Abstract
Myotonic dystrophy type 1 (DM1) is caused by the expansion of (CTG)n in the 3′ untranslated region of the dystrophia myotonica-protein kinase (DMPK) gene, which is transcribed as (CUG)n repeats that accumulate in the nucleus. The RNA repeats specifically sequester or change the expression levels of several RNA-binding proteins, leading to aberrant splicing of many target genes. In this study, we developed artificial site-specific RNA endonucleases (ASREs) that specifically bind and cleave (CUG)n repeats RNA. We have generated one ASRE that can target the expanded RNA repeats in DM1 patient cells and specifically degrade the pathogenic DMPK messenger RNAs with minimal effect on wild-type alleles. Such ASRE treatment significantly decreased the number of nuclear foci in DM1 patient cells and can reverse the missplicing of many genes affected in DM1 patients. Taken together, the application of ASRE provides a new route of gene therapy for DM1 treatment.
Introduction
Myotonic dystrophy type 1 (DM1), the most common form of adult muscular dystrophy with a prevalence of ~1:8,000 worldwide, is an autosomal dominant disease with multisystemic symptoms, including myotonia, muscle wasting, cardiac conduction defects, insulin resistance, cataracts, and cognitive dysfunction (reviewed in ref. 1). DM1 is caused by the progressive expansion of (CTG) triplet in the 3′ untranslated region of dystrophia myotonica-protein kinase (DMPK) gene on chromosome 19q13.3.2,3 According to the prevailing model of DM1 pathogenesis, the triplet expansion is transcribed as toxic (CUG)n repeats RNA that accumulate in the nucleus as RNA–protein complex, which changes the levels of several RNA-binding proteins, including some important splicing factors.4,5
Two classes of splicing factors are well known to be affected by (CUG)n repeats: the Muscleblind-like family (MBNL1-3)6,7,8 and CUG-BP, Elav-like family (CELF).9,10 The members of MBNL family (e.g., MBNL1) have a high affinity for (CUG)n repeats and thus are sequestered in the nuclear foci, leading to a functional deficiency of these proteins.6,7,8 The levels of CELF proteins (e.g., CUGBP1) are upregulated in DM1 through a PKC-mediated phosphorylation event that stabilizes the protein.9,10 It was recently reported that a double-stranded RNA-binding protein, staufen1, is also increased in DM1 skeletal muscle and contributes to the splicing changes in DM1 cells.11 The changes of these splicing regulators interfere with the alternative splicing of hundreds of genes,12 including insulin receptor (IR),13 cardiac troponin T (cTNT),9,14 bridging integrator-1 (BIN1),15 sarcoplasmic/endoplasmic reticulum Ca2+-ATPase 1 (SERCA1)16,17, and more. Generally, the aberrant splicing caused the expression of embryonic isoforms in adult tissues of DM1 patients, resulting in symptoms that resemble developmental abnormality.18
Although some symptoms of DM1 can be alleviated, there is currently no cure for this disease. Specific therapy of DM1 is difficult because the toxic RNA repeats affect multiple factors that subsequently cause dysregulation of splicing in more than 100 proteins.12 Several therapeutic approaches of DM1 are currently under investigation. The first is to restore the misregulated splicing factors with gene therapy approaches using adeno-associated virus (AAV) vectors.19 The strength of such protein-based approaches is the efficient delivery of protein to muscle and heart by available gene therapy vectors (such as AAV). However, because multiple splicing factors are affected either positively or negatively in DM1 patients, conventional gene therapy approach cannot restore the normal level of all protein factors. The second strategy includes various antisense oligonucleotides–based methods to cleave (CUG)n repeats. This strategy has produced some promising results in cultured cells20,21 and in animal models.21,22,23,24 However, the delivery of oligonucleotides in the muscle and heart of DM1 patients, together with the balance of delivery efficiency and toxicity, will be a major challenge. Another therapeutic route is to identify small molecules that inhibit the binding of (CUG)n to MBNL1.25,26,27,28 Since these small molecules all bind to structured RNA repeats to release the MBNL1 sequestered by (CUG)n, they are fairly toxic, and additional work is needed to increase their selectivity to specific RNA repeats.
Here, we report a novel therapeutic strategy that uses artificial site-specific RNA endonucleases (ASREs) containing a specific RNA recognition domain (PUF domain of human PUM1) and an RNA endonuclease domain (PIN domain of human SMG6) to specifically bind and cleave (CUG)n repeats. We demonstrated that ASREs can decrease the levels of messenger RNA (mRNA) harboring long (CUG)n repeats and can significantly decrease the number of nuclear foci in DM1 patient cells. Expression of ASRE also reversed the missplicing of genes affected in DM1 patients. Taken together, this work represented a proof of principle for a new therapy that combines the strength of both protein- and oligonucleotide-based approaches.
Results
Generate the PUF domains that bind (CUG)n repeats
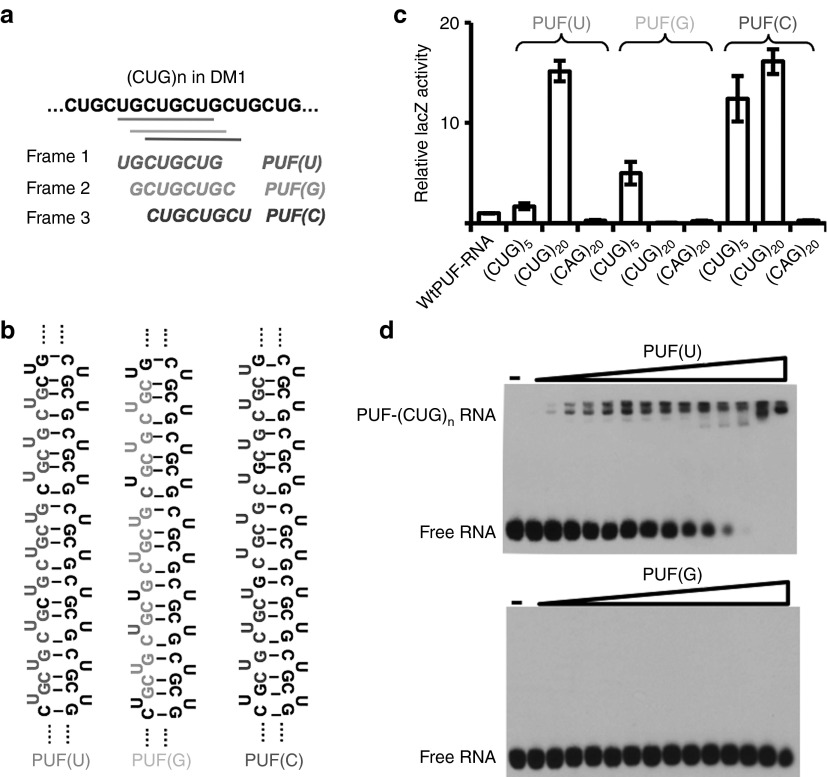
The ASREs were designed by combining the unique RNA recognition domain (PUF domain of human pumilio1) and a general RNA endonuclease domain (PIN domain of human SMG6).29 The PUF domain contains eight repeats that bind consecutive RNA bases in an antiparallel fashion. Each repeat uses two amino acids to recognize the Watson–Crick edge of the corresponding base and a third amino acid (Tyr, His, or Arg) to stack between adjacent RNA bases, generating a specific binding between a PUF repeat and the cognate base.30,31 The PUF repeat in natural protein can recognize the bases A, U, and G in a modular fashion.30 In addition, we and another group recently identified the amino acid combination of PUF to specifically recognize cytosine using a yeast three-hybrid screen.32,33 Completion of the unique RNA-binding code enabled us to reprogram the specificity of each PUF repeat to generate a designer domain that specifically recognizes any 8-nt RNA. The (CUG)n repeats contain three different RNA octamers (UGCUGCUG, GCUGCUGC, and CUGCUGCU) from different frames. Through stepwise mutagenesis on each PUF repeat, we generated three PUF domains, each recognizing one of the possible 8-nt in (CUG)n (Figure 1a; the first base of each sequence was used to name the PUF domain). Because the expanded (CUG)n repeats form a long hairpin in vitro based on the studies using electron microscopy and crystallography,34,35 the designer PUFs may recognize a series of 8-nt targets tandemly positioned along the hairpin structure (Figure 1b).
