Abstract
The hepatitis B virus (HBV) is a DNA virus that can cause chronic hepatitis B (CHB) in humans. Current therapies for CHB infection are limited in efficacy and do not target the pre-existing viral genomic DNA, which are present in the nucleus as a covalently closed circular DNA (cccDNA) form. The transcription activator-like (TAL) effector nucleases (TALENs) are newly developed enzymes that can cleave sequence-specific DNA targets. Here, TALENs targeting the conserved regions of the viral genomic DNA among different HBV genotypes were constructed. The expression of TALENs in Huh7 cells transfected with monomeric linear full-length HBV DNA significantly reduced the viral production of HBeAg, HBsAg, HBcAg, and pgRNA, resulted in a decreased cccDNA level and misrepaired cccDNAs without apparent cytotoxic effects. The anti-HBV effect of TALENs was further demonstrated in a hydrodynamic injection-based mouse model. In addition, an enhanced antiviral effect with combinations of TALENs and interferon-α (IFN-α) treatment was observed and expression of TALENs restored HBV suppressed IFN-stimulated response element-directed transcription. Taken together, these data indicate that TALENs can specifically target and successfully inactivate the HBV genome and are potently synergistic with IFN-α, thus providing a potential therapeutic strategy for treating CHB infection.
Introduction
Chronic hepatitis B virus (HBV) infection is a serious health problem that affects ~350 million people worldwide.1The mechanism by which chronic infection is maintained remains obscured but is thought to be due, in part, to the persistence of the HBV covalently closed circular DNA (cccDNA). The cccDNA plays a key role in viral persistence, reactivation after treatment withdrawal, and drug resistance. It is the template for transcription of the pregenomic RNA (pgRNA) and other subgenomic RNAs that are translated to produce the viral proteins. The pgRNA can be also reverse-transcribed by the viral polymerase into relaxed circular (rc) DNA. Some of the rc-DNA is packaged, and some goes to the nucleus where it is converted to additional cccDNA.2 The current treatments of HBV infection with interferon α (IFN-α) or nucleot(s)ide analogues block the production of viral RNA and new viral DNA, but do not directly target the pre-existing viral genomic DNAs. This may be one of the reasons that the therapeutics are not very effective in elimination of HBV infections in chronic hepatitis B (CHB) patients. Control of the cccDNA level or its transcriptional activity is thought to be critical for successful treatment of HBV infection.3,4 However, the cccDNA accumulates and persists in the cell nucleus as a very stable episome with an ~30–50 days half-life5,6 and there is still no therapeutic available to specifically target this nuclear viral genomes.
The transcription activator-like effector (TALE) nucleases (TALEN), composed of a TALE DNA binding domain fused to the nonspecific FokI nuclease (N), are newly developed sequence-specific nucleases that could be used to introduce targeted double-stranded breaks in mammalian cells with high efficiency. The DNA specificity of the TALE proteins is mediated by a variable number (often 12–20) of tandem repeats of a 33–35-aminoacid repeat sequence. The repeats are nearly identical, with the exception of two variable amino acids at position 12 and 13, also referred to as the repeat variable diresidues, that referentially recognize one of the four DNA bases in the target site have been determined. Engineered TALENs function in pairs, that is, the two highly sequence-specific TAL effector nuclease units bind the target DNA and create a spacer region (often 12–19 bp) to allow the endonuclease domains to dimerize and create the double-stranded break.7,8,9,10,11 Hence, TALENs appears to have tremendous potential as molecular tools for cleavage of any desired DNA sequences.
Here, we have designed TALENs targeting the conserved regions of HBV genomic DNA. The effect of TALENs on the production of viral proteins, pgRNA and the intracellular cccDNA levels were examined in cells transfected with linear full-length HBV DNA and in a hydrodynamic injection-based mouse model. In addition, we explored the possibility of TALENs and IFN-α combination therapy for an enhanced antiviral effect. These studies may provide new insights into therapeutic approaches for treating chronic HBV infection.
Results
Design of TALENs specifically targeting to HBV genome
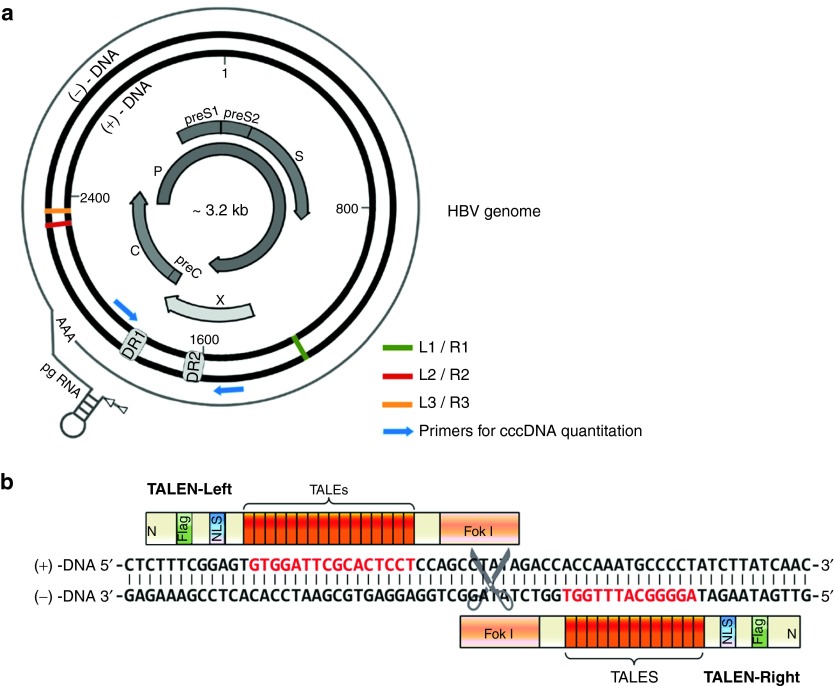
As a small hepatotropic DNA virus, HBV contains a small (~3.2 kb), circular, double-stranded DNA genome (Figure 1a). HBV is divided into different genotypes (A–H) according to the homogeneity of the viral sequence, which is based on an intergenotype divergence of >8% in the complete nucleotide sequence. HBV genotype A and D are more common in western European, whereas genotypes B and C are predominant in Asia.2,12 To ensure that the HBV genome-targeting TALENs are theoretical fit for multiple HBV genotypes and having a beneficial impact on the prevention of drug resistance, we performed sequence alignment among HBV isolates of genotypes A–D and TALENs were designed to target the highly conserved regions (Supplementary Figure S2), though it was not easy to find pairs of regions that are both conserved among all the four isolates with different genotypes. The schematic representation of a pair of TALENs targeting sequences on HBV genome were shown in Figure 1b. The pair of TALENs recognizing the regions around RNaseH (one out of the four domains of the viral polymerase protein) was named as L1/R1. In addition, the L2/R2 and L3/R3 recognize the DNA sequences in the core protein-coding region.
Figure 1.
TAL effector nucleases designed for specific targeting HBV genome. (a) Schematic representation of the HBV genome and the target sites of the TALENs. (b) Schematic of a pair of TALENs (L2/R2) bound to the target HBV DNA. HBV, hepatitis B virus; TALEN, transcription activator-like effector nucleases.
