temperatures (CMT, Tt,bulk) of ELP diblock copolymers can be directly related to the phase behavior of their monoblocks. To estimate the CMT and Tt,bulk for any ELP diblock the following relationships were evaluated:
| Eq. 3 |
| Eq. 4 |
Where Tt1 and Tt2 are the respective transition temperatures of the hydrophobic and hydrophilic ELP monoblocks, and ΔTt1 and ΔTt2 account for the respective shifts associated with incorporating the monoblock (Table 2) into a diblock copolymer (Table 1). Eq 1 was substituted into Eqs 3 and 4 and rearranged to yield the following:
| Eq. 5 |
| Eq. 6 |
Where, ΔTc1, Δm1, Δn1, Δk1, ΔTc2, Δm2, Δn2, Δk2 are the parameter shifts between those describing the monoblock (Eq. 1) and the respective (CMT, Tt,bulk) phase behavior of related diblock ELPs.
Phase separation of the hydrophobic block of an ELP diblock qualitatively predicts the formation of the core of the nanoparticle, while the properties of the hydrophilic block are expected to dominate the interaction between nanoparticles. In this manuscript, Yaa=Ile was chosen as the hydrophobic block as its monoblock has Tt near room temperature (supplementary Fig. s1d), and this resulted in diblock copolymers that assemble below physiological temperatures (Fig. 2b). In contrast, monoblock libraries of Xaa=Ser, Ala both showed phase separation well above physiological temperature (supplementary Fig. s1), and so were chosen as the hydrophilic blocks. Based on these combined datasets, multiple linear regressions revealed that ΔTc and Δn significantly (p=2×10-11) predict deviations from the monoblock transition temperature, ΔTt. Interestingly, Δn for the ELP diblock copolymers with two hydrophilic blocks (Xaa= Ser, Ala) were similar; furthermore, a simultaneous fit enabled the estimation of ΔTc= -0.94 ± 0.50 °C and Δn = 247 ± 22 °C pentamers (Table 3). This finding supports the hypothesis that the CMT for an ELP nanoparticle with a hydrophobic (Xaa=Ile) block can be predicted using Eq. 5 (Fig. 5a,c).
Table 3. Fit parameters relating the critical micelle temperature of ELP diblock copolymers the hydrophobic monoblock.
| **Diblock libraries [Xaa:Yaa] | ELPs fit | ΔTc1 [°C] | Δn1 [°C pentamers ] | Δk1 [°C pentamers/ln(μM)] | data points | R2 | ||
|---|---|---|---|---|---|---|---|---|
| Ile:Ser | I24S24, I36S36, I48S48, I96S96 | -1.1 | ±0.8 | 255 | ±31 | 0 | 18 | 0.813 |
| Ile:Ala | I48A48, I96A96 | 0.0 | ±0.6 | 185 | ±34 | 0 | 10 | 0.789 |
| Ile:Yaa | I24S24, I36S36, I48S48, I96S96, I48A48, I96A96 | -0.94 | ±0.50 | 247 | ±22 | 0 | 28 | 0.829 |
Multiple linear regression was used to fit CMT datasets to Eq. 5. Non-predictive parameters are indicated with a value of 0; therefore Δm1 = 0. Values indicated are the mean ± standard error.
Ile:Yaa indicates a simultaneous fit to both Yaa=Ser, Ala libraries.
Figure 5.
Quantitative model of the critical micelle concentration and the bulk phase transition temperature for ELP nanoparticles. a, b) Model fit (line) of observed (symbols) CMT and Tt,bulk respectively of I48S48, I48A48, A96A96 and I96S96 are indicated. c) The model based on Eq. 3 was used to predict the CMT for the library of ELP diblock copolymers over a range of concentration (5 to 100 μM). Linear regression between the predicted and observed values (solid line) show a significant correlation (r2 = 1.00, slope = 0.998 ±3.73×10-9, intercept = 0.063 ±1.28×10-7 °C. d) The model based on Eq. 4 was used to predict Tt,bulk also shows a high degree of correlation to the observed values. (r2 = 1.00, slope = 0.999 ±4.62×10-9, intercept = 0.068 ±2.96×10-7 °C). e) Eq. 3 was used to predict the CMT for ELPs as a function of length when the hydrophobic ELP monoblock was composed from Xaa=Val. These predicted values were in relative agreement with those reported previously by Dreher and coworkers16.
While the hydrophobic block of the ELP copolymer is largely responsible for the observed CMT, we further hypothesized that the hydrophilic block dominates the second phase transition, Tt,bulk. As seen in Fig. 3a, there is a large difference in the observed Tt,bulk between ELP diblock copolymers with Yaa=Ser vs. Ala as the hydrophilic component. Further, the Tt,bulk of Yaa=Ala shows little length dependence, while a substantial upwards shift in Tt,bulk is observed when Yaa=Ser. A multivariate fit (Table 4) relating Tt,bulkto the hydrophilic monoblock showed dependence on the parameter shifts Δn, Δk, and ΔTc2. While the model fit of Tt,bulk observed vs. predicted is good (Fig. 5b,d), the model parameters relating nanoparticle bulk phase separation to the behavior of the Yaa=Ser, Ala monoblocks were substantially different (Table 4). To conclude, it is not yet possible to predict the bulk phase separation of ELP nanoparticles without first creating a library of diblock copolymers. Despite this limitation, when fit (Table 4) to a library of diblock copolymers, this model (Eq. 4) does fit the observed dataset (Fig. 5b,d) and enables the engineering of the Tt,bulk across various ELP lengths and concentrations.
Table 4. Fit parameters relating the bulk phase transition temperature of ELP diblock copolymers compared to the hydrophilic monoblock.
| Diblock libraries [Xaa:Yaa] | ELPs fit | ΔTc2 [°C] | Δn2 [°C pentamers ] | Δk2 [°C pentamers/ln(μM)] | data points | R2 | |||
|---|---|---|---|---|---|---|---|---|---|
| Ile:Ser | I48S48, | I96S96 | 3.5 ±1.1 | 1,434 | ±93 | -60.8 | ±20 | 10 | 0.980 |
| Ile:Ala | I48A48, | I96A96 | 4.0 ±0.2 | -6,563 | ±20 | 811 | ±4.3 | 10 | 0.999 |
Multiple linear regression was used to fit Tt,bulk to Eq. 6 with Δm2= 0. Values indicated are the mean ± standard error
Discussion
This manuscript demonstrates that by modulating the length and amino acid composition of ELP diblock copolymers, it is possible to control the onset of self-assembly (CMT, Tt,bulk) and the size of the nanostructure formed. These properties are highly relevant determinants for the disposition of nanostructures in physiological environments51, 52. Varying the amino acid sequence of the hydrophilic block has little effect on nanoparticle-assembling properties; however, it has a significant influence on their bulk phase separation. Further investigation into the changes that occur during the assembly process using circular dichroism (CD) revealed that variations of the hydrophilic block have remarkably little influence on shift in secondary structure found in the more hydrophobic block.
The most significant finding is that the phase behavior for both the CMT and Tt,bulk for ELP diblock copolymers can be approximated by the behavior of their monoblocks. This approximation has immediate applications in designing ELPs for biological applications that assemble at target temperatures, concentrations, and molecular weights. For new ELP diblock copolymers with approximately a 1:1 ratio in molecular weight between the hydrophobic and hydrophilic domain, to predict the CMT the Tc,n, and k can be picked for the hydrophobic monoblock from Table 1, adjusted using ΔTc= -0.94 °C, and Δn = 247 °C pentamers, and predicted using Eq. 3. Using this approach, the results for the CMT prediction using Eq. 3 were found to closely agree with the observed CMTs as a function of concentration and length (Fig. 5a). All of the diblock copolymers (Table 3) showed an excellent correlation between the observed and predicted CMT (Fig. 5c). To further validate this approach, Eq. 3 was used to predict the CMTs for ELP diblock copolymers with a different hydrophobic core (Xaa=Val), as previously reported16. Despite a slight downward shift in CMT, to a reasonable approximation, these predictions followed the same trend and magnitude as those previously reported (Fig. 5e). Thus we conclude that the CMT of an ELP diblock copolymer can be predicted based on the transition temperature of its hydrophobic monoblock primarily by adjusting its dependence on length, Δn. To the best of our knowledge, this is the first time that a predictive model has been used to estimate the CMT of an ELP diblock polymer.
These findings are consistent with the hypothesis that the hydrophilic block comprises the outer portion or corona of the formed nanostructures. As the corona is usually in contact with the cellular environment its composition imparts the characteristics that allow for prolong circulation of the particle by preventing the opsonization and removal by the reticuloendothelial system (RES). Polyethylene glycol (PEG) has thus far been the polymer of choice for endowing nanoparticles/polymers with stealth properties to protect them from premature clearance53-56. This polymeric brush afforded by the PEG is thought to adopt a splayed appearance, which sterically suppresses the binding of opsonins57. Similarly the hydrophilic corona of the ELP micelle may take the place of PEG in reducing opsonization and ensuring extended circulation time. More work is required to determine what steric stabilization properties can be attained by controlling the amino acid sequence of the hydrophilic ELP and any resulting changes in its secondary structure. Certainly, future in vivo characterization will be required to better determine how to optimize their properties for use as therapeutic platforms58, 59.
Conclusion
A series of protein polymers of various lengths and sequence have been recombinantly prepared that assemble stable nanoparticles at physiological temperatures. The hydrophobic block length has a profound effect on the ability of the block copolymers to form stable nanoparticles, whereby approximately 48 ELP repeat units were necessary for stability. A quantitative model was developed that fits the phase diagrams (length, concentration, temperature) of both nanoparticle assembly and bulk phase separation. This library of biopolymers has future applications as a platform for coassembly and delivery within living organisms.
Supplementary Material
Acknowledgments
This work was made possible by the University of Southern California, the National Institute of Health R21EB012281 to J.A.M., and P30 CA014089 to the Norris Comprehensive Cancer Center, the USC Molecular Imaging Center, the USC Nanobiophysics Core Facility, the Translational Research Laboratory at the School of Pharmacy, the American Cancer Society IRG-58-007-48, the Stop Cancer Foundation, the USC Ming Hsieh Institute, and the USC Whittier Foundation. SMJ is a recipient of Malaysian Public Services Department scholarship.
Abbreviations
- ELP
Elastin like polypeptides
- CMT
critical micelle temperature
- ITC
inverse transition cycling
- DLS
dynamic light scattering
- Tt
transition temperature
- Tt,bulk
bulk phase transition temperature
- Rh
hydrodynamic radius
- PBS
phosphate buffered saline
- RDL
recursive directional ligation
References
- 1.DiMarco RL, Heilshorn SC. Advanced Materials. 2012;24:3923–3940. doi: 10.1002/adma.201200051. [DOI] [PubMed] [Google Scholar]
- 2.MaHam A, Tang Z, Wu H, Wang J, Lin Y. Small. 2009;5:1706–1721. doi: 10.1002/smll.200801602. [DOI] [PubMed] [Google Scholar]
- 3.Prince JT, McGrath KP, DiGirolamo CM, Kaplan DL. Biochemistry. 1995;34:10879–10885. doi: 10.1021/bi00034a022. [DOI] [PubMed] [Google Scholar]
- 4.Arcidiacono S, Mello C, Kaplan D, Cheley S, Bayley H. Applied Microbiology and Biotechnology. 1998;49:31–38. doi: 10.1007/s002530051133. [DOI] [PubMed] [Google Scholar]
- 5.Winkler S, Szela S, Avtges P, Valluzzi R, Kirschner DA, Kaplan D. International Journal of Biological Macromolecules. 1999;24:265–270. doi: 10.1016/s0141-8130(98)00088-9. [DOI] [PubMed] [Google Scholar]
- 6.Frank S, Kammerer RA, Mechling D, Schulthess T, Landwehr R, Bann J, Guo Y, Lustig A, Bächinger HP, Engel J. Journal of Molecular Biology. 2001;308:1081–1089. doi: 10.1006/jmbi.2001.4644. [DOI] [PubMed] [Google Scholar]
- 7.Lee TAT, Cooper A, Apkarian RP, Conticello VP. Advanced Materials. 2000;12:1105–1110. [Google Scholar]
- 8.Meyer DE, Shin BC, Kong GA, Dewhirst MW, Chilkoti A. Journal of Controlled Release. 2001;74:213–224. doi: 10.1016/s0168-3659(01)00319-4. [DOI] [PubMed] [Google Scholar]
- 9.Welsh ER, Tirrell DA. Biomacromolecules. 2000;1:23–30. doi: 10.1021/bm0002914. [DOI] [PubMed] [Google Scholar]
- 10.Aluri S, Janib SM, Mackay JA. Adv Drug Deliv Rev. 2009;61:940–952. doi: 10.1016/j.addr.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mackay JA, Callahan DJ, Fitzgerald KN, Chilkoti A. Biomacromolecules. 2010 doi: 10.1021/bm100571j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah M, Hsueh PY, Sun G, Janib SM, MacKay JA. Protein Science. 2012 doi: 10.1002/pro.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreher MR, Simnick AJ, Fischer K, Smith RJ, Patel A, Schmidt M, Chilkoti A. J Am Chem Soc. 2008;130:687–694. doi: 10.1021/ja0764862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simnick AJ, Valencia CA, Liu R, Chilkoti A. ACS Nano. 2010;4:2217–2227. doi: 10.1021/nn901732h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright ER, Conticello VP. Adv Drug Deliv Rev. 2002;54:1057–1073. doi: 10.1016/s0169-409x(02)00059-5. [DOI] [PubMed] [Google Scholar]
- 16.Dreher MR, Simnick AJ, Fischer K, Smith RJ, Patel A, Schmidt M, Chilkoti A. Journal of the American Chemical Society. 2007;130:687–694. doi: 10.1021/ja0764862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fairman R, Åkerfeldt KS. Current Opinion in Structural Biology. 2005;15:453–463. doi: 10.1016/j.sbi.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Chow D, Nunalee ML, Lim DW, Simnick AJ, Chilkoti A. Materials Science and Engineering: R: Reports. 2008;62:125–155. doi: 10.1016/j.mser.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W, MacKay JA, Dreher MR, Chen M, McDaniel JR, Simnick AJ, Callahan DJ, Zalutsky MR, Chilkoti A. Journal of controlled release:official journal of the Controlled Release Society. 2010;144:2–9. doi: 10.1016/j.jconrel.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janib SM, Liu S, Park R, Pastuszka MK, Shi P, Moses AS, Orosco MM, Lin YA, Cui H, Conti PS, Li Z, Mackay JA. Integr Biol (Camb) 2012 doi: 10.1039/c2ib20169k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huemmerich D, Scheibel T, Vollrath F, Cohen S, Gat U, Ittah S. Current biology : CB. 2004;14:2070–2074. doi: 10.1016/j.cub.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Lyons RE, Lesieur E, Kim M, Wong DCC, Huson MG, Nairn KM, Brownlee AG, Pearson RD, Elvin CM. Protein Engineering Design and Selection. 2007;20:25–32. doi: 10.1093/protein/gzl050. [DOI] [PubMed] [Google Scholar]
- 23.Bellingham CM, Lillie MA, Gosline JM, Wright GM, Starcher BC, Bailey AJ, Woodhouse KA, Keeley FW. Biopolymers. 2003;70:445–455. doi: 10.1002/bip.10512. [DOI] [PubMed] [Google Scholar]
- 24.Daamen WF, Veerkamp JH, van Hest JCM, van Kuppevelt TH. Biomaterials. 2007;28:4378–4398. doi: 10.1016/j.biomaterials.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 25.Xu C, Kopeček J. Pharmaceutical Research. 2008;25:674–682. doi: 10.1007/s11095-007-9343-z. [DOI] [PubMed] [Google Scholar]
- 26.Kopecek J. European Journal of Pharmaceutical Sciences. 2003;20:1–16. doi: 10.1016/s0928-0987(03)00164-7. [DOI] [PubMed] [Google Scholar]
- 27.Hartgerink JD, Beniash E, Stupp SI. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 28.McPherson DT, Morrow C, Minehan DS, Wu J, Hunter E, Urry DW. Biotechnology Progress. 1992;8:347–352. doi: 10.1021/bp00016a012. [DOI] [PubMed] [Google Scholar]
- 29.Fukushima Y. Biopolymers. 1998;45:269–279. doi: 10.1002/(SICI)1097-0282(19980405)45:4<269::AID-BIP1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 30.Nagarsekar A, Crissman J, Crissman M, Ferrari F, Cappello J, Ghandehari H. Journal of Biomedical Materials Research. 2002;62:195–203. doi: 10.1002/jbm.10272. [DOI] [PubMed] [Google Scholar]
- 31.McMillan RA, Lee TAT, Conticello VP. Macromolecules. 1999;32:3643–3648. [Google Scholar]
- 32.Meyer DE, Chilkoti A. Biomacromolecules. 2002;3:357–367. doi: 10.1021/bm015630n. [DOI] [PubMed] [Google Scholar]
- 33.McDaniel JR, Mackay JA, Quiroz FG, Chilkoti A. Biomacromolecules. 2010;11:944–952. doi: 10.1021/bm901387t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urry DW. Journal of Physical Chemistry B. 1997;101:11007–11028. [Google Scholar]
- 35.Sun G, Hsueh PY, Janib SM, Hamm-Alvarez S, MacKay JA. Journal of controlled release:official journal of the Controlled Release Society. 2011;155:218–226. doi: 10.1016/j.jconrel.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cappello J, Crissman JW, Crissman M, Ferrari FA, Textor G, Wallis O, Whitledge JR, Zhou X, Burman D, Aukerman L, Stedronsky ER. Journal of Controlled Release. 1998;53:105–117. doi: 10.1016/s0168-3659(97)00243-5. [DOI] [PubMed] [Google Scholar]
- 37.Haider M, Leung V, Ferrari F, Crissman J, Powell J, Cappello J, Ghandehari H. Molecular Pharmaceutics. 2005;2:139–150. doi: 10.1021/mp049906s. [DOI] [PubMed] [Google Scholar]
- 38.Fluegel S, Buehler J, Fischer K, McDaniel JR, Chilkoti A, Schmidt M. Chemistry – A European Journal. 2011;17:5503–5506. doi: 10.1002/chem.201100436. [DOI] [PubMed] [Google Scholar]
- 39.Haghpanah JS, Yuvienco C, Civay DE, Barra H, Baker PJ, Khapli S, Voloshchuk N, Gunasekar SK, Muthukumar M, Montclare JK. ChemBioChem. 2009;10:2733–2735. doi: 10.1002/cbic.200900539. [DOI] [PubMed] [Google Scholar]
- 40.van Eldijk MB, Wang JCY, Minten IJ, Li C, Zlotnick A, Nolte RJM, Cornelissen JJLM, van Hest JCM. Journal of the American Chemical Society. 2012;134:18506–18509. doi: 10.1021/ja308132z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christensen T, Hassouneh W, Trabbic-Carlson K, Chilkoti A. Biomacromolecules. 2013;14:1514–1519. doi: 10.1021/bm400167h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Golemis E, Adams PD. Protein-protein interactions:a molecular cloning manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2005. [Google Scholar]
- 43.Wang J, Byrne JD, Napier ME, DeSimone JM. Small. 2011;7:1919–1931. doi: 10.1002/smll.201100442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meyer DE, Chilkoti A. Nat Biotechnol. 1999;17:1112–1115. doi: 10.1038/15100. [DOI] [PubMed] [Google Scholar]
- 45.Yamaoka T, Tamura T, Seto Y, Tada T, Kunugi S, Tirrell DA. Biomacromolecules. 2003;4:1680–1685. doi: 10.1021/bm034120l. [DOI] [PubMed] [Google Scholar]
- 46.Nicolini C, Ravindra R, Ludolph B, Winter R. Biophysical Journal. 2004;86:1385–1392. doi: 10.1016/S0006-3495(04)74209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kountouris P, Hirst JD. BMC bioinformatics. 2010;11:407. doi: 10.1186/1471-2105-11-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamaoka T, Tamura T, Seto Y, Tada T, Kunugi S, Tirrell DA. Biomacromolecules. 2003;4:1680–1685. doi: 10.1021/bm034120l. [DOI] [PubMed] [Google Scholar]
- 49.Marcelino AM, Gierasch LM. Biopolymers. 2008;89:380–391. doi: 10.1002/bip.20960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meyer DE, Chilkoti A. Biomacromolecules. 2004;5:846–851. doi: 10.1021/bm034215n. [DOI] [PubMed] [Google Scholar]
- 51.Duan X, Li Y. Small. 2012:n/a–n/a. [Google Scholar]
- 52.Zhu M, Perrett S, Nie G. Small. 2012:n/a–n/a. [Google Scholar]
- 53.Harris JM, Martin NE, Modi M. Clinical pharmacokinetics. 2001;40:539–551. doi: 10.2165/00003088-200140070-00005. [DOI] [PubMed] [Google Scholar]
- 54.Klibanov AL, Maruyama K, Torchilin VP, Huang L. FEBS Letters. 1990;268:235–237. doi: 10.1016/0014-5793(90)81016-h. [DOI] [PubMed] [Google Scholar]
- 55.Gref R, Minamitake Y, Peracchia MT, Trubetskoy V, Torchilin V, Langer R. Science. 1994;263:1600–1603. doi: 10.1126/science.8128245. [DOI] [PubMed] [Google Scholar]
- 56.Joralemon MJ, McRae S, Emrick T. Chemical Communications. 2010;46:1377–1393. doi: 10.1039/b920570p. [DOI] [PubMed] [Google Scholar]
- 57.Vonarbourg A, Passirani C, Saulnier P, Benoit JP. Biomaterials. 2006;27:4356–4373. doi: 10.1016/j.biomaterials.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 58.Janib SM, Liu S, Park R, Pastuszka MK, Shi P, Moses AS, Orosco MM, Lin YA, Cui H, Conti PS, Li Z, MacKay JA. Integrative Biology. 2013;5:183–194. doi: 10.1039/c2ib20169k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi P, Aluri S, Lin YA, Shah M, Edman-Woolcott M, Dhandhukia J, Cui H, MacKay JA. Journal of Controlled Release. doi: 10.1016/j.jconrel.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.