Figure 1.
Generation of PUF domains to recognize CUG repeats. (a) Diagram of PUF domain and their target 8-nt in CUG repeats. The (CUG)n repeats can be recognized as 8-nt by three different frames (different shades of gray coded for clarity), thus three possible PUF domains (named by the first base of their targets) can be generated to recognize three cognate targets. (b) The long (CUG)n repeats in myotonic dystrophy type 1 patients forms a double-stranded hairpin structure. The target 8-nt sequences for three PUF domains were shown in gray in the context of the RNA hairpin. (c) Measurement of RNA–protein interactions using yeast three-hybrid system. The binding of wild-type (WT) PUF-cognate RNA was used as a control. We tested each PUF with a short repeat of (CUG)5, a long repeat of (CUG)20, and a control repeat (CAG)20. (d) Electrophoretic mobility shift assay using biotin-labeled 12 CUG repeats. The PUF (U), but not PUF (G), can form stable complex with RNA repeats to causing the mobility shift of RNA.
We used a yeast three-hybrid system to assay for the specific binding between the PUF domains with their cognate RNA targets. We cotransfected each PUF with a short RNA repeat (CUG)5, a long repeat (CUG)20, and a control repeat (CAG)20 into the YBZ-1 strains and assayed for the binding affinity between different PUF–RNA pairs as relative β-galactosidase activity.32,36 The (CUG)5 contains one to two copies of the 8-nt targets in an unpaired conformation, whereas the (CUG)20 contains multiple 8-nt targets and forms into a hairpin structure that resembles the toxic RNA in DM1 cells. We found that all PUFs can specifically bind (CUG)5 RNA with an affinity of ~2–10 fold of the wild-type (WT) PUF–RNA pair, suggesting that these PUFs indeed can recognize the target as designed. As controls, none of three PUFs bound to noncognate (CAG)20 RNA with similar structure, indicating that the binding between the designer PUFs and the toxic (CUG)n repeat is specific. In addition, the binding affinity between (CUG)20 repeats and PUF(U) or PUF (C) is much higher (>15-fold) than the binding between WT PUF and its target (sample 1 in Figure 1c) as judged by LacZ activity.
However, PUF(U) and PUF(C), but not PUF(G), can bind to the long (CUG)20 repeats with high affinity. This observation suggested that the local structure of RNA target probably affects the recognition of RNA by PUF domain. PUF repeat recognizes its cognate RNA through direct contact of amino acids to the Watson–Crick edge of RNA bases.30,31,32 Therefore, this interaction is probably influenced by the accessibility of base: an RNA base in double-stranded RNA region is a poor PUF target because it is already paired with another base (i.e., the Watson–Crick edge is occupied), thus there may be a maximal number of paired bases that PUF can tolerate. The (CUG)n repeats form a hairpin structure in vitro with the U being unpaired,34 our results may suggest that the binding between PUF and RNA requires at least three unpaired nucleotides in the 8-nt target sequence (i.e., the maximal five paired–based can be tolerated; Figure 1b). In addition, there might be other possibilities, such as alternative structures of (CUG)n repeats or the binding of the repeats by other proteins or helicase, which are partially responsible for the difference observed.
The binding between PUFs and (CUG)n repeats RNA was further confirmed with electrophoretic mobility shift assay using the purified protein and in vitro transcribed (CUG)12 sequences. We found that PUF(U), but not PUF(G), can form stable complex with RNA repeats to cause the mobility shift of RNA (Figure 1d), consistent with the finding from yeast three-hybrid system (Figure 1c). The expression and purification of PUF(C) were attempted multiple times, but the protein was expressed poorly for unknown reason. This PUF(C) was expressed in extremely low level in both Escherichia coli and mammalian cells, thus we mainly used PUF(U) and PUF(G) for most of the remaining analyses.
Generation of ASREs to cleave (CUG)n repeats
To generate ASREs for (CUG)n repeats, we linked the designer PUF domains with the PIN domain using a short peptide linker. We also included a FLAG epitope tag for protein detection and a nuclear localization sequence (PKKKRKV) to facilitate the import of ARSEs into the nucleus where the (CUG)n repeats accumulate. The PIN domain of human SMG6 was chosen because it has a well-defined molecular architecture and requires only a divalent metal cation for RNA cleavage.37,38 As a control, we fused PUF(U) with an inactive PIN harboring a D1353A mutation in its active site.29,37 The resulting ASRE-(U), mutated ASRE-(U), ASRE-(G), and ASRE-(C) were used for further assays (Supplementary Figure S1).
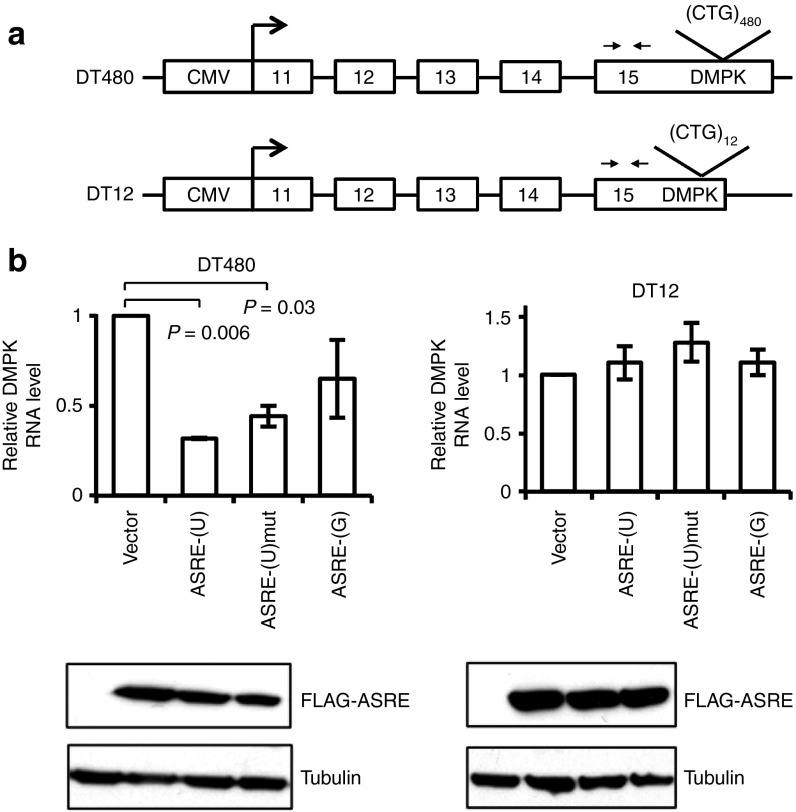
To determine whether ASREs can degrade (CUG)n repeats RNA in cultured cells, we first tested the ASREs in COS7 cells transiently transfected with a plasmid reporter DT480 containing DMPK exon 11–15 and 480 interrupted CTG repeats in exon 15 (Figure 2a).9,21 We found that ASRE-(U) caused >60% decrease in DT480 mRNAs by real-time reverse transcriptase–polymerase chain reaction (RT-PCR) and mutated ASRE-(U) with an inactive PIN domain led to a small decrease, whereas the ASRE-(G) had little effects (Figure 2b). Because of extremely low expression level, ASRE-(C) had little effects on the level of DT480-containing mRNAs (not shown).
Figure 2.
Artificial site-specific RNA endonucleases (ASREs) degrade CUG repeats in COS7 cells. (a) Diagram of the DT480 and DT12 minigene constructs. The minigenes contain exons 11–15 of human dystrophia myotonica-protein kinase gene and interrupted CTG repeats (DT480, 480 CTG repeats; DT 12, 12 CTG repeats) expressed by a cytomegalovirus (CMV) promoter. Primer pairs for reverse transcriptase–polymerase chain reaction (RT-PCR) are indicated by arrows. (b) The minigene construct and ASRE expression plasmid were cotransfected into COS7 cells, and the RNA levels were measured with real-time RT-PCR at 48 hours after transfection. The messenger RNA levels of GAPDH were also measured as internal controls, and the P values were calculated according to t-test (only significant difference was shown). The expressions of ASREs were detected using western blot and showed below each sample.
We also examined the effect of ASREs on the level of DMPK mRNAs containing normal number of (CUG)n repeats. We transiently transfected COS7 cells with ASREs and the DT12 reporter that contains DMPK exon 11–15 with 12 interrupted CTG repeats in exon 15 (Figure 2a). The real-time RT-PCR analysis showed that ASREs did not significantly affect the mRNA level of DT12, suggesting that the ASRE-catalyzed RNA degradation may be fairly slow and can only be observed with a large number of targets (Figure 2b).
Expression of ASREs degraded expanded (CUG)n repeats in DM1 cells
We further examined the cellular effect of designer ASREs on the (CUG)n repeats in DM1 patient cells. To convert broblasts into myoblasts, we transfected DM1 broblasts (GM04602 and GM04033), as well as a normal human foreskin fibroblast with a lentiviral vector encoding MyoD gene.39 The stably transfected cells were further transfected with ASRE-(U), ASRE-(G), ASRE-(C), or a mutated ASRE-(U) with an inactive PIN domain, and the resulting myoblasts were induced into myocytes with differentiation medium containing horse serum, insulin, and transferrin (Figure 3a). After a 10-day differentiation period, we analyzed the expression of ASREs using western blot (Figure 3b) and immunofluorescence (Figure 3c) with anti-FLAG antibody. We found that ASRE-(U), ASRE-(G), and mutated ASRE-(U) were robustly expressed in ~70% of cells, whereas ARSE-(C) has very low level of expression in patient-derived myocytes. As expected, the ASREs were predominantly expressed in the nucleus (Figure 3c). In addition, the expression of ASRE-(U), but not the control ASRE-(G) or mutated ASRE-(U), reduced the endogenous DMPK mRNA level by 40% in DM1 cells (Figure 3d). If assuming that DMPK is transcribed equally from both WT and the CUG-expansion mutation alleles, we could roughly estimate that at least 80% of the mutated DMPK transcripts were degraded. In the control experiments, ASRE-(U) has limited effects on the DMPK mRNA level in myocytes derived from normal fibroblasts through the transfection of MyoD followed by differentiation (Supplementary Figure S2).
Figure 3.
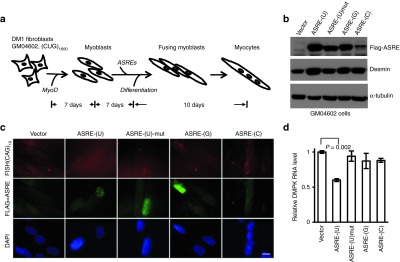
Artificial site-specific RNA endonuclease (ASRE) specifically reduces dystrophia myotonica-protein kinase (DMPK) messenger RNA (mRNA) levels with CUG repeats in myotonic dystrophy type 1 (DM1) cells. (a) Schematic representation of ASRE treatment in DM1 myocytes derived from patient fibroblasts. The DM1 patient fibroblasts (GM04602) were stably transfected with human MyoD and further cultured for 7 days to convert them into myoblasts. The resulting cells were stably transfected with ASREs, after incubation for 10 days, the myoblasts were induced into myocytes with differentiation medium, and the cells were harvested in another 10 days for further analyses. (b) The protein levels of ASREs were examined by western blot. The differentiation levels of myoblasts were determined by the expression of Desmin. As an internal control, the α-tubulin was detected. (c) The (CUG)n repeats in differentiated GM04602 cells were assayed by fluorescence in situ hybridization (FISH) using (CAG)10 probes (red), and the localization of ASRE was detected with immunofluorescence (green). As a negative control, the cells were transfected with expression vector only. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bar = 10 µm. (d) For the same cells described above, the mRNA levels of DMPK were examined with real-time reverse transcriptase–polymerase chain reaction. P values were calculated according to t-test (only significant difference was shown).
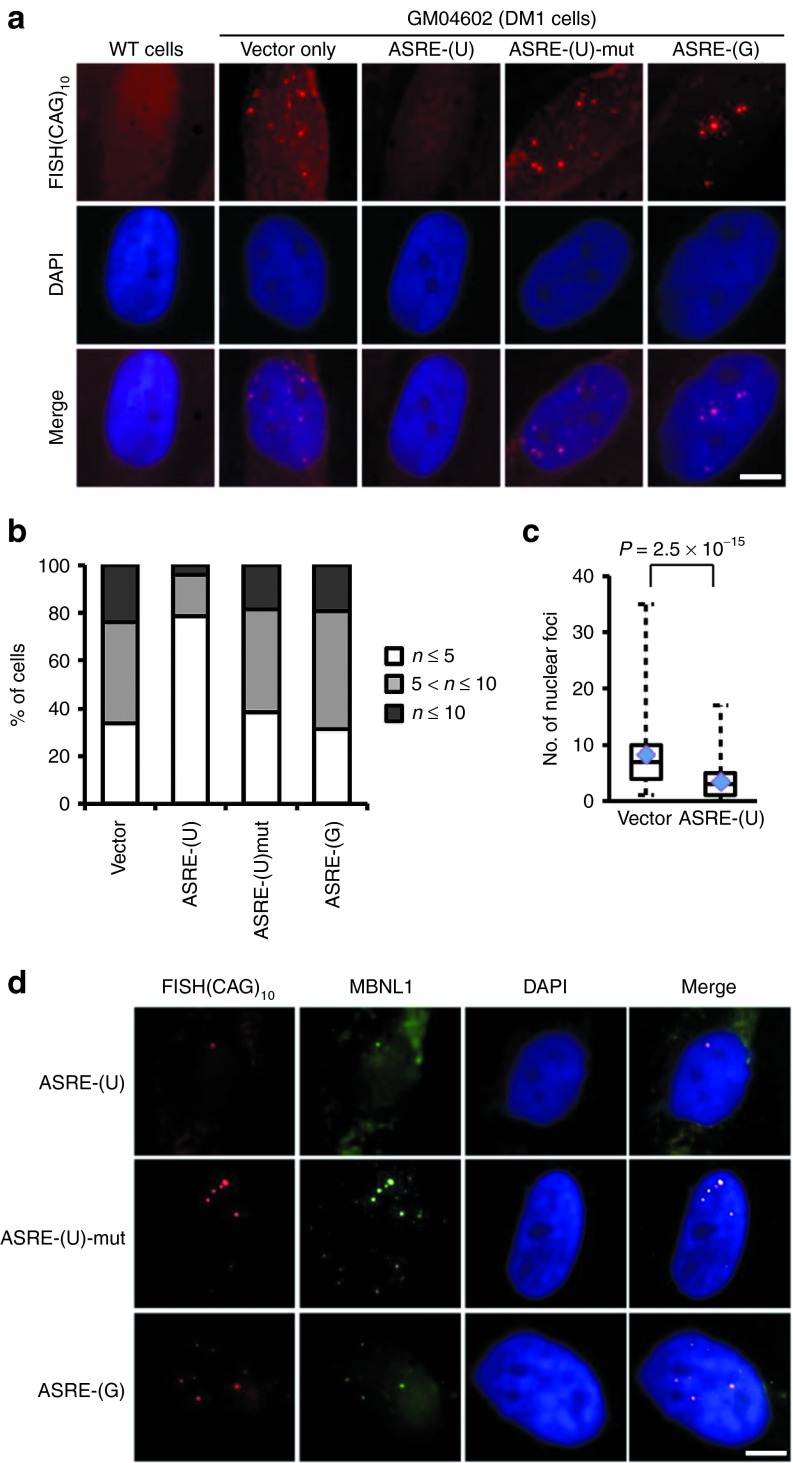
We also used an in situ hybridization with a Cy5-labeled (CAG)10 probe to detect (CUG)n repeats in DM1 cells. As expected, the DM1 cells had an average of 8–9 nuclear foci containing (CUG)n accumulation, whereas the normal cells had no detectable nuclear accumulation of (CUG)n (Figure 4a). When transfected with ASRE-(U), the number of nuclear foci in DM1 cells was significantly decreased, whereas the mutated ASRE-(U) had no detectable effect (Figure 4a). Importantly, the numbers of cells with at least five nuclear foci were decreased from 66 to 20% by ASRE-(U) treatment, and the majority of ASRE-treated cells had 3–4 foci (Figure 4b). In comparison, the expression of mutated ASRE-(U) or ASRE-(G) had no obvious effect on the numbers of (CUG)n containing nuclear foci (Figure 4a,b), consistent with the finding that PUF(G) did not bind long repeat of (CUG)n (Figure 1c,d). The reduction of nuclear foci by ASRE-(U) was statistically significant (P = 2.5 × 10−15, t-test), suggesting that this ASRE can indeed reduce the accumulation of toxic RNAs as designed. A small but statistically significant reduction of nuclear foci was also observed in another DM1 cell line (GM04033; Supplementary Figure S3), which further supported the effectiveness of ASRE as a general therapeutic reagent.
Figure 4.
Cellular effects of artificial site-specific RNA endonucleases (ASREs) in myotonic dystrophy type 1 patient cells. (a) GM04602 cells and normal human fibroblast cells were induced into myocytes, and then examined by by fluorescence in situ hybridization (FISH) using (CAG)10 probes. GM04602 cells were transfected with different ASREs or vector control. 4′,6-Diamidino-2-phenylindole (DAPI) was used to stain the nuclei. Scale bar = 5 µm. (b) The differentiated GM04602 cells transfected with vector or different ASREs were selected from random fields and counted for the number of nuclear foci. The percentages of cells with different number of nuclear foci were represented as a histogram. From left to right, n = 133, 126, 81, and 67. (c) The number of nuclear foci of differentiated GM04602 cells transfected with vector or ASRE-(U) were counted and represented as box plot. The average numbers were indicated by diamonds, and a significant decrease (P = 2. 5 × 10−15, t-test) of foci number were observed in ASRE-(U) treated cells. (d) For the same cells described above, the localization of MBNL1 proteins was detected with immunofluorescence (green). The (CUG)n repeats and the nuclear DNA were also detected with FISH and DAPI staining. Scale bar = 5 µm.
In addition, we also found that the expression of ASRE-(U) reduced the nuclear sequestration of MBNL1, as judged by immunofluorescence using MBNL1 antibody. The MBNL1 in DM1 cells usually are localized as nuclear foci with (CUG)n repeats, and we found that ASRE-(U), but not ASRE-(G) or mutated ASRE-(U), reduced such localization and made MBNL1 defused inside cells (Figure 4d).
ASREs reversed abnormal regulation of alternative splicing in human DM1 cells
The toxic RNA repeats in DM1 can affect multiple splicing factors (e.g., MBNL1 and CUGBP) that subsequently cause deregulated splicing in multiple mRNAs, and such splicing shift contributes to most of DM1 phenotypes. Given that we have succeeded in partially degrading mutant DMPK mRNAs and reducing the number of nuclear foci, we examined the effect on the deregulated splicing of several genes that are responsible for many DM1 phenotypes, including insulin resistance,13 T-tubule alterations and muscle weakness15, and cardiac conduction defects.9,14
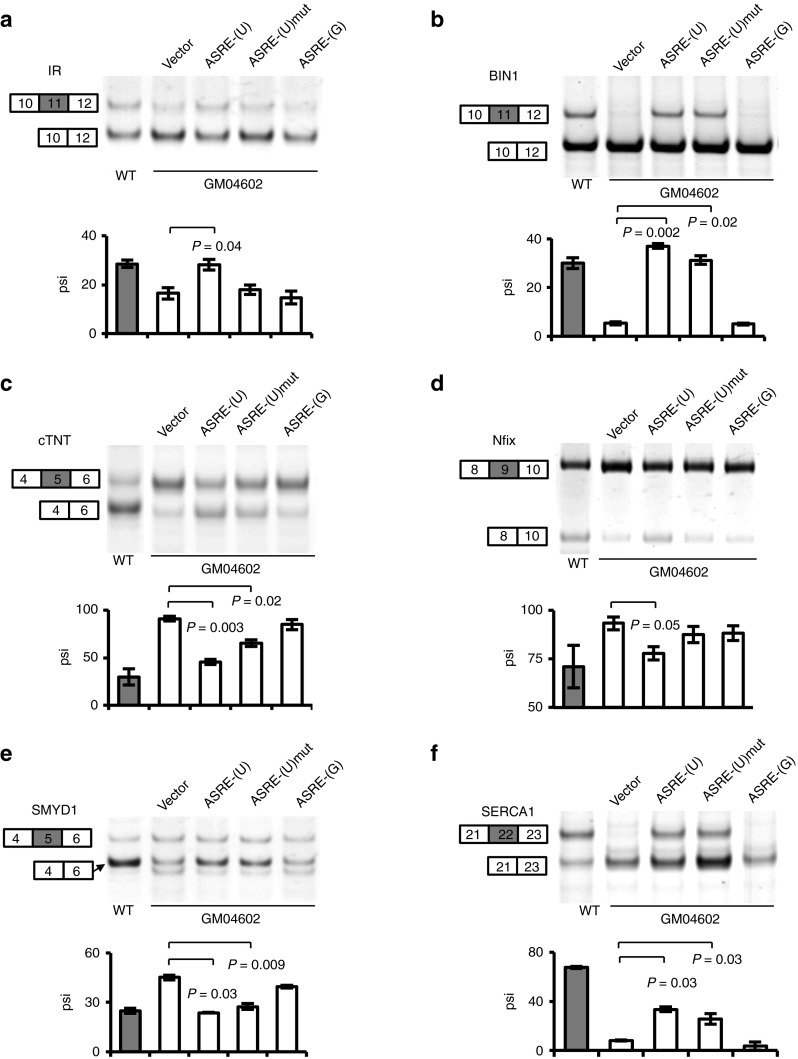
The insulin receptor can produce two isoforms through alternative splicing of exon 11 of α-subunit: IR-A (lacking exon 11) and IR-B (including exon 11). In DM1 cells, exon 11 of IR gene is largely skipped to generate isoform A that has a lower signaling capacity40 (Figure 5a, first two lanes). The expression of ASRE-(U) partially reversed such splicing defect by promoting the inclusion of exon 11 (Figure 5a). Similar effect of splicing restoration was also observed in a DM1 cell line derived from another patient (GM04033), suggesting that the ASRE has the potential to reverse the phenotypes caused by IR missplicing in different DM1 patients (Supplementary Figure S4a).
Figure 5.
The reverse of aberrant alternative splicing by artificial site-specific RNA endonucleases (ASREs) expression in myotonic dystrophy type 1 (DM1) cells. Semi-quantitative reverse transcriptase–polymerase chain reaction was used to detect different alternative splicing isoforms for (a) insulin receptor (a), (b) bridging integrator-1, (c) cTNT, (d) Nfix, (e) SMYD 1, and (f) SERCA1 in differentiated DM1 cells (GM04602). We used differentiated normal human fibroblast and DM1 cells transfected with vector only as controls. At least two independent experiments were carried out, with the means and SD of the percent-splice-in (psi) shown below a representative gel. In all panels, the P values were calculated by t-test comparing with vector only (only significant difference was shown).
BIN1 is another gene that is misregulated in DM1. This gene is involved in the formation of T tubule in skeletal muscles, and its exon 11 encodes a phosphoinositide-binding domain and is mostly skipped in DM1.15 The missplicing of BIN1 caused the expression of an inactive BIN1 that lacks phosphatidylinositol 5-phosphate binding and membrane-tubulating activities.15 The expression of ASRE-(U) restored the normal splicing pattern of BIN1 to produce BIN1 mRNA that contains exon 11 (Figure 5b). Interestingly, the mutated ASRE-(U) with an inactive PIN also reversed the splicing of BIN1, suggesting that the binding of artificial protein alone may disperse the MBNL proteins from nