Abstract
As a powerful research tool, siRNA's therapeutic and target validation utility with leukemia cells and long-term gene knockdown is severely restricted by the lack of omnipotent, safe, stable, and convenient delivery. Here, we detail our discovery of siRNA-containing lipid nanoparticles (LNPs) able to effectively transfect several leukemia and difficult-to-transfect adherent cell lines also providing in vivo delivery to mouse spleen and bone marrow tissues through tail-vein administration. We disclose a series of novel structurally related lipids accounting for the superior transfection ability, and reveal a correlation between expression of Caveolins and successful transfection. These LNPs, bearing low toxicity and long stability of >6 months, are ideal for continuous long-term dosing. Our discovery represents the first effective siRNA-containing LNPs for leukemia cells, which not only enables high-throughput siRNA screening with leukemia cells and difficult-to-transfect adherent cells but also paves the way for the development of therapeutic siRNA for leukemia treatment.
Introduction
Small interfering RNA (siRNA) technology has become a powerful research tool for probing gene function and a promising therapeutic for previously undruggable targets.1,2,3,4 However, efficient and safe delivery of small siRNAs into desired cells or tissues has proven challenging, especially for hard-to-transfect suspension cells. Over the past decade, many siRNA delivery approaches have been developed to enhance cellular uptake of siRNAs.5 Among them, a family of lipid nanoparticles (LNPs) known as stabilized nucleic acid lipid particles (SNALPs), represent the most clinically advanced approach for siRNA delivery due to their high siRNA-encapsulation efficiency and low immunogenic properties, and are currently being evaluated in clinical trials.6,7,8,9
In SNALPs, siRNA is encapsulated in a mixture of fusogenic cationic lipids and helper neutral lipids, coated with diffusible polyethylene glycol (PEG)-lipids to stabilize the nascent particles and reduce the association with serum proteins.10 The cationic lipids in SNALPs play key roles for the potency of SNALPs, because they participate in the encapsulation of negatively charged siRNAs, mediate endocytosis by interacting with the negatively charged cell plasma membrane and facilitate subsequent endosomal escape of the siRNA cargo.8,9 Despite SNALP's successful in vivo siRNA delivery into mouse livers, their ability to deliver siRNA into suspension cells, especially leukemia cells, has not been reported.
Suspension cells, such as primary blood cells and leukemia cells are well known to be difficult to transfect by conventional lipid and polymer-based nucleic acid delivery approaches, although the exact cellular mechanism has remained elusive. So far, siRNA delivery to suspension cells has relied on electroporation, such as the Nucleofector Technology from Lonza,11 which is challenging to use on a large scale and whose conditions need to be carefully tailored for individual cell lines to avoid cell damage. Recently, Accell siRNAs, a novel type of chemically modified siRNAs developed by Thermo Scientific were claimed to be able to silence gene expression in difficult-to-transfect cells without transfection reagents. However, high concentration of siRNAs (1 µmol/l) are required in conjunction with serum-free growth media for at least 48 hours, affecting normal cell growth and compromising subsequent biological assays. When tested in leukemia cells, Accell siRNAs only modestly knockdown target genes (30–60%) although they seems to enter cells quite efficiently.12 Therefore, although high-throughput siRNA screening on adherent cell lines has been widely applied to explore new biology, its application on suspension cells has been prohibitive.
Efficient and safe delivery of siRNAs into suspension blood cells also holds significant therapeutic value. Development of therapeutic siRNAs has focused on solid tumors given the relatively successful in vitro siRNA delivery over suspension blood cancer cells.13,14 However, in order for siRNAs to work in vivo for solid tumors, they have to survive in the blood stream, extravasate and move through the complex extracellular matrix before reaching the tumor cells. In contrast, blood cancer cells have the advantage of direct exposure to intravenously delivered siRNAs. In this sense, treating blood cancers with siRNAs holds a distinct in vivo advantage. The discovery of a siRNA delivery system to efficiently and safely deliver siRNAs into suspension blood cells will not only advance our research tools but also transform current therapeutic strategies for leukemia treatment.
Here, we reported the discovery of a series of novel structurally related cationic lipids that break the barrier of siRNA delivery for suspension leukemia cells. SNALP-like LNPs (SLPs) incorporating a small percentage of these novel lipids efficiently transfect a variety of leukemia cell lines as well as hard-to-transfect adherent cell lines, where leading brands of commercial siRNA transfection reagents fail. These SLPs, bearing low toxicity and long stability of >6 months, performed well in leukemia cell–based biological assays. They are also able to deliver siRNAs into mouse hematopoietic organs, namely spleen and bone marrow, leading to in vivo targeted mRNA knockdown. Furthermore, comparative microarray analysis reveals that the expression of endosomal processing pathway genes, Caveolin 1 and 2, correlate with successful cell transfection.
Results
Discovery of novel LNPs able to transfect siRNA into leukemia cells
To develop an effective siRNA delivery system for suspension leukemia cells, we started by evaluating a panel of commercially available siRNA transfection reagents in a commonly used suspension leukemia cell line, K562. However, even at 100 nmol/l siRNA concentrations, none of the tested reagents efficiently knock down the targeted gene, KIF11 (<30%), a kinesin essential for bipolar spindle formation,15 suggesting that commercial reagents are not a good starting point for further optimization (Supplementary Figure S1).
Recent progress has been made for siRNA delivery using SNALPs.8,9 However, their use on suspension leukemia cells has not been reported. In order to develop LNPs able to efficiently deliver siRNAs into leukemia cells, we started by designing, preparing and screening a large variety of KIF11 siRNA-encapsulated SNALP-like particles (SLPs) made of different cationic lipids. Initial formulation of SLPs was based on established SNALPs,9 with 1,2-dilinoleyl-4-(2-dimethylaminoethyl)-[1,3]-dioxolane (DLin-KC2-DMA) as cationic lipid, cholesterol/dipalmitoylphosphatidylcholine (DPPC) or distearoylphosphatidylcholine (DSPC) as helper neutral lipids and R-3-[(ω-methoxy poly(ethylene glycol)2000)carbamoyl)]-1,2-dimyristyloxl-propyl-3-amine (PEG-c-DOMG) as PEGylsated lipids (Figure 1a).
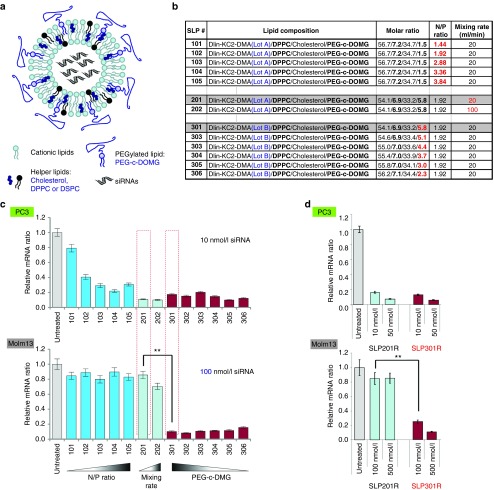
Figure 1.
Newly discovered SNALP-like lipid nanoparticles (SLPs) efficiently deliver siRNA into suspension leukemia cells. (a) Schematic representation of SLPs. (b) Composition of optimized SLPs with different N/P ratios (SLP100s), mechanical mixing rates (SLP200s), and percentages of PEG-c-DMG components (SLP300s). The differences among formulations were colored in blue or red. (c) Discovery of SLP300s able to efficiently knockdown KIF11 in suspension Molm13 leukemia cells. SLP100s-SLP300s were tested in easy-to-transfect adherent PC3 cells (upper panel) and hard-to-transfect suspension Molm13 cells (lower panel). KIF11 mRNA knockdown was measured by quantitative RT-PCR after 24 hours of siRNA transfection. Error bars represent mean ± SD. P value is calculated using a Student's t-test. SLP201 and SLP301, prepared from the same formula, are boxed in red. (d) Only SLP301R is able to efficiently knockdown KIF11 in Molm13 cells. SLP201R, repeat of SLP201 with Lot A of Dlin-KC2-DMA; SLP301R, repeat of SLP301 with Lot B of Dlin-KC2-DMA.