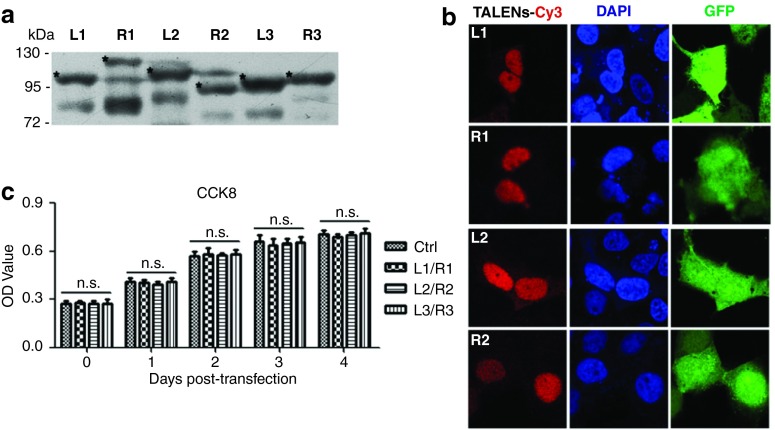
To examine the expression of the designed TALENs, the constructed plasmids encoding TALENs were transfected into the Huh7 cells. The expression of Flag-tagged TALENs was detected by immunoblotting (Figure 2a). Moreover, cells co-transfected with TALENs and pEGFP were subjected to the immunofluorescent analyses to observe the cellular localization of TALENs. The results showed that the designed TALENs, which contain a nuclear localization signal, were predominantly localized in the nucleus as expected while the green fluorescent protein (GFP) distributed in both cytoplasm and nucleus (Figure 2b).
Figure 2.
Expression of TALENs in cells. (a) Huh7 cells transfected with the plasmids encoding indicated Flag-tagged TALENs were lysed 48 hours post-transfection and the lysates were analyzed by immunoblotting with monoclonal antibody against Flag. (b) Huh7 cells were co-transfected with the plasmids encoding TALENs along with pEGFP. Immunofluorescence assay was performed 48 hours post-transfection. (c) The viability of the control cells and the cells expressing TALENs was measured by CCK8 assay every day for 4 days. TALEN, transcription activator-like effector nucleases.
We then examined the cytotoxicity of the TALENs-expression in cells using CCK8 assays. There was no obvious difference in cell viability and growth rates between the control cells and TALENs-expressing cells (Figure 2c). In addition, we blasted the viral sequence that TALENs targeted against human genomic DNA database and the results suggested that the TALEN-targeted sequences were highly specific for HBV and had almost no homology with the human DNAs, which also ruled out the possibility of off-target effects of the TALENs.
The effect of TALENs on the production of viral proteins and pgRNA
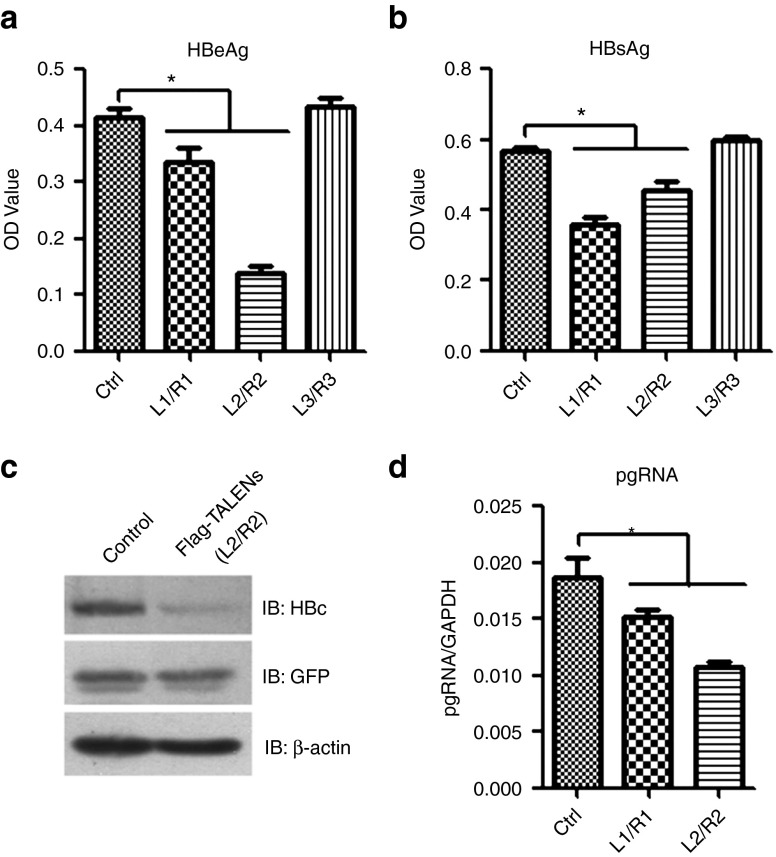
To evaluate the effect of TALENs on the viral protein production and the HBV pregenomic RNA transcription, we co-transfected Huh7 cells with the monomeric linear full-length HBV DNA (genotype A) and the plasmids expressing the TALENs pair. The transfection of plasmid-free linear HBV DNA into Huh7 cell lines allows to fully recapitulate the HBV replication cycle, including the nuclear generation of viable cccDNA.6 The transfection efficiency among different groups was stable as determined by the expression of co-transfected GFP (data not shown). As shown in Figure 3a,b, two out of the three pairs of designed TALENs significantly reduced the HBeAg and HBsAg production. Moreover, the degree of inhibition depends on the expression level of TALENs (Supplementary Figure S3a). We then examined the effect of TALENs on HBcAg production and the results showed that the HBcAg expression level was drastically decreased in TALENs-L2/R2 group compared with the control group (Figure 3c). We further compared the pgRNA production in control and TALENs-expressing cells and found that the expression of TALENs-L1//R1 and L2/R2 led to an inhibition of 20–50% in the level of pgRNA (Figure 3d).
Figure 3.
The effect of TALENs on the production of viral proteins and pgRNA. (a–d) Huh7 cells were co-transfected with the linear full-length HBV DNA (genotype A) and the plasmids encoding indicated TALENs. Forty-eight hours post-transfection, the supernatants were collected to detect HBeAg (a) and HBsAg (b) by ELISA and the cells were harvested for western blotting to detect the HBcAg expression (c) or for RT real-time PCR analysis to identify the level of pgRNA (d). ELISA, enzyme-linked immunosorbent assay; HBV, hepatitis B virus; TALEN, transcription activator-like effector nucleases.
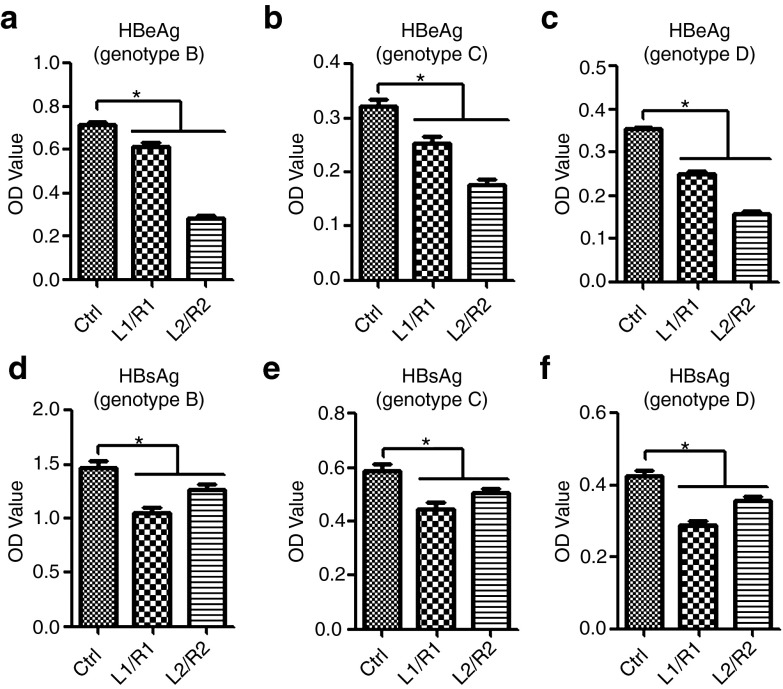
To compare the antiviral effect of TALENs among the different genotypes, we co-transfected cells with plasmids that contain complete HBV genome with different genotypes (B, C, or D) and plasmids encoding TALENs and then examined the level of HBeAg and HBsAg in the cell culture media. As expected, suppression of HBeAg and HBsAg production by TALENs was observed in all of the three groups transfected with plasmids containing HBV of different genotypes (Figure 4). These results suggested that TALENs targeting the certain regions of the HBV genome are effective in inhibiting viral RNA transcription and proteins production in cultured hepatic cells.
Figure 4.
Suppression of HBeAg and HBsAg production from different HBV genotypes by TALENs. (a–f) Huh7 cells were co-transfected with the plasmids encoding indicated TALENs and pHBV1.3-AF100309-genotype B (a,d), pHBV1.3-AF411411-genotype C (b,e), or pCMV-HBV-ayw-genotype D (c,f). Forty-eight hours post-transfection, the supernatants were collected to detect HBeAg and HBsAg by ELISA. ELISA, enzyme-linked immunosorbent assay; HBV, hepatitis B virus; TALEN, transcription activator-like effector nucleases.
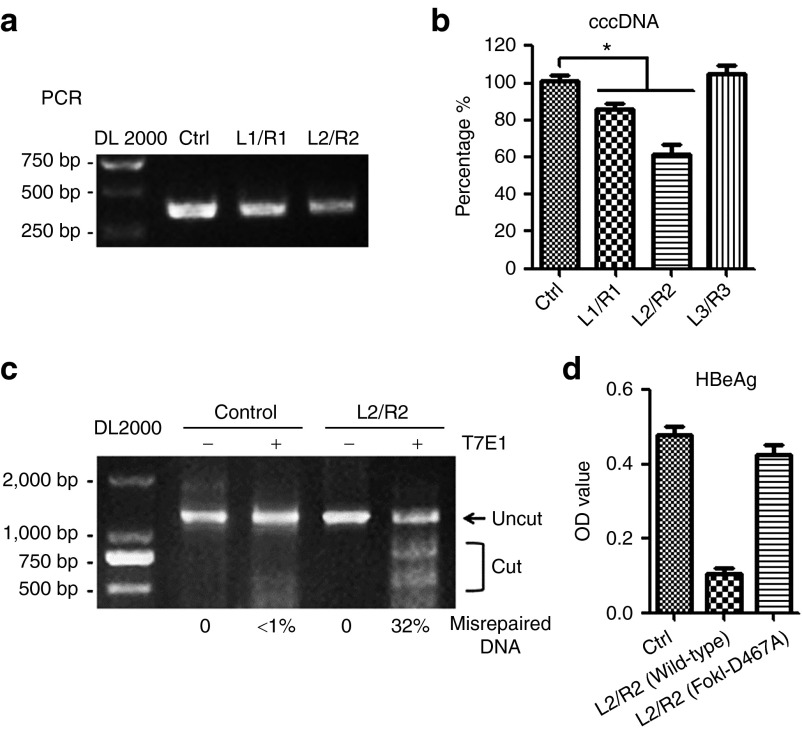
TALENs result in a decreased cccDNA level and misrepaired cccDNA
As the HBV cccDNA accumulates in cell nuclei and acts as a template for the transcription of viral genes and plays a key role in the development of persistent infection by HBV, we further assessed the effect of TALENs on the intracellular cccDNA levels. The amounts of the cccDNA in Huh7 cells transfected with the linear HBV DNA were measured by a semi-quantitative PCR (Figure 5a) and the real-time PCR (Figure 5b) using a pair of primers specific for cccDNA that across two gaps of HBV relaxed circular DNA. The results showed that the level of cccDNA decreased ~10–20% in TALENs-L1/R1- and twofold in TALENs-L2/R2-expressing cells. The inhibitory effect of TALENs-L2/R2 on cccDNA-based viral HBeAg production was confirmed in HBV-replicating HepAD38 cells (Supplementary Figure S3b), in which the HBV expression is under the control of tetracycline-off promoter and the HBeAg production was suggested to be dependent upon cccDNA.13
Figure 5.
TALENs result in a decreased cccDNA level and misrepaired cccDNA. Huh7 cells were co-transfected with the linear full-length HBV DNA and the plasmids encoding indicated TALENs. (a,b) The levels of intracellular cccDNA 48 hours post-transfection were analyzed by PCR (a) and real-time PCR (b) using primers for cccDNA quantitation. (c) The cccDNA was extracted from the control and TALENs-expressing cells as described in methods. The DNA segments encompassing the recognition sites of TALENs-L2/R2 were PCR-amplified, and the DNA amplicons were melted and annealed. The annealed DNA was treated with five units of the mismatch-sensitive T7 endonuclease 1 (T7E1) for 15 minutes at 37 °C and then assessed by agarose gel electrophoresis. (d) Huh7 cells were co-transfected with the linear full-length HBV DNA and indicated plasmids. Forty-eight hours post-transfection, the supernatants were collected to detect HBeAg by ELISA. ELISA, enzyme-linked immunosorbent assay; HBV, hepatitis B virus; TALEN, transcription activator-like effector nucleases.
We further tested whether the inhibition of the viral transcription by TALENs is through its directly binding to the viral genomic DNA. We introduced a point mutation to the TALE fused endonuclease FokI (D467A) to eliminate its ability to cleave substrate DNA.14 The data showed that TALENs with no endonuclease activity hardly inhibited the HBeAg and HBcAg expression (Figure 5d, Supplementary Figure S5), suggesting that the inhibitory effect of TALENs is largely dependent on the FokI endonuclease activity.
To assess the endonuclease-dependent genome editing activities of TALENs, cells co-transfected with HBV genome and TALENs-L2/R2 were lysed to extract cccDNA and tested in the T7E1 assay. As shown in Figure 5c, amplified DNA fractions from TALENs-treated cells were cleaved by T7E1, indicating that TALENs did target the sites.
Evaluation of the anti-HBV activity of TALENs in a hydrodynamic-based mouse model
We further substantiated the anti-HBV activity of TALENs in a mouse model. C3H/HeN mice were hydrodynamically-injected with monomeric linear full-length HBV DNA and control vectors or plasmids encoding the TALENs-L2/R2.The levels of serum HBeAg and HBsAg at 3 and 6 days post-transfection were determined and the results showed that TALENs significantly decreased both HBeAg and HBsAg production (Figure 6a,b). In addition, reduced level of the serum HBV DNA and liver HBV pgRNA was observed in the TALENs group compared with the control group (Figure 6c–d). Immunoblotting and immunohistochemical staining analysis in livers of mice further revealed that less core proteins were produced in TALENs group (Figure 6e, Supplementary Figure S4a) while no significant difference in liver histology was observed between the control and TALENs groups (Supplementary Figure S4b), suggesting that TALENs effectively interfere with viral transcription and replication in the livers without apparent cytotoxic effects.
Figure 6.
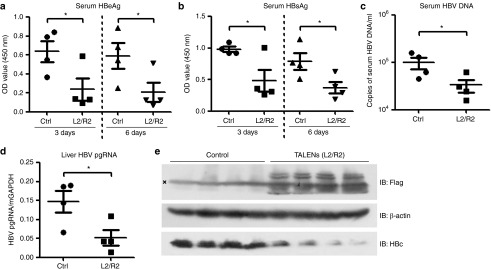
Evaluation of the anti-HBV activity of TALENs in a hydrodynamic-based mouse model. The mice were hydrodynamically-injected with the linear full-length HBV DNA and the control vector (n = 4) or the plasmids encoding TALENs-L2 and -R2 (n = 4). (a,b) The levels of serum HBeAg (a) and HBsAg (b) of the mice were determined by ELISA at 3 and 6 days post-transfection. (c,d) The levels of serum HBV DNA (c) and intrahepatic pgRNA (d) were measured by quantitative real-time PCR. (e) Lysates of the mice liver samples were analyzed by western blotting to detect the expression of Flag-TALENs and the HBcAg. HBV, hepatitis B virus; TALEN, transcription activator-like effector nucleases.
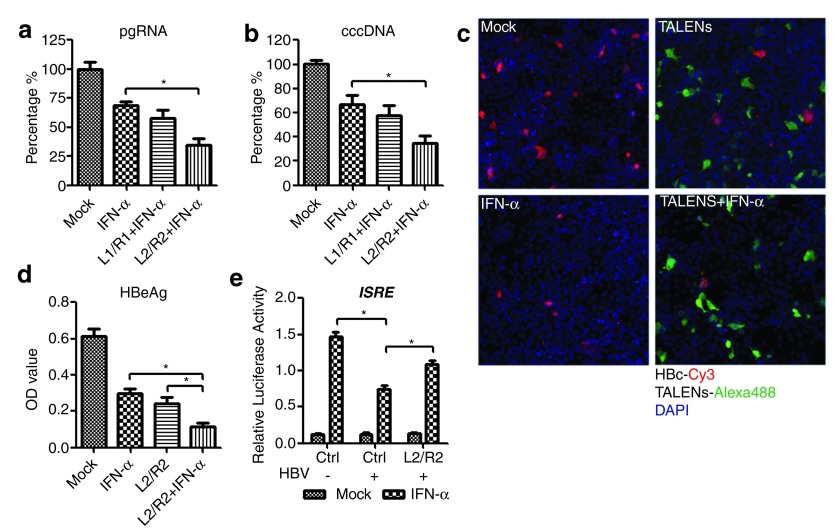
Enhanced antiviral effect with combinations of TALENs and IFN treatment
As a cytokines with antiviral activities, IFN-α is able to inhibit HBV replication and has been approved for treatment of patients with chronic HBV infection. Up to now, attempts to increase the rate of viral suppression and HBsAg loss by administering a combination of nucleot(s)ide analogues and pegIFN-α have been unsuccessful.15 To determine whether combinations of TALENs and IFN-α treatment would elicit a synergistic effect in inhibiting HBV transcription and reducing the cccDNA level, we added IFN-α to cells that co-transfected with HBV DNA and TALENs at 36 hours post-transfection and collected the cells to analyze the pgRNA production at 72 hours post-transfection. The results showed that IFN-α treatment alone has already reduced the level of pgRNA in cells while a roughly twofold enhancing effect on pgRNA reduction was observed in TALENs co-treatment cells (Figure 7a). The level of cellular cccDNA was also significantly decreased in cells transfected with TALENs followed by IFN-α treatment for 36 hours (Figure 7b). Moreover, we found that the TALENs-L2/R2 had comparable, even better, efficacy than IFN-α in reducing the HBcAg and HBeAg production and the combination of TALENs and IFN-α is more effective than using either of them individually (Figure 7c,d). Considering that we previously found that HBV-encoded polymerase proteins are able to suppress the IFN-α signaling pathway16 and TALENs were shown to reduce the production of pgRNA that encodes the polymerase, we compared the IFN-α-induced responses between cells co-transfected with control plasmids and plasmids encoding TALENs and the results showed that the IFN-α-induced IFN-stimulated response element (ISRE) promoter activation was inhibited in the presence of HBV but was restored by ectopic expression of TALENs-L2/R2 (Figure 7e). Taken together, these results confirm the anti-HBV activity of TALENs and suggest a synergistic effect of TALENs and IFN-α on inhibiting HBV transcription.
Figure 7.
Enhanced antiviral effect with combinations of TALENs and IFN treatment. (a–b) Huh7 cells were co-transfected with the linear full-length HBV DNA and the plasmids encoding indicated TALENs. 1,000 IU/ml IFN-α was added to the cells at 36 hours post-transfection. At 72 hours post-transfection, the cells were extracted for pgRNA quantitation (a), or the cells from indicated group were lysed for cccDNA extraction followed by real-time PCR using primers for cccDNA quantitation (b). (c–d) Huh7 cells were co-transfected with the linear full-length HBV DNA and the plasmids encoding L2/R2 or the control vectors. 1,000 IU/ml IFN-α was added to the cells at 36 hours post-transfection. At 72 hours post-transfection, the cells were fixed to perform immunofluorescence staining (c) and the supernatants of the cells were collected to detect HBeAg by ELISA (d). (e) Huh7 cells co-transfected with pISRE-Luc and the indicated plasmids were treated with 1,000 IU/ml IFN-α- or mock-treated for 12 hours at 48 hours post-transfection and then harvested for the luciferase assay using the Dual-Luciferase Assay Kit (Promega). ELISA, enzyme-linked immunosorbent assay; HBV, hepatitis B virus; TALEN, transcription activator-like effector nucleases.
Discussion
Current therapies for CHB infection with nucleot(s)ide analogues and IFN-α can block the formation of new viral DNAs, but do not directly affect the pre-existing cccDNA pool.17,18,19 In this study, we have presented an alternative approach to target the HBV genomic DNAs by using the sequence-specific cleavage enzymes TALENs.8,9,10,11 The expression of TALENs targeting the conserved regions of the viral DNA in HBV-replicating cells significantly reduced the production of pgRNA and proteins and the amounts of cccDNA. Moreover, an enhanced antiviral effect was observed when TALENs were used in combination with IFN-α. These data collectively suggest a therapeutic potential of TALENs in treatment of chronic HBV infection.
Theologically, there are three possible mechanisms for the TALENs to work: (i) cleavage of the HBV DNA, (ii) inducing mutation of HBV DNA, and (iii) binding to HBV DNA to disrupt its transcription and replication. Our results showed that a TALEN mutant with no endonuclease activity lost most of its ability to inhibit the HBeAg and HBcAg expression (Figure 5d and Supplementary Figure S5). The possibility (i) and (ii) thus appears to be the main mechanisms for the TALENs-mediated inhibition of the viral transcription and replication. As it has been reported that the nuclease-mediated DNA cleavage will either lead to rapid degradation of the episomal DNA or, alternatively, to error-prone DNA repair by the non-homologous end-joining and homology directed repair pathway,8,11 it will be interesting to examine what is the situations in the TALENs-expressed HBV-replicating cells.
The inhibitory effect of TALENs that targeting conserved region of the HBV genomic DNA was found to be similar among four HBV genotypes (A–D). However, we found that the three pairs of TALENs had different effects on HBeAg/HBsAg production. This may be explained by the differences in the way of HBeAg and HBsAg transcription and the sites that targeted by TALENs, that is the viral 3.5- and 2.4/2.1-kb transcripts encode the core protein/HBeAg and HBsAg, respectively,2 and the L1/R1-targeting regions do not include the viral sequences encoding the HBeAg protein, while the targeting sites of L2/R2 located outside of the region of HBV DNA for the 2.4/2.1-kb viral RNA transcription (Figure 1a). Intriguingly, the target sites of the L3/R3 located near to that of the L2/R2 (Supplementary Figure S2), however, L3/R3 has little effect on neither HBeAg nor HBsAg production. We speculated that this may be due to the unique structure of the cccDNA minichromosomes.6 Since it was reported that there is a nucleosome distribution pattern on DHBV cccDNA and the nucleosomes along viral cccDNA in the minichromosomes are not random but sequence-specifically positioned,20 it is possible that the target sites of the TALENs-R3 were overlapped with the nucleosome binding to the cccDNA and thus the binding of R3 proteins to the cccDNA is blocked. These results suggest that the efficacy of a given pair of TALENs highly depends on its target sequences on the HBV DNA and it requires further design and test to optimize the most efficient TALENs against HBV.
In addition to the cell model, we confirmed the anti-HBV activity of TALENs in the hydrodynamic injection-based mouse model. Interestingly, the decreased level of viral proteins and pgRNA was much higher in the mouse model than in the cell lines. The better effect of TALENs in vivo may be due to hydrodynamic injection delivering plasmids and the HBV DNA into the nucleus more efficiently than in vitro transfection does. In addition, since the HBV-encoded viral proteins including HBsAg and HBeAg and others were reported to be able to counteract the pattern recognition receptors-mediated innate immune responses,21,22,23,24,25 we speculate that the suppression of HBeAg and HBsAg production by TALENs may restore the host innate immune response in vivo, which will contribute to the increased clearance of HBV. It should be noted that both of the cell and mice model used in this study are somewhat different from the real conditions of chronic HBV infection. It is thus necessary to study whether TALENs could bind to the viral minichromosomes and to evaluate the efficiency of TALENs under more natural infection conditions in the future.
Here we also showed an enhanced antiviral activity with combination TALENs and IFN-α treatment. The mechanism by which IFN-α inhibits the HBV replication has not been clearly clarified but is thought to be dependent on the induction of IFN-α-stimulated genes such as APOBEC3G,26,27 MxA,28 and MyD88.29 However, viruses, including HBV, have evolved various strategies for circumventing the IFN response.30 We and others have previously showed that the HBV-polymerase suppresses the IFN responses16 and the HBV-core can inhibit the IFN-induced MxA production.31,32 In this study, we found that the expression of TALENs-L2/R2 partially restored the suppression of IFN-α-stimulated ISRE activation by HBV (Figure 7e), indicating that the decreased production of pgRNA that encodes both of the polymerase and core proteins in TALENs-expressing cells may contribute to reactivate the type I IFN system. In addition, on one hand, TALENs have been suggested to make the double-stranded break on DNA and reduce the intracellular cccDNA level; on the other hand, IFNs, besides direct antiviral effect, could epigenetically regulate the transcriptional activity of the cccDNA,18 augment the DNA double-strand breaks33 which might promote the decay of the cleaved viral DNA, and IFN therapy for CHB patients with lower intrahepatic cccDNA and HBV DNA level may achieve a higher sustained virological response.34 These investigations collectively suggest a highly potential synergistic effect of combination treatment of TALENs and IFN-α on inhibiting and eliminating HBV infection.
Since the current treatments for CHB infection has obvious limitations, there have developed some new antiviral strategies for treating HBV infection. RNA interference- and RNaseP-associated external guide sequence-based approaches have been investigated35,36,37 but are limited because they do not directly eliminate the pre-existing HBV genomic DNAs. Recently, many techniques have been developed for selectively targeting genes.4,38,39 Zinc finger proteins and zinc finger nucleases have been used to target HBV genome. Designed ZFPs that binds to duck HBV DNA resulted in substantial reductions in viral RNA, proteins, and viral progeny in cell culture, however zinc finger proteins do not cleave DNA due to their lack of endonuclease activity.40 Designed zinc finger nucleases that directly target the viral genome showed site-specific cleavage leading to a reduction in pregenomic RNA levels.41 However, a side-by-side comparison between zinc finger nucleases and TALENs suggested that TALENs can be as effective as zinc finger nucleases in terms of genome editing activity but significantly less cytotoxic. Moreover, TALEN platform enables the design of nucleases with single-nucleotide specificity.11
Whatever anti-HBV strategies are once developed, they are all faced with the problem of delivery. Fortunately, the liver appears to be an organ favoring the induction of immune tolerance42 and is accessible to a number of nucleic acid delivery methods.43,44 Therefore, any novel anti-HBV therapy has an inherent advantage. The most-common gene delivery systems for hepatic gene transfer are recombinant adenovirus and adeno-associated virus vectors,45,46 which may also be used for delivering TALENs into the hepatocytes. Nevertheless, it requires much detailed studies to improve the TALENs-based anti-HBV efficiency47 and to explore the use of the combination of TALENs and current antivirals for the cure of chronic HBV infection.
Materials and Methods
Plasmids and regents. pCR.HBV.A.EcoRI was kindly provided by Prof. Massimo Levrero, which contains the ~3.2 kb complete HBV genome (genotype A, adw).6 To prepare the monomeric linear full-length HBV DNA for cell transfection, the plasmid was double digested with EcoRI and BglI (Fermentas) at 37 °C for 1 hour. The unit-length HBV linear DNA was gel-purified with a TIANgelMaxi purification Kit (Tiangen, China). The pHBV1.3-AF100309 (genotype B, adw)29 and pHBV1.3-AF411411 (genotype C),48 both of which contain a 1.3-fold-overlength genome of HBV under the control of the authentic HBV promoter, were constructed and provided by Prof. Yumei Wen and Dr. Yongxiang Wang. The pCMV-HBV (genotype D, ayw), a CMV promoter-controlled HBV expression construct, was a generous gift of Prof. Jianming Hu. The pEGFP vector encoding enhanced green fluorescent protein was from Invitrogen China. The reporter plasmid with a firefly luciferase gene under the control of ISRE promoter pISRE-Luc and the Renilla luciferase encoding plasmid pRL-TK were purchased from Stratagene and Promega China, respectively. SIDANSAI Biotechnology (China) helped to construct the three designed pairs of TALENs-expression plasmids, which were then identified by double endonuclease digestion (Supplementary Figure S1) and sequencing (Biosune, China).
The antibodies used for immunoblotting, immunofluorescence, and immunohistochemistry were as follows: anti-Flag (Sigma, China), anti-HBcAg for immunoblotting (Santa Cruz, China), anti-HBcAg for immunohistochemistry (Shanghai Long Island Biotech, China), anti-β-actin (Sigma), and peroxidase conjugated secondary goat anti-mouse antibodies (Amersham Biosciences, China). The CCK8 assay kit was obtained from Dojindo (Japan) to evaluate the cell viability. The levels of HBeAg and HBsAg in culture supernatants and in the sera of mice were determined using ELISA Kit from Kehua Biotech (China). Human recombinant IFN-α was purchased from Calbiochem China.
Cells and transfection. The human hepatoma cell line Huh7 was obtained from the Cell Bank of the Chinese Academy of Sciences. The cells were routinely cultured as described and were plated 12 hours before transfection and transfected with the indicated plasmids using Fugene HD (Roche) according to the manufacturer's instructions. The amounts of plasmids per well of 12-well plate included in each transfection were 300 ng of monomeric linear full-length HBV DNA, 300 ng of pTALEN-Left, 300 ng of pTALEN-Right, and 100 ng of pEGFP as a transfection marker.
HBV pgRNA and plasma viral load quantification. Total cellular RNA was extracted using the TRIzol kits (Tiangen, China) and reverse-transcribed using Toyobo Ace reverse transcriptase according to the manufacturer's instructions. The cDNA samples were subjected to real-time PCR using the following pgRNA specific primers: the forward primer 5′- GCCTTAGAGTCTCCTGAGCA-3′; the reverse primer 5′- GAGGGAGTTCTTCTTCTAGG-3′. The relative amount of pgRNA in each sample was normalized to that of GAPDH.
The amounts of HBV DNA in the sera of mice were determined by quantitative real-time PCR using the commercial test kits from Piji Biotech (China).
Extraction and quantification of HBV cccDNA. The extraction and quantification of the cccDNA was performed by using the method described by Belloni et al.18 with minor modifications. Briefly, Huh7 cells at 48 hours post-transfection with monomeric linear HBV DNA and TALENs or control vector were harvested and lysed in lysis buffer A (50 mmol/l Tris-HCl, pH 7.4, 1 mmol/l EDTA, 1% NP-40) and incubated 15 minutes at 4 °C. Lysates were centrifuged at 10,000g for 1 minute. Pelleted nuclei were resuspended in lysis buffer B (10 mmol/l Tris-HCl, 10 mmol/l EDTA, 150 mmol/l NaCl, 0.5 % SDS, 0.5 mg/ml proteinase K) and incubated overnight at 37 °C. Lysates were centrifuged at 10,000g for 20 minutes and the nucleic acids in supernatant were purified by phenolchloroform (1:1) extraction and ethanol precipitation. The DNA samples were treated with DpnI (Fermantas) at 37 °C for 1 hour and at 80 °C for 20 minutes. The DpnI-treated DNAs were further digested with Plasmid-safe DNase I (Epicentre, China ) at 37 °C for 45 minutes and at 70 °C for 30 minutes (Supplementary Figure S6).
Real-time PCR was performed in StepOneTM (Applied Biosystems, China) and the PCR reaction mixture (20 μl) contained 10 μl real-time PCR Master Mix (TOYOBO), 20 ng DNA, 0.3 μl (20 μmol/l) forward and reverse primers, and 0.06 μl FAM-5′-labeled probes (Invitrogen). The forward primer was 5′-CTCCCCGTCTGTGCCTTCT-3′ (nt 1548–1566); the reverse primer was 5′-GCCCCAAAGCCACCCAAG-3′ (nt 1903–1886); and the taqman probe was 5′-FAM-AGCGAAGTGCACACGGACCGGCAGA-TAMRA-3′. Amplification was performed as follows: 95 °C for 10 minutes, then 45 cycles of 95 °C for 10 seconds, 62 °C for 10 seconds, and 72 °C for 20 seconds. Serial dilutions of the plasmid pCR-HBV.A.EcoRI were used as quantification standards.
T7E1 assay. Huh7 cells cultured in the 12-well plates were transfected with plasmids encoding TALENs and the monomeric linear full-length HBV DNA. After 48 hours of incubation, the cccDNA was extracted from the cells. The DNA region encompassing the TALENs-L2/R2 target site (1548–2853) was amplified and then melted and annealed to form heteroduplex DNA. The annealed DNA was treated with five units of T7 endonuclease 1 (New England BioLabs, China) for 20 minutes at 37 °C and then analyzed by agarose gel electrophoresis. If the DNA amplicons contain both wild-type and mutated DNA sequences, heteroduplexes would be formed. T7E1 recognizes and cuts heteroduplexes, but not homoduplexes. The intensity of the DNA bands were quantitated using PhotoShop and the percentage of the TALENs-induced cleavage were calculated using the formula: fractional modification = 1 − (1 − (fraction cleaved))0.5 as described.49
Immunofluorescence. Cells transfected with indicated plasmids were plated into Lab-Tek coverglass chamber slides. 60–72 hours post-transfection, the cells were fixed in 4% paraformaldehyode, permeabilized by the addition of 0.1% Triton X-100 and blocked with 10% FBS for 2 hours. After incubation with primary antibodies, the cells were incubated with Alexa 488- or Cy3- coupled secondary Abs (Jackson Immunologicals, China) for 1 hour and then stained with the nuclear marker DAPI (Invitrogen) for 1 minute. Fluorescence was observed with a Leica SP5 II confocal microscope.
Animal studies. Male C3H/HeN mice were purchased from and maintained under specific pathogen-free conditions at the Center of Laboratory Animals, Shanghai Public Health Clinical Center. All mouse experiments were conducted in accordance with the guide for the care and use of medical laboratory animals (Ministry of Health, China). Mice were used at 7 weeks of age. Six micrograms of the monomeric linear full-length HBV DNA, 3+3 μg of plasmids encoding TALENs-Left+Right or equal amounts of control vector were diluted in 2 ml of PBS and injected into the tail veins of mice within 5 seconds. Three days after hydrodynamic injection, mice were bled by retro-orbital puncture, and the serum was assayed for HBeAg and HBsAg. On day 6, mice were bled for HBeAg, HBsAg, and viral load analyses and killed. Liver specimens from mice were either snap-frozen in liquid nitrogen for total RNA and protein extraction (extraction kit for tissue, Tiangen) or 4% formaldehyde fixed for HE- and immuno-staining (Pathology Department, Shanghai Public Health Clinical Center).
Statistics. Data were presented as mean ± SD. Statistical analyzes were performed by Student's t-test for comparison between two groups using GraphPad Prism software. A value of P < 0.05 (*) was considered statistically significant.
Author Contributions
J.C. designed research, collected data, and drafted the manuscript; W.Z. prepared the linear HBV DNA and did the qPCR analysis; J.L. helped to design and construct the TALENs; F.W. performed the immunofluorescence studies; M.W, Y.Z., and X.P. helped to perform the animal experiments; C.C. helped to culture the cells; J.L. revised the draft; and Z.Y. supervised the study.
SUPPLEMENTARY MATERIAL Figure S1. Construction of plasmids carrying the coding sequence of designed TALENs. Figure S2. Designed TALENs for conserved regions of viral DNA among different genotypes. Figure S3. Suppression of HBeAg production by TALENs. Figure S4. HBcAg expression and immunohistochemical analysis in the mice liver sections. Figure S5. The inhibitory effect of TALENs is largely dependent on the FokI endonuclease activity. Figure S6. The main steps of extraction and quantification of the intracellular HBV cccDNA.
Acknowledgments
We are grateful to Massimo Levrero, Yumei Wen, Yongxiang Wang and Jianming Hu for their providing plasmids. We also thank Xiaoyu Yu and Xiaohui Zhou for their assistance with the mice experiments. This study was supported by the National Key Basic Research Program of China (2012CB519000) and the “Twelfth Five-Year” National Key Technology Research and Development Programs of China (2012ZX10002007). The authors declared no conflict of interest.
Supplementary Material
References
- Lok AS, McMahon BJ. Chronic hepatitis B. Hepatology. 2007;45:507–539. doi: 10.1002/hep.21513. [DOI] [PubMed] [Google Scholar]
- Lee JY, Locarnini S. Hepatitis B virus: pathogenesis, viral intermediates, and viral replication. Clin Liver Dis. 2004;8:301–320. doi: 10.1016/j.cld.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Levrero M, Pollicino T, Petersen J, Belloni L, Raimondo G, Dandri M. Control of cccDNA function in hepatitis B virus infection. J Hepatol. 2009;51:581–592. doi: 10.1016/j.jhep.2009.05.022. [DOI] [PubMed] [Google Scholar]
- Schiffer JT, Aubert M, Weber ND, Mintzer E, Stone D, Jerome KR. Targeted DNA mutagenesis for the cure of chronic viral infections. J Virol. 2012;86:8920–8936. doi: 10.1128/JVI.00052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Yamamoto T, Cullen J, Saputelli J, Aldrich CE, Miller DS, et al. Kinetics of hepadnavirus loss from the liver during inhibition of viral DNA synthesis. J Virol. 2001;75:311–322. doi: 10.1128/JVI.75.1.311-322.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belloni L, Pollicino T, De Nicola F, Guerrieri F, Raffa G, Fanciulli M, et al. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci USA. 2009;106:19975–19979. doi: 10.1073/pnas.0908365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, 2nd, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cade L, Reyon D, Hwang WY, Tsai SQ, Patel S, Khayter C, et al. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 2012;40:8001–8010. doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowyer SM, Sim JG. Relationships within and between genotypes of hepatitis B virus at points across the genome: footprints of recombination in certain isolates. J Gen Virol. 2000;81 Pt 2:379–392. doi: 10.1099/0022-1317-81-2-379. [DOI] [PubMed] [Google Scholar]
- Zhou T, Guo H, Guo JT, Cuconati A, Mehta A, Block TM. Hepatitis B virus e antigen production is dependent upon covalently closed circular (ccc) DNA in HepAD38 cell cultures and may serve as a cccDNA surrogate in antiviral screening assays. Antiviral Res. 2006;72:116–124. doi: 10.1016/j.antiviral.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Waugh DS, Sauer RT. Single amino acid substitutions uncouple the DNA binding and strand scission activities of Fok I endonuclease. Proc Natl Acad Sci USA. 1993;90:9596–9600. doi: 10.1073/pnas.90.20.9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoulim F. Are novel combination therapies needed for chronic hepatitis B. Antiviral Res. 2012;96:256–259. doi: 10.1016/j.antiviral.2012.09.006. [DOI] [PubMed] [Google Scholar]
- Chen J, Wu M, Zhang X, Zhang W, Zhang Z, Chen L, et al. Hepatitis B virus polymerase impairs interferon-a-induced STA T activation through inhibition of importin-a5 and protein kinase C-d. Hepatology. 2013;57:470–482. doi: 10.1002/hep.26064. [DOI] [PubMed] [Google Scholar]
- Doong SL, Tsai CH, Schinazi RF, Liotta DC, Cheng YC. Inhibition of the replication of hepatitis B virus in vitro by 2',3'-dideoxy-3'-thiacytidine and related analogues. Proc Natl Acad Sci USA. 1991;88:8495–8499. doi: 10.1073/pnas.88.19.8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belloni L, Allweiss L, Guerrieri F, Pediconi N, Volz T, Pollicino T, et al. IFN-a inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J Clin Invest. 2012;122:529–537. doi: 10.1172/JCI58847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas J, Schorr O, Jamard C, Gibbs C, Trépo C, Hantz O, et al. Inhibitory effect of adefovir on viral DNA synthesis and covalently closed circular DNA formation in duck hepatitis B virus-infected hepatocytes in vivo and in vitro. Antimicrob Agents Chemother. 2002;46:425–433. doi: 10.1128/AAC.46.2.425-433.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Li S, Shen F, Li H, Qian S, Lee DH, et al. Characterization of nucleosome positioning in hepadnaviral covalently closed circular DNA minichromosomes. J Virol. 2012;86:10059–10069. doi: 10.1128/JVI.00535-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Meng Z, Jiang M, Pei R, Trippler M, Broering R, et al. Hepatitis B virus suppresses toll-like receptor-mediated innate immune responses in murine parenchymal and nonparenchymal liver cells. Hepatology. 2009;49:1132–1140. doi: 10.1002/hep.22751. [DOI] [PubMed] [Google Scholar]
- Wang S, Chen Z, Hu C, Qian F, Cheng Y, Wu M, et al. Hepatitis B virus surface antigen selectively inhibits TLR2 ligand-induced IL-12 production in monocytes/macrophages by interfering with JNK activation. J Immunol. 2013;190:5142–5151. doi: 10.4049/jimmunol.1201625. [DOI] [PubMed] [Google Scholar]
- Revill P, Yuan Z. New insights into how HBV manipulates the innate immune response to establish acute and persistent infection. Antivir Ther (Lond) 2013;18:1–15. doi: 10.3851/IMP2542. [DOI] [PubMed] [Google Scholar]
- Yu S, Chen J, Wu M, Chen H, Kato N, Yuan Z. Hepatitis B virus polymerase inhibits RIG-I- and Toll-like receptor 3-mediated beta interferon induction in human hepatocytes through interference with interferon regulatory factor 3 activation and dampening of the interaction between TBK1/IKKepsilon and DDX3. J Gen Virol. 2010;91 Pt 8:2080–2090. doi: 10.1099/vir.0.020552-0. [DOI] [PubMed] [Google Scholar]
- Chang J, Block TM, Guo JT. The innate immune response to hepatitis B virus infection: implications for pathogenesis and therapy. Antiviral Res. 2012;96:405–413. doi: 10.1016/j.antiviral.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Bonvin M, Achermann F, Greeve I, Stroka D, Keogh A, Inderbitzin D, et al. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology. 2006;43:1364–1374. doi: 10.1002/hep.21187. [DOI] [PubMed] [Google Scholar]
- Li J, Liu K, Liu Y, Xu Y, Zhang F, Yang H, et al. Exosomes mediate the cell-to-cell transmission of IFN-a-induced antiviral activity. Nat Immunol. 2013;14 8:793–803. doi: 10.1038/ni.2647. [DOI] [PubMed] [Google Scholar]
- Gordien E, Rosmorduc O, Peltekian C, Garreau F, Bréchot C, Kremsdorf D. Inhibition of hepatitis B virus replication by the interferon-inducible MxA protein. J Virol. 2001;75:2684–2691. doi: 10.1128/JVI.75.6.2684-2691.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lin S, Chen Q, Peng L, Zhai J, Liu Y, et al. Inhibition of hepatitis B virus replication by MyD88 involves accelerated degradation of pregenomic RNA and nuclear retention of pre-S/S RNAs. J Virol. 2010;84:6387–6399. doi: 10.1128/JVI.00236-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89 Pt 1:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Fernández M, Quiroga JA, Carreño V. Hepatitis B virus downregulates the human interferon-inducible MxA promoter through direct interaction of precore/core proteins. J Gen Virol. 2003;84 Pt 8:2073–2082. doi: 10.1099/vir.0.18966-0. [DOI] [PubMed] [Google Scholar]
- Rosmorduc O, Sirma H, Soussan P, Gordien E, Lebon P, Horisberger M, et al. Inhibition of interferon-inducible MxA protein expression by hepatitis B virus capsid protein. J Gen Virol. 1999;80 Pt 5:1253–1262. doi: 10.1099/0022-1317-80-5-1253. [DOI] [PubMed] [Google Scholar]
- Houghton JA, Morton CL, Adkins DA, Rahman A. Locus of the interaction among 5-fluorouracil, leucovorin, and interferon-alpha 2a in colon carcinoma cells. Cancer Res. 1993;53:4243–4250. [PubMed] [Google Scholar]
- Sung JJ, Wong ML, Bowden S, Liew CT, Hui AY, Wong VW, et al. Intrahepatic hepatitis B virus covalently closed circular DNA can be a predictor of sustained response to therapy. Gastroenterology. 2005;128:1890–1897. doi: 10.1053/j.gastro.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Uprichard SL, Boyd B, Althage A, Chisari FV. Clearance of hepatitis B virus from the liver of transgenic mice by short hairpin RNAs. Proc Natl Acad Sci USA. 2005;102:773–778. doi: 10.1073/pnas.0409028102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z, Xu Y, Wu J, Tian Y, Kemper T, Bleekmann B, et al. Inhibition of hepatitis B virus gene expression and replication by endoribonuclease-prepared siRNA. J Virol Methods. 2008;150:27–33. doi: 10.1016/j.jviromet.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia C, Chen YC, Gong H, Zeng W, Vu GP, Trang P, et al. Inhibition of hepatitis B virus gene expression and replication by ribonuclease P. Mol Ther. 2013;21:995–1003. doi: 10.1038/mt.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon MA, Rahdar M, Porteus M. Gene editing: not just for translation anymore. Nat Methods. 2012;9:28–31. doi: 10.1038/nmeth.1811. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman KA, Fischer KP, Joyce MA, Tyrrell DL. Zinc finger proteins designed to specifically target duck hepatitis B virus covalently closed circular DNA inhibit viral transcription in tissue culture. J Virol. 2008;82:8013–8021. doi: 10.1128/JVI.00366-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradick TJ, Keck K, Bradshaw S, Jamieson AC, McCaffrey AP. Zinc-finger nucleases as a novel therapeutic strategy for targeting hepatitis B virus DNAs. Mol Ther. 2010;18:947–954. doi: 10.1038/mt.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispe IN, Giannandrea M, Klein I, John B, Sampson B, Wuensch S. Cellular and molecular mechanisms of liver tolerance. Immunol Rev. 2006;213:101–118. doi: 10.1111/j.1600-065X.2006.00435.x. [DOI] [PubMed] [Google Scholar]
- Cuestas ML, Mathet VL, Oubiña JR, Sosnik A. Drug delivery systems and liver targeting for the improved pharmacotherapy of the hepatitis B virus (HBV) infection. Pharm Res. 2010;27:1184–1202. doi: 10.1007/s11095-010-0112-z. [DOI] [PubMed] [Google Scholar]
- Wooddell CI, Rozema DB, Hossbach M, John M, Hamilton HL, Chu Q, et al. Hepatocyte-targeted RNAi therapeutics for the treatment of chronic hepatitis B virus infection. Mol Ther. 2013;21:973–985. doi: 10.1038/mt.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Sun CP, Ma HI, Fang CC, Wu PY, Xiao X, et al. Comparative study of anti-hepatitis B virus RNA interference by double-stranded adeno-associated virus serotypes 7, 8, and 9. Mol Ther. 2009;17:352–359. doi: 10.1038/mt.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Kay MA, Finegold M, Stratford-Perricaudet LD, Woo SL. Assessment of recombinant adenoviral vectors for hepatic gene therapy. Hum Gene Ther. 1993;4:403–409. doi: 10.1089/hum.1993.4.4-403. [DOI] [PubMed] [Google Scholar]
- Schiffer JT, Swan DA, Stone D, Jerome KR. Predictors of hepatitis B cure using gene therapy to deliver DNA cleavage enzymes: a mathematical modeling approach. PLoS Comput Biol. 2013;9:e1003131. doi: 10.1371/journal.pcbi.1003131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YX, Wen YM, Nassal M. Carbonyl J acid derivatives block protein priming of hepadnaviral P protein and DNA-dependent DNA synthesis activity of hepadnaviral nucleocapsids. J Virol. 2012;86:10079–10092. doi: 10.1128/JVI.00816-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Zhao P, Mariano A, Han R. Targeted Myostatin Gene Editing in Multiple Mammalian Species Directed by a Single Pair of TALE Nucleases. Mol Ther Nucleic Acids. 2013;2:e112. doi: 10.1038/mtna.2013.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.