For the initial optimization, SLPs with increasing N/P ratio (SLP100 series, molar ratio of amine group N (cationic lipid) to phosphate group P (siRNA)), increasing mechanical mixing rate (SLP200 series, affecting particle size), and decreasing PEG-c-DOMG lipid content (SLP300 series) (Figure 1b), were tested in a hard-to-transfect suspension leukemia cell line Molm13, as well as a highly transfectable adherent PC3 prostate cancer cell line as positive control. Although most of these SLPs yielded high transfection efficiency in PC3 cells with >80% KIF11 knockdown at 10 nmol/l siRNA (Figure 1c, upper panel), when used on the hard-to-transfect suspension Molm13 leukemia cells, only the SLP300 series generated a profound ~90% KIF11 knockdown (Figure 1c, lower panel).
To understand why only the SLP300 series was able to transfection Molm13 leukemia cells, we carefully examined the composition of our SLP formulations. As shown by Figure 1b, SLP201 and SLP301 were actually prepared from the exact same formula except that the cationic lipid, Dlin-KC2-DMA (DMA for short, hereafter), originated from two different lot preparations. These two lots were synthesized using different protocols yielding different lipid purities. DMA used in SLP201 (preparation Lot-A) was much purer than the preparation used for SLP301 (preparation Lot-B), suggesting the impurities in DMA Lot-B contained the important component contributing to the strong Molm13 transfection. When we repeated SLP201 and SLP301 preparations (SLP201R and SLP301R) from these two different DMA lots and then tested on PC3 and Molm13 cells, the same result was obtained (Figure 1d), further supporting Lot-B DMA impurities contributed to the strong Molm13 transfection.
Identification of novel lipids critical for leukemia cell siRNA transfection
To identify the lipid contaminants responsible for the successful transfection of Molm13 cells, both DMA lots were subjected to detailed chemical analysis. Owing to the different synthetic approach, DMA Lot-B specifically contained about 1.5% Dlin-KC2-ClMDMA (ClMDMA for short, hereafter), a synthetic alkylated DMA byproduct (Figure 2a). To validate whether ClMDMA plays a causative role in the Molm13 transfection, we synthesized ClMDMA and prepared SLPs with different molar ratios of ClMDMA/DMA. As shown in Figure 2b, no significant KIF11 knockdown in Molm13 cells was observed from SLPs without ClMDMA. By contrast, significant KIF11 knockdown was obtained from SLPs containing 4.2–8.4% ClMDMA, a ratio similar to that in the original Lot-B, indicating that effective siRNA delivery into Molm13 by SLPs depended on the presence of ClMDMA. Different from DMA whose charge is dependent on pH, ClMDMA is permanently positively charged, which may account for enhanced transfection efficiency. However, increasing amounts of ClMDMA was detrimental to the SLP transfection efficiency (Figure 2b). It is possible that excessive positive charges and the slightly bulkier head group of ClDMA interfere with the assembly and stability of SLPs.
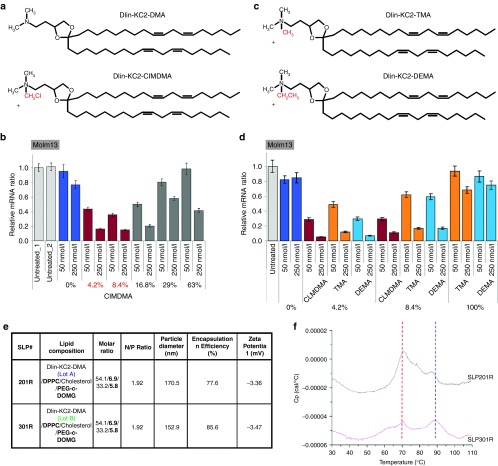
Figure 2.
Alkylated Dlin-KC2-DMA lipids are the key components of novel SNALP-like lipid nanoparticles (SLPs) for efficient siRNA transfection into leukemia cells. (a) Chemical structures of Dlin-KC2-DMA (DMA) and Dlin-KC2-ClMDMA (ClMDMA). (b) Addition of ClMDMA into DMA-based SLPs leads to potent siRNA transfection of suspension Molm13 leukemia cells. ClMDMA was mixed with DMA at indicated ratios, and resulting SLPs were tested on Molm13 cells at indicated siRNA concentrations. KIF11 mRNA knockdown was measured by quantitative RT-PCR after 24 hours of siRNA transfection. (c) Chemical structures of Dlin-KC2-TMA (TMA) and Dlin-KC2-DEMA (DEMA). (d) Addition of TMA- or DEMA- to DMA-based SLPs leads to the similar siRNA transfection efficiency as addition of ClMDMA. ClMDMA, TMA, or DEMA was mixed with DMA at indicated ratio, and resulting SLPs were tested on Molm13 cells at indicated siRNA concentrations. (e) Comparison of biophysical properties of SLP201R and SLP301R. (f) Differential scanning calorimetry measurement of SLPs as indicated. In ClMDMA-containing SLP301R, the peak of the lamellar-to-inverted hexagonal phase transition shifted from lower temperature (~70 °C indicated by read line) to higher temperature (~90 °C indicated by blue line) compared with SLP201R.
To test whether the Molm13 transfection enhancement property is unique to ClMDMA, we synthesized two alkylated DMA molecules similar to ClMDMA by replacing the chloromethyl group with methyl (Dlin-KC2-TMA, TMA for short, hereafter)) or ethyl groups (Dlin-KC2-DMEA, DMEA for short, hereafter) (Figure 2c), and prepared SLPs with different ratios of these two lipids. As shown in Figure 2d, both TMA and DMEA lipids at the optimal ratios displayed a similar ability to transfect Molm13 cells as ClMDMA, demonstrating that a subclass of alkylated DMA cooperating with DMA greatly improve SLP transfection efficiency of Molm13 cells.
To further investigate why SLPs with alkylated DMA lipids gained the ability to transfect suspension cells, we compared the biophysical properties of SLPs with or without alkylated DMA lipids. No significant differences were seen on particle size or zeta potential between SLP201R and SLP301R (Figure 2e). However, differential scanning calorimetry (DSC) measurement suggested that ClMDMA-containing SLP301R has increased particle order and stability, as shown by a higher lamellar-to-inverted hexagonal phase transition temperature (~90 °C for SLP301R versus ~70 °C for SLP201R) (Figure 2f).
Superiority of alkylated DMA-containing LNPs as in vitro research tools
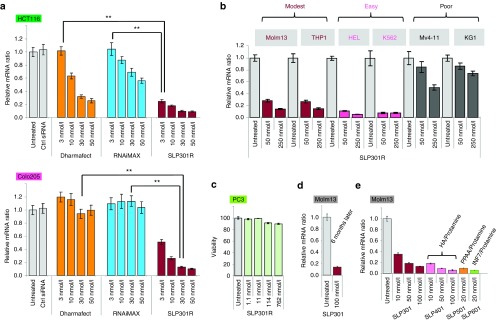
An ideal siRNA transfection reagent for research should have the following three properties: (i) high transfection efficiency of diverse cell lines, (ii) low toxicity, and (iii) excellent stability. To demonstrate the superior transfection efficiency of our alkylated DMA-containing SLPs, SLP301R was compared with two leading-brand siRNA transfection reagents, Dharmafect (Thermo Scientific) and RNAiMAX (Invitrogen) for transfection efficiency on HCT116 and Colo205 colorectal cancer cells, representing an easily transfected and a poorly transfected adherent cell line, respectively. Throughout the comparison, a constant N/P ratio was maintained for all siRNA concentrations tested, which means that a stock solution of siRNA transfection mix was prepared as recommended by each manufacture, then a serial dilution was made and applied to cells. The same serial dilution approach was performed for SLP301R. Cells were seeded at higher density in order to make the transfection more challenging so as to better differentiate the transfectability of these reagents. Under the above condition, SLP-301R performed at least 10 times more efficient than Dharmafect and RNAiMAX in easily transfected HCT116 cells (Figure 3a, upper panel, 75% knockdown reached by 3 nmol/l siRNA with SLP301R compared with 50 nmol/l siRNA with Dharmafect). In poorly transfected Colo205 cells, only SLP-301R was able to generate significant KIF11 knockdown, >90% knockdown at 30–50 nmol/l siRNA (Figure 3a, lower panel). These data indicate that alkylated DMA-containing SLPs possess high siRNA transfection efficiency not only in suspension leukemia cells, but also in poorly transfected adherent cells.
Figure 3.
Alkylated DMA-containing SNALP-like lipid nanoparticles (SLPs) displayed superior in vitro siRNA transfection efficiency, low cellular toxicity and high stability in both adherent cells and suspension leukemia cells. (a) ClMDMA-containing SLP301R showed superior transfection efficiency compared with leading commercial siRNA transfection reagents. Control siRNA or increasing concentration of KIF11 siRNA were transfected using Dharmafect (Thermo Fisher), RNAiMAX (Invitrogen), or SLP301R into easily transfected HCT116 (upper panel) and poorly transfected Colo205 adherent cells (lower panel). KIF11 mRNA knockdown was measured by quantitative RT-PCR after 24 hours of siRNA transfection. Error bars represent mean ± SD. P value is calculated using a Student's t-test. (b) SLP301R is able to potently transfect multiple leukemia cell lines. Increasing concentration of KIF11 siRNA were transfected using SLP301R in six leukemia cell lines as indicated. (c) SLP301R encapsulating siRNA is not toxic for cells. Increasing concentration of luciferase siRNA as indicated was transfected into PC3 cells using SLP301R, and cell viability was measured by CellTiter-Glo at 24 hours after transfection. (d) SLP301R maintained Molm13 transfection efficiency after storage in 4 °C for 6 months. The efficiency of knockdown is measured by quantitative RT-PCR. (e) Incorporating biocompatible polymers into alkylated DMA-containing SLPs (Supplementary Table S1) as indicated leads to more than fivefold enhancement of Molm13 transfection efficiency. HA, hyaluronic acid; INF7, a glutamic acid-enriched cell penetrating peptide derived from influenza virus; PPAA, poly(propylacrylic acid).
We further tested the breadth of transfection ability of alkylated DMA-containing SLPs across leukemia cell lines. Five commonly used leukemia cell lines were tested along with Molm13 line using SLP301R. Besides Molm13, three more cell lines, THP1, HEL and K562, also yielded about 90% KIF11 knockdown (Figure 3b), in stark contrast to a lack of KIF11 knockdown in K562 cells by commercial siRNA transfection reagents as shown previously (Supplementary Figure S1). As far as we know, this is the first report that siRNA can be efficiently transfected into multiple suspension leukemia cells using untargeted LNPs.
Lastly, alkylated DMA-containing SLPs show limited cellular toxicity and good stability. As shown in Figure 3c, even with treatment up to 762 nmol/l of luciferase siRNA encapsulated with SLP301R, little change in the viability of PC3 cells was observed. In addition, alkylated DMA-containing SLPs are very stable at 4 °C. During our studies, we routinely used the original SLP301 as our positive control. Even after 6 months, SLP301 still generated similar KIF11 knockdown in Molm13 cells as we first observed (Figures 1c and 3d). The diameter of these particles remained unchanged (123 nm when newly prepared versus 121 nm after 6 months)
Improved efficiency of alkylated DMA-containing SLPs following incorporation of cationic and anionic biopolymers
Although it is significant for alkylated DMA-containing SLPs to deliver siRNAs into Molm13 cells, the siRNA concentration required to obtain a significant knockdown remains relatively high (~100 nmol/l for 90% knockdown, Figure 1c, bottom panel). Currently with commercial siRNA transfection reagents, 20–50 nmol/l of siRNA is routinely used as starting concentration for adherent cells. In order to further improve transfection efficiency of SLPs, we incorporated biocompatible polymers based on success in the nucleotide delivery field. It has been reported that biocompatible polymers, such as cationic protamine sulfate and anionic hyaluronic acid, improve siRNA nanoparticle formation and particle stability, increasing siRNA delivery.16 As shown in Figure 3e, SLPs with optimal amounts of hyaluronic acid and protamine sulfate (SLP401) (Supplementary Table S1), the transfection efficiency was improved by fivefold (about 80% KIF11 knockdown reached by 10 nmol/l siRNA of SLP401 compared with 50 nmol/l siRNA of SLP301). Endosomal escape is another well-known efficiency-limiting step for nucleotide delivery. Many pH-sensitive anionic membrane-disrupting peptides and polymers, including PPAA (poly(propylacrylic acid))17 and INF7 (a glutamic acid-enriched cell penetrating peptide derived from influenza virus),18 have been used to enhance endosomal escape. When optimal ratios of these polymers were used in SLP501 and SLP601 (Supplementary Table S1), siRNA transfection of Molm13 cells was also significantly improved (Figure 3f, >80% KIF11 mRNA knockdown at 20 nmol/l siRNA).
These improved SLPs generated 80–90% knockdown in Molm13 cells with only 10–20 nmol/l siRNA, which is comparable with siRNA concentration routinely used for adherent cell studies.
Alkylated DMA-containing SLPs as convenient tools for in vitro siRNA screening and subsequent biological assays
siRNA is widely used in research to study gene function and validate new drug targets. siRNA mediated high-throughput screening has proven to be a powerful screening method. However, it has been prohibitive in leukemia cells due to poor transfection methods. Given that our improved SLP401-SLP601 are now able to generate 80–90% target knockdown with 10–20 nmol/l siRNAs in previously modestly transfected Molm13 leukemia cells, our SLPs hold great potential for future high-throughput siRNA screening of leukemia cells.
To demonstrate this potential, we chose to identify potent siRNAs against FLT3 (fms-like tyrosine kinase 3) from a number of in-house designed and synthesized siRNAs. FLT3 is a critical receptor tyrosine kinase expressed in hematopoietic stem cells and progenitors. Mutations in FLT3 lead to constitutive receptor activation in about 30% of acute myeloid leukemia, making FLT3 an attractive target for acute myeloid leukemia therapy.19,20 Molm13 cells which carry a FLT3 mutation are ideal for FLT3 siRNA screening and subsequent functional studies. As shown in Figure 4a, 20 FLT3 siRNAs were encapsulated into SLP401 and applied directly to Molm13 cells. These SLPs generated FLT3 gene knockdown at both 40 nmol/l and 100 nmol/l concentrations. Potent siRNAs can be easily selected from this experiment for further biological characterization and optimization.
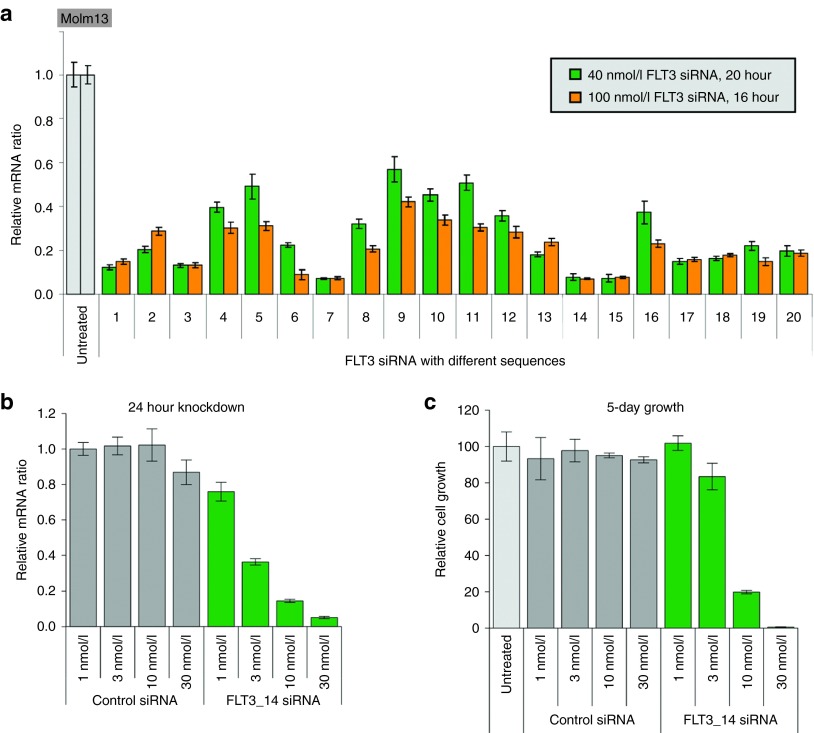
Figure 4.
Alkylated DMA-containing SNALP-like lipid nanoparticles (SLPs) are efficient and convenient in vitro siRNA screening tools for suspension leukemia cells. (a) Screening of FLT3 siRNAs in Molm13 cells. Twenty FLT3 siRNAs were transfected into Molm13 cells at two concentrations as indicated by SLP401. FLT3 knockdown was measured by quantitative RT-PCR at 24 hours after transfection. (b) Compared with control siRNA, a potent FLT3 siRNA, FLT3_14, encapsulated in SLP401 showed dose-dependent FLT3 knockdown. (c) FLT3_14 siRNAs encapsulated in SLP401 showed dose-dependent growth inhibition in a 5-day growth assay. Cell growth was measured by CellTiter-Glo.
To demonstrate that our SLPs are suitable for long-term cellular studies, we selected #14, a potent FLT3 siRNA (Figure 4a), for a 5-day growth inhibition study in Molm13 cells. In Figure 4b, FLT3_14 siRNA generated a nice dose-dependent knockdown in Molm13 cells after 24 hours. When these cells were maintained for 5 days without medium change, control siRNA transfected cells and untreated cells grew at a similar rate, confirming the low toxicity of our SLPs (Figure 4c). Furthermore, FLT3_14 siRNA-treated cells displayed a dose-dependent growth inhibition corresponding to FLT3 knockdown (Figure 4c), reproducing the growth inhibition demonstrated by a small molecule FLT3 inhibitor.21 These results indicate that alkylated DMA-containing SLPs are powerful tools allowing easy siRNA screening and subsequent biological assays in suspension leukemia cells and other hard-to-transfect cells.
In vivo siRNA delivery to mouse hematopoietic tissues using alkylated DMA-containing SLPs
The efficient in vitro siRNA transfection of leukemia cells by alkylated DMA-containing SLPs led us to profile them for in vivo delivery. Efficient in vivo siRNA delivery by SNALPs has only been demonstrated in livers, limiting their therapeutic potential. When tested, our SLPs efficiently delivered siRNAs into mouse livers (Supplementary Figure S3). We further tested whether SLPs were able to deliver siRNA to hematopoietic tissues, namely spleen and bone marrow, where blood tumors usually reside. Successful in vivo siRNA transfection to these tissues would serve as a stepping stone for development of alkylated DMA-containing SLPs as therapeutics for blood tumors.
SLPs encapsulating KIF11 siRNAs or control luciferase siRNAs were then given to mice at 4 mg/kg dosage by tail-vein injection. Forty-eight hours later, spleens and bone marrows were harvested and analyzed. It has been well known that KIF11 knockdown results in the mitotic arrest phenotype with syntelically oriented chromosomes that can be easily observed in proliferating cells (Supplementary Figure S2).15 Indeed, when spleen and bone marrow tissue samples were fixed and stained by hematoxylin and eosin (H&E), the characteristic mitotic arrest phenotype from KIF11 knockdown could be clearly observed in many cells (Figure 5a), allowing rapid evaluation of in vivo KIF11 siRNA delivery.
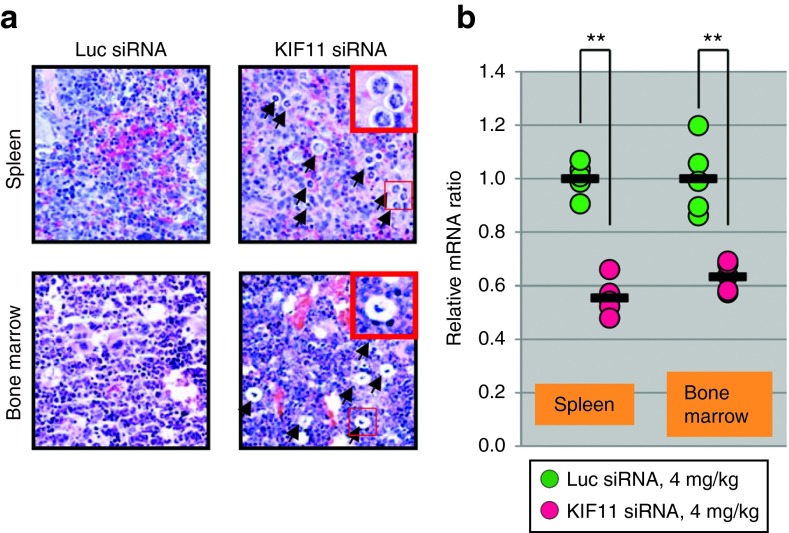
Figure 5.
Efficient siRNA delivery to mouse hematopoietic tissues in vivo with alkylated DMA-containing SNALP-like lipid nanoparticles (SLPs). (a) Examination of syntelically oriented chromosomes, characteristic of KIF11 knockdown, from representative spleen and bone marrow sections of mice at 48 hours after receiving one tail-vein injection of KIF11 siRNA encapsulated in SLP301R. Arrows point to irregular mitotic figures resulted from KIF11 knockdown. Some part of sections from spleen and bone marrow were enlarged to provide a better view. (b) Quantitative RT-PCR analysis of mouse KIF11 mRNA expression in spleens and bone marrows (n = 5) from mice 48 hours after receiving one tail-vein injection of control luciferase siRNA and KIF11 siRNA encapsulated with SLP401. KIF11 mRNA knockdown was measured by quantitative RT-PCR. P value is calculated using a Student's t-test.
To further quantify in vivo KIF11 gene knockdown by SLPs, spleen and bone marrow tissue samples were subjected to qRT-PCR analysis. As shown in Figure 5b, we were able to detect modest knockdown in both spleen (45%, P < 0.01) and bone marrow (37%, P < 0.01) tissues.
A correlation between Caveolin expression and siRNA cell entry
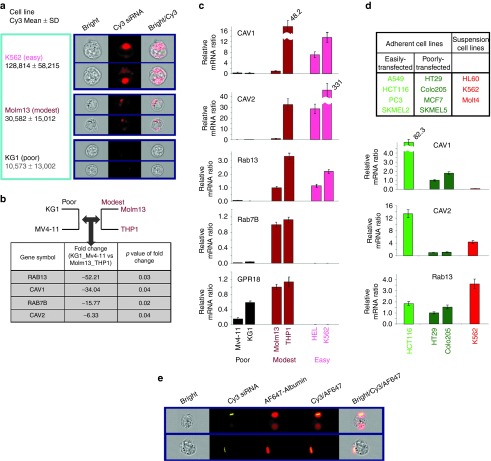
As shown in Figure 3b, two leukemia cell lines Mv4-11 and KG1 are insensitive to transfection by SLP-301R. They are also refractory to transfection by improved SLP401-SLP601 (data not shown), implicating a cell specific transfection barrier. There are two critical events for siRNAs to knockdown target mRNA in cells, siRNA entry into cells through endocytosis and siRNA release from endosomes. To understand which of these two events may account for the poor transfection in SLP-insensitive leukemia cell lines, we first measured the cellular uptake of Cy3-labeled siRNAs encapsulated in SLP301R across three representative leukemia cell lines (Figure 6a). The mean Cy3 florescent intensity inside the cell correlates well with transfection ability of these three cell lines (Figure 3b), with highest intensity in K562 cells (easily transfected by SLPs), fourfold less in Molm13 cells (modestly transfected by SLPs) and about 12-fold less in KG1 cells (poorly transfected by SLPs) (Figure 6a, left panel). Two representative images of Cy3-siRNA uptake by each cell line are shown in Figure 6a right panel. This result suggests that siRNA cellular entry through endocytosis could be the limiting event for siRNA knockdown by our SLPs.
Figure 6.
Expression of Caveolin 1 and 2 correlates with efficient siRNA transfection with alkylated DMA-containing SNALP-like lipid nanoparticles (SLPs). (a) Transfection efficiency of three tested leukemia cell lines, K562 (easily transfected), Molm13 (modestly transfected) and KG1 (poorly transfected), correlates with the amount of siRNAs entering into cells. Cy3-labeled control luciferase siRNAs were transfected into K562, Molm13 and KG1 cells using SLP301R. The cellular entry of siRNA was measured by quantitative fluorescent imaging (ImageStreamX, Amnis) at 2 hours after transfection. Mean Cy3 florescent intensity with SD was shown on the left. Two representative images from each cell line were shown on the right. (b) Four endocytosis-related genes were identified to be significantly underexpressed in poorly transfected KG1 and Mv4-11 cells compared with modestly transfected Molm13 and THP1 cells by comparative microarray analysis (Supplementary Table S2). (c) The expression levels of candidate genes identified in microarray were confirmed by quantitative RT-PCR in cell lines as indicated. Cav1, Cav2, and Rab13 were confirmed as underexpressed genes in poorly transfected KG1 and Mv4-11 cells compared with modestly and easily transfectd cell lines Molm13, THP1, HEL, and K562. (d) Upper panel, three groups of cell lines including easily transfected and poorly transfected adherent cell lines as well as hardest-to-transfect suspension leukemia cells, were subjected to comparative microarray analysis (Supplementary Tables S3 and S4). Lower panels, Cav1 and Cav2 were confirmed by quantitative RT-PCR as overexpressed genes in easily transfected adherent cell line HCT116, as compared with poorly transfected adherent cell lines HT29 and Colo205, and suspension leukemia cell line K562. (e) Colocalization of siRNA and Caveoloae. Cy3-labeled control luciferase siRNAs encapsulated in SLP301R were coadministered into K562 cells with Alexa647-labeled Albumins, which have been known to enter cells through Caveolae-mediated endocytosis. The cellular entry of siRNAs and Albumins was measured by quantitative fluorescent imaging (ImageStreamX, Amnis) at 30 minutes after administration. Two representative colocalization images were shown.
To search for endocytosis-related genes that may account for different SLP transfection efficiency, a comparative microarray analysis was performed between poorly transfected leukemia cell lines Mv4-11 and KG1, and modestly transfected leukemia cell lines Molm13 and THP1. There are about 170 genes that demonstrate at least twofold difference in their expression level between these two cell sets (Supplementary Table S2). Among them, four endocytosis-related genes, Caveolin 1 (Cav1) and 2 (Cav2), Rab13 and Rab7B (Figure 6b), and seven lipid metabolism-related genes, GPR18, GPR183, LGR7, SMPDL3A, SPTLC2, CPVL, and C13ORF18, were picked for further confirmation by qRT-PCR across all six leukemia cells we tested previously, including easily transfected K562 and HEL (Figure 3b). As shown in Figure 6c, the expression level of three genes, Cav1, Cav2, and Rab13, but none of the other genes, correlated to siRNA delivery efficiency, with higher average expression in modestly and easily transfected Molm13, THP1, K562, and HEL cells, but less expression in poorly transfected Mv4-11 and KG1 cells.
To further validate the correlation between expression of Cav1, Cav2, and Rab13 and SLP transfection efficiency, we expanded our microarray analysis on three more groups of cell lines, including easily transfected and poorly transfected adherent cell lines as well as suspension leukemia cells which are generally hardest-to-transfect (Figure 6d, upper panel). Cav1 and Cav2 are highly expressed in easily transfected group (Supplementary Tables S3 and S4). qRT-PCR results on representative cell lines from each group confirmed that the sum expression of Cav1 and Cav2, but not Rab13, correlates well with SLP transfection efficiency (Figure 6d, lower panels). Primary human umbilical vein endothelial cells are well documented to express high level of caveolins.22 As shown in Supplementary Figure S4, when these cells were tested for SLP transfection in comparison with Molm13 leukemia cells, they are at least 10 times more transfectable than Molm13 by SLPs, agreeing with our prediction.
Blood and neuronal cells are well known as poorly transfected. Using the tissue gene expression dataset published from Zlotnik's group,23 we found that Cav1 and Cav2 expression in bone marrow and all brain regions is much lower than that in the other tissues (Supplementary Figure S5), further supporting the correlation between Cav1 and Cav2 expression levels and transfection responsiveness.
Caveolins are the major protein component of caveolae, a subset of lipid rafts located at the cell surface. They have been implicated in the endocytosis of albumin and other proteins.24 To investigate the possibility that siRNAs may be endocytosed by caveolaes, we took advantage of fluorescently labeled albumins. It has been well documented that albumins enter cells through caveolae-mediated endocytosis. When caveolin-abundant K562 cells were treated with both Cy3-labeled siRNAs and Alexa647-labeled albumins, clear colocalization of siRNA and albumin fluorescence was observed near the cell surface membranes, where caveolae locate. Therefore, siRNAs encapsulated in SLPs can enter cells not only through well-known clathrin-mediated endocytosis, but also through caveolae-mediated endocytosis. To further elucidate the roles of Caveolins during SLP transfection, we attempted to modulate Caveolin expression by siRNA knockdown or gene overexpression in easily transfected K562 and modestly transfected Molm13 leukemia cells. However, any significant change of Caveolin expression level in these two cells led to massive cell death, consistent with a previous report.25 Therefore, a deep understanding of the role of Caveolins during SLP transfection awaits further studies in Caveolin knockout mice and MEF cells.
Discussion
Although lipid-containing reagents have been widely used for transfecting siRNAs into adherent cells, success in suspension cells has been limited. Therefore, use of RNAi in suspension cells relies on electroporation or viral delivery.26,27,28,29 Recently, several reports have outlined limited success in delivering siRNAs into suspension blood cells, including use of cell penetrating peptides,30 antibody-targeted nanoparticles,31,32 exosomes,33 CpG-conjugated siRNA,34 β1,3-D-glucan-encapsulated siRNA particles,35 and minicells.36 However, these particles are often challenging to make and suffer from potency, stability, and toxicity issues. At least a few hundred nmol/l of siRNA is required for in vitro knockdown. Therefore, the overall performance requires several areas of improvement.
Here, we report the discovery of a novel LNP-based siRNA delivery system, SLP, to efficiently deliver siRNAs into leukemia cell lines in vitro. When tested in adherent cells, SLP shows superiority over commercial reagents. Our SLPs are straightforward to make, stable, and produce little toxicity to cultured cells. They serve as important tools for in vitro biological studies across both adherent and suspension cell lines, as demonstrated by our FLT3 siRNA study. In addition, our mice studies demonstrate that these SLPs deliver siRNAs in vivo into mouse hematopoietic tissues, namely spleen and bone marrow. These data provide an important starting point and rationale for improvements aimed at siRNA therapies. Interestingly, we established that the inclusion of alkylated-DMA lipids enables dramatic improvements in the SLP performance. At the optimal concentration, alkylated lipids are able to significantly modulate the biophysical properties of DMA-containing SLPs. It will be interesting to see in the future whether the similar strategy can be applied to LNPs made from different cationic lipids. When we were preparing this manuscript, Leuschner et al. reported that LNPs made of a previously optimized cationic lipid are also able to deliver siRNAs in vivo into mouse splenic monocytes.37,38 It will be important to test whether these LNPs are able to efficiently deliver siRNAs into leukemia cells.
In the past, several hypotheses have been proposed to explain why blood cells are harder to transfect than adherent cells including potential differences in the lipid composition of plasma membranes and the volume of cytoplasm.12 However, it has been difficult to study any mechanism without a positive transfection result in blood cells. Using comparative microarray analysis of SLP-responsive and SLP-resistant leukemia cell lines combined with qRT-PCR validation, we found that elevated Cav1 and Cav2 mRNA expression in leukemia cell lines correlated with SLP transfection success. We subsequently showed evidence to support the correlation between Caveolin expression and successful siRNA delivery. First, Cav1 and Cav2 expression is >100-fold higher in routinely used adherent cells compared with hard-to-transfect leukemia cells, correlating with the robust siRNA transfection in adherent cells. Second, Cav1 and Cav2 are expressed much higher in easily transfected than modestly transfected adherent cell lines. Third, neuronal cells, although adherent, are well known as poorly transfected. They have very low Cav1 and Cav2 levels, similar to blood cells. Fourth, siRNAs are able to enter cells through caveolae-mediated endocytosis as indicated by colocalization studies.
Caveolae-mediated endocytosis, together with clathrin-mediated endocytosis, macropinocytosis and phagocytosis, are previously well-studied endocytotic pathways in the gene delivery field.24,39 A given formulation may carry plasmid DNAs into cells by multiple endocytotic pathways, but only one of them may yield functional gene expression.40 Functional use of these pathways varies depending on formulation compositions and cell types.41 Generally, functional DNA transfection by PEI (cationic polymer polyethyleneimine) polyplexes seems mainly mediated by caveolae,42 whereas lipoplexes transfection is mediated by clathrin.43 Regarding siRNA delivery, the functional transfection mechanism for LNPs is likewise complex. The involvement of different endocytotic pathways is dependent on the LNP formulation compositions. For example, siRNAs encapsulated in LNPs based on a pH-sensitive cationic lipid enter cells through both clathrin-mediated endocytosis and macropinocytosis,44 whereas siRNAs encapsulated in LNPs based on a cationic lipidoid enter cells through macropinocytosis.45 In this study the transfection efficiency of siRNAs encapsulated in alkylated-DMA-containing SLPs correlates markedly with Cav1 and Cav2 mRNA expression. In supporting the role of caveolae in functional siRNA delivery, Lu et al. showed earlier that siRNAs encapsulated in the commercial Dharmafect1 lipoplex reagent are able to enter cells through both clathrin-mediated and caveolae-mediated endocytosis. However, depleting cholesterol from the plasma membrane, but not blocking clathrin-mediated endocytosis, abolished target knockdown. Given that Caveolins are well-known cholesterol-binding proteins, caveolae-mediated endocytosis is the likely candidate responsible for functional siRNA delivery by the Dharmafect lipoplex reagent.46 Several recent reports also support caveolae-mediated endocytosis playing the major role in functional siRNA delivery by using PEI polyplexes, magnetic nanoparticles, and polysorbitol-based polyplexes.47,48,49 Therefore, increasing the efficiency for caveolae-mediated endocytosis of siRNAs may enable more effective siRNA delivery approaches.
Currently, the majority of efforts around siRNA therapy have been centered on liver-related diseases because most formulated siRNAs are quickly trapped in the liver, leading to target gene knockdown in hepatocytes and accessory cells of the liver.14 In phase I clinical trials recently completed by Alnylam Pharmaceuticals, LNPs encapsulating VEGF and KIF11 siRNAs demonstrated promising target knockdown and antitumor activity in liver cancers and liver metastases.50 Our alkylated DMA-containing SLPs, by potentially engaging Caveolin, can efficiently deliver siRNA into suspension leukemia cells and mouse hematopoietic tissues. These SLPs will serve as valuable tools for future biological research, provide a strong platform of in vivo optimization paving the way for the development of siRNA therapeutics against leukemia, as well as understand the underlying biology for siRNA cellular delivery.
Materials and Methods
SLP preparation. Dlin-KC2-DMA: 2-(2,2-di((9Z,12Z)-octadeca-9,12-dienyl)-1,3-dioxolan-4-yl)-N,N-dimethylethanamine; DLin-KC2-ClMDMA: N-(chloromethyl)-2-(2,2-di((9Z,12Z)-octadeca-9,12-dienyl)-1,3-dioxolan-4-yl)-N,N-dimethylethanamonium chloride; DLin-KC2-TMA: 2-(2,2-di((9Z,12Z)-octadeca-9,12-dienyl)-1,3-dioxolan-4-yl)-N,N,N-trimethylethanamonium iodide; DLin-KC2-DMEA: 2-(2,2-di((9Z,12Z)-octadeca-9,12-dienyl)-1,3-dioxolan-4-yl)-N-ethyl-N,N-dimethylethanamonium iodide; PEG-c-DOMG: R-3-[(ω-methoxy-poly(ethyleneglycol)2000)carbamoyl]-1,2-dimyristyloxy-propyl-3-amine, were synthesized in Roche Chemistry Department. DPPC: 1,2-dipalmitoyl-sn-glycero-3-phosphocholine; and Cholesterol, were purchased from Avanti Polar Lipids.
Lipid portion of each formulation was dissolved in absolute ethanol. In a separate vial (borosilicate, Schott Fiolax Clear) with magnetic stirring bar, the siRNA portion was diluted to an appropriate concentration using either nuclease-free water (Qiagen, Valencia, CA), citrate buffer (20 mmol/l, pH 4.0), or 5% Dextrose. The lipid-ethanol solution was then added with rapid stirring to the aqueous siRNA-containing solution resulting in a final ethanol concentration of 10–50%. The injection rate of the lipid-ethanol solution is controlled by a syringe pump (Harvard Apparatus). After standing at room temperature for 3–24 hours, under argon and shielded from the light, the formed LNP suspension was transferred to 10,000 MWCO dialysis cassettes (Slide-A-Lyzer, Thermo Scientific) and dialyzed against either phosphate-buffered saline or 5% Dextrose for 12–24 hours at room temperature. The LNPs were filtered using a 0.22 micron syringe filter (Millex-GV, Millipore) before use.
Measurement of particle size and Zeta potential. Particle size and Zeta potential were determined using a Malvern Zetasizer Nano-ZS Instrument. For particle size measurements, the LNPs were diluted by a factor of 1 to 100 (20 µl diluted in 2,000 µl buffer) using either phosphate-buffered saline, 5% dextrose (D5W), or 20 mmol/l citrate buffer (pH 4.0). Light scattering measurements were performed at 25 °C in polystyrene cuvettes. For Zeta potential measurements, the LNPs were diluted using nuclease-free water using a dilution factor of 1–100 (20 µl diluted in 2,000 µl water).
Cell culture. The human adherent cancer cell lines, PC3, Colo205, HCT116, and HT-29 (ATCC, Manassas, VA); human leukemia cell lines, MV-4;11, K562, KG1, HEL, THP1 (ATCC); and MOLM13 (DSMZ, Braunschweig, Germany), were maintained in corresponding media (DMEM for adherent cell lines and RPMI for leukemia cell lines) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen). Human umbilical vein endothelial cells were purchased from Lonza and cultured for 6 days in the Lonza-optimized medium.
Transfection. PC3 (2 × 104 cells/ml), HCT116 (3 × 105 cells/ml), Colo205 (3 × 105 cells/ml), or leukemia cells (2 × 105 cells/ml) were seeded 24 hours before transfection in 24-well plates. siRNAs were formulated in SLPs and directly added to the medium for transfection at indicated siRNA concentrations. Cells were then collected for RNA quantification.
SiRNA sequences. Human KIF11 siRNA : ucGAGAAucuAAAcuAAcudTsdT (sense strand) and AGUuAGUUuAGAUUCUCGAdTsdT (antisense strand).
Firefly luciferase siRNA : cuuAcGcuGAGuAcuucGAdTsdT (sense strand) and UCGAAGuACUcAGCGuAAGdTsdT (antisense strand).
Lower case letters are 2′-O-methyl nucleotides; upper case letters are RNA nucleotides; s indicates a phosphorothioate linkage; dT is deoxythymidine. These siRNAs showed no detectable stimulation of IFN-α, TNF-α, and IL-6 secretion when formulated and incubated with human peripheral blood mononuclear cells.
Gene knockdown assessment. Sample collection and mRNA purification for in vitro studies were performed as follows. Adherent cells were lysed directly on the plate with RNA lysis buffer (Qiagen). Suspension cells were collected into tubes and spun down at 2,000 rpm for 1 minute, and then the cell pellets were lysed with RNA lysis buffer (Qiagen). Sample collection and mRNA purification for in vivo studies were performed as follows. Tissues or tumors were collected from mice and immediately frozen in liquid nitrogen. Then these samples were lysed with RNA lysis buffer (Qiagen) by homogenization.
Total RNA from all collected samples was purified using Qiagen RNeasy Kit following the manufacturer's protocol. Relative quantification of target mRNA and 18S ribosomal RNA gene expression was carried out with cDNA Reverse Transcription Reagents from Applied Biosystems followed by Taqman Gene Expression Assays (Applied Biosystems, Foster City, CA) using the manufacturer's protocol. The catalog numbers for each probe set were human KIF11 (Hs00946303_m1), mouse KIF11 (Mm01204225_m1), and 18S (4319413E).
Comparative gene microarray analysis. For the statistical analysis of the expression measurements, RMA algorithm was used to perform the background correction, normalization, and signal summarization. The gene expression profile for each cell line (Molm13, THP1, Mv4-11, and KG1) represents the average value of four independent replicates. The differential gene expression list between the “easily transfected” and “modestly transfected” is obtained by two sample t-test. In addition, three groups of cell lines, “easily transfected adherent” (A549, HCT116, PC3, SKMEL2), “poorly transfectd adherent” (HT29, Colo205, MCF7, SKMEL5), and “leukemia” (HL60, K562, Molt4) are assembled from NCI-60 panel based on their siRNA transfection properties we experienced. The comparisons among these groups are conducted using one-way analysis of variance and the resulting gene lists are filtered by the following criteria: fold change >2, P value <0.05, signal intensity of lower group <100 and that of higher group >250.
In vivo administration. Athymic nude mice were purchased from Charles River Laboratories (Sulzfeld, Germany). All animal experiments were done in accordance with protocols approved by the Institutional Animal Care and Use Committees. SLPs were dosed at 4 mg/kg SLPs via tail-vein injection. Five mice were used in each dosing group. For histopathology assessment, the indicated tissues were harvested at 48 hours after injection and fixed in 10% neutral buffered formalin overnight, processed, paraffin embedded, sectioned at 5 µm, and stained with hematoxylin and eosin. For mRNA knockdown assessment, the indicated tissues were harvested at 48 hours after injection and frozen in liquid Nitrogen.
SUPPLEMENTARY MATERIAL Figure S1. None of the major commercial siRNA transfection reagents tested successfully delivers siRNA into suspension K562 leukemia cells. Figure S2. KIF11 knockdown leads to characteristic syntelically oriented chromosomes in proliferating cells. Figure S3. Efficient siRNA delivery to mouse livers in vivo with alkylated DMA-containing SLPs. Figure S4. Caveolin-abundant HUVEC cells are highly transfectable by SLP501-formulated siRNAs in comparison to Caveolin-low Molm13 leukemia cells. Figure S5. mRNA expression level of Cav1, Cav2, and Rab13 across multiple normal human tissues based on the tissue gene expression dataset published from Zlotnik's group. Table S1. Composition of SLP401-SLP601 with biocompatible polymers. Table S2. Genes differentially expressed between modestly transfected leukemia cell lines (THP1 and Molm13) and poorly transfected leukemia cell lines (Mv4-11 and KG1). Table S3. Genes differentially expressed between easily transfected adherent cell lines (A549, HCT116, PC3, and SKMEL2) and leukemia cell lines (HL60, K562, and Molt4). Table S4. Genes differentially expressed between easily transfectd adherent cell lines (A549, HCT116, PC3, and SKMEL2) and poorly transfected adherent cell lines (HT29, Colo205, MCF7, and SKMEL5).
Acknowledgments
We thank Giuseppe Ciccone, Preeti Yadava, Farooq Qureshi, Jiakui Li, and Denise Biondi for technical assistance and helpful discussions. All authors were employees of Hoffmann-La Roche Inc. when performing the work reported in this manuscript.
Supplementary Material
References
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Iorns E, Lord CJ, Turner N, Ashworth A. Utilizing RNA interference to enhance cancer drug discovery. Nat Rev Drug Discov. 2007;6:556–568. doi: 10.1038/nrd2355. [DOI] [PubMed] [Google Scholar]
- Dykxhoorn DM, Lieberman J. Running interference: prospects and obstacles to using small interfering RNAs as small molecule drugs. Annu Rev Biomed Eng. 2006;8:377–402. doi: 10.1146/annurev.bioeng.8.061505.095848. [DOI] [PubMed] [Google Scholar]
- Gindy ME, Leone AM, Cunningham JJ. Challenges in the pharmaceutical development of lipid-based short interfering ribonucleic acid therapeutics. Expert Opin Drug Deliv. 2012;9:171–182. doi: 10.1517/17425247.2012.642363. [DOI] [PubMed] [Google Scholar]
- Rossi JJ. RNAi therapeutics: SNALPing siRNAs in vivo. Gene Ther. 2006;13:583–584. doi: 10.1038/sj.gt.3302661. [DOI] [PubMed] [Google Scholar]
- Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- MacLachlan I.2007Liposomal formulations for nucleic acid delivery. Crooke ST.(ed.). Antisense Drug Technology Principles, Strategies, and Applications, 2nd edn CRC Press [Google Scholar]
- Merkerova M, Klamova H, Brdicka R, Bruchova H. Targeting of gene expression by siRNA in CML primary cells. Mol Biol Rep. 2007;34:27–33. doi: 10.1007/s11033-006-9006-x. [DOI] [PubMed] [Google Scholar]
- Larsen HØ, Roug AS, Nielsen K, Søndergaard CS, Hokland P . Nonviral transfection of leukemic primary cell lines by siRNA-a direct comparison between nucleofection and accell delivery. Exp Hematol. 2011;39:1081–1089. doi: 10.1016/j.exphem.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett JC, Rossi JJ. RNA-based therapeutics: current progress and future prospects. Chem Biol. 2012;19:60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan JB, et al. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol Cell. 2005;16:3187–3199. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chono S, Li SD, Conwell CC, Huang L. An efficient and low immunostimulatory nanoparticle formulation for systemic siRNA delivery to the tumor. J Control Release. 2008;131:64–69. doi: 10.1016/j.jconrel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed ME, Hoffman AS, Stayton PS. Smart polymeric carriers for enhanced intracellular delivery of therapeutic macromolecules. Expert Opin Biol Ther. 2005;5:23–32. doi: 10.1517/14712598.5.1.23. [DOI] [PubMed] [Google Scholar]
- Plank C, Oberhauser B, Mechtler K, Koch C, Wagner E. The influence of endosome-disruptive peptides on gene transfer using synthetic virus-like gene transfer systems. J Biol Chem. 1994;269:12918–12924. [PubMed] [Google Scholar]
- Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- Walters DK, Stoffregen EP, Heinrich MC, Deininger MW, Druker BJ. RNAi-induced down-regulation of FLT3 expression in AML cell lines increases sensitivity to MLN518. Blood. 2005;105:2952–2954. doi: 10.1182/blood-2004-07-2758. [DOI] [PubMed] [Google Scholar]
- Kiyoi H, Shiotsu Y, Ozeki K, Yamaji S, Kosugi H, Umehara H, et al. A novel FLT3 inhibitor FI-700 selectively suppresses the growth of leukemia cells with FLT3 mutations. Clin Cancer Res. 2007;13 15 Pt 1:4575–4582. doi: 10.1158/1078-0432.CCR-07-0225. [DOI] [PubMed] [Google Scholar]
- Lisanti MP, Scherer PE, Vidugiriene J, Tang Z, Hermanowski-Vosatka A, Tu YH, et al. Characterization of caveolin-rich membrane domains isolated from an endothelial-rich source: implications for human disease. J Cell Biol. 1994;126:111–126. doi: 10.1083/jcb.126.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth RB, Hevezi P, Lee J, Willhite D, Lechner SM, Foster AC, et al. Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics. 2006;7:67–80. doi: 10.1007/s10048-006-0032-6. [DOI] [PubMed] [Google Scholar]
- Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- Sunaga N, Miyajima K, Suzuki M, Sato M, White MA, Ramirez RD, et al. Different roles for caveolin-1 in the development of non-small cell lung cancer versus small cell lung cancer. Cancer Res. 2004;64:4277–4285. doi: 10.1158/0008-5472.CAN-03-3941. [DOI] [PubMed] [Google Scholar]
- Tyner JW, Walters DK, Willis SG, Luttropp M, Oost J, Loriaux M, et al. RNAi screening of the tyrosine kinome identifies therapeutic targets in acute myeloid leukemia. Blood. 2008;111:2238–2245. doi: 10.1182/blood-2007-06-097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K, Elledge SJ, Hannon GJ. Lessons from Nature: microRNA-based shRNA libraries. Nat Methods. 2006;3:707–714. doi: 10.1038/nmeth923. [DOI] [PubMed] [Google Scholar]
- Bernards R, Brummelkamp TR, Beijersbergen RL. shRNA libraries and their use in cancer genetics. Nat Methods. 2006;3:701–706. doi: 10.1038/nmeth921. [DOI] [PubMed] [Google Scholar]
- Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods. 2006;3:715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- Eguchi A, Meade BR, Chang YC, Fredrickson CT, Willert K, Puri N, et al. Efficient siRNA delivery into primary cells by a peptide transduction domain-dsRNA binding domain fusion protein. Nat Biotechnol. 2009;27:567–571. doi: 10.1038/nbt.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer D, Zhu P, Carman CV, Lieberman J, Shimaoka M. Selective gene silencing in activated leukocytes by targeting siRNAs to the integrin lymphocyte function-associated antigen-1. Proc Natl Acad Sci USA. 2007;104:4095–4100. doi: 10.1073/pnas.0608491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer D, Park EJ, Morishita Y, Carman CV, Shimaoka M. Systemic leukocyte-directed siRNA delivery revealing cyclin D1 as an anti-inflammatory target. Science. 2008;319:627–630. doi: 10.1126/science.1149859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- Kortylewski M, Swiderski P, Herrmann A, Wang L, Kowolik C, Kujawski M, et al. In vivo delivery of siRNA to immune cells by conjugation to a TLR9 agonist enhances antitumor immune responses. Nat Biotechnol. 2009;27:925–932. doi: 10.1038/nbt.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aouadi M, Tesz GJ, Nicoloro SM, Wang M, Chouinard M, Soto E, et al. Orally delivered siRNA targeting macrophage Map4k4 suppresses systemic inflammation. Nature. 2009;458:1180–1184. doi: 10.1038/nature07774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDiarmid JA, Amaro-Mugridge NB, Madrid-Weiss J, Sedliarou I, Wetzel S, Kochar K, et al. Sequential treatment of drug-resistant tumors with targeted minicells containing siRNA or a cytotoxic drug. Nat Biotechnol. 2009;27:643–651. doi: 10.1038/nbt.1547. [DOI] [PubMed] [Google Scholar]
- Leuschner F, Dutta P, Gorbatov R, Novobrantseva TI, Donahoe JS, Courties G, et al. Therapeutic siRNA silencing in inflammatory monocytes in mice. Nat Biotechnol. 2011;29:1005–1010. doi: 10.1038/nbt.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci USA. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon C, Billiet L, Midoux P. Chemical vectors for gene delivery: uptake and intracellular trafficking. Curr Opin Biotechnol. 2010;21:640–645. doi: 10.1016/j.copbio.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Rejman J, Conese M, Hoekstra D. Gene transfer by means of lipo- and polyplexes: role of clathrin and caveolae-mediated endocytosis. J Liposome Res. 2006;16:237–247. doi: 10.1080/08982100600848819. [DOI] [PubMed] [Google Scholar]
- Douglas KL, Piccirillo CA, Tabrizian M. Cell line-dependent internalization pathways and intracellular trafficking determine transfection efficiency of nanoparticle vectors. Eur J Pharm Biopharm. 2008;68:676–687. doi: 10.1016/j.ejpb.2007.09.002. [DOI] [PubMed] [Google Scholar]
- van der Aa MA, Huth US, Häfele SY, Schubert R, Oosting RS, Mastrobattista E, et al. Cellular uptake of cationic polymer-DNA complexes via caveolae plays a pivotal role in gene transfection in COS-7 cells. Pharm Res. 2007;24:1590–1598. doi: 10.1007/s11095-007-9287-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billiet L, Gomez JP, Berchel M, Jaffrès PA, Le Gall T, Montier T, et al. Gene transfer by chemical vectors, and endocytosis routes of polyplexes, lipoplexes and lipopolyplexes in a myoblast cell line. Biomaterials. 2012;33:2980–2990. doi: 10.1016/j.biomaterials.2011.12.027. [DOI] [PubMed] [Google Scholar]
- Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- Sahay G, Querbes W, Alabi C, Eltoukhy A, Sarkar S, Zurenko C, et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat Biotechnol. 2013;31:653–658. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JJ, Langer R, Chen J. A novel mechanism is involved in cationic lipid-mediated functional siRNA delivery. Mol Pharm. 2009;6:763–771. doi: 10.1021/mp900023v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MA, Shin JY, Firdous J, Park TE, Choi YJ, Cho MH, et al. The role of osmotic polysorbitol-based transporter in RNAi silencing via caveolae-mediated endocytosis and COX-2 expression. Biomaterials. 2012;33:8868–8880. doi: 10.1016/j.biomaterials.2012.08.049. [DOI] [PubMed] [Google Scholar]
- Lim J, Clements MA, Dobson J. Delivery of short interfering ribonucleic acid-complexed magnetic nanoparticles in an oscillating field occurs via caveolae-mediated endocytosis. PLoS ONE. 2012;7:e51350. doi: 10.1371/journal.pone.0051350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaZebnik M.2012Endocytosis Mechanism of Poly- and Lipoplexes in siRNA Delivery in Hela Cells.Thesis, Graduate College of the University of Illinois at Urbana-Champaign.
- Tabernero J, Shapiro GI, LoRusso PM, Cervantes A, Schwartz GK, Weiss GJ, et al. First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov. 2013;3:406–417. doi: 10.1158/2159-8290.CD-12-0429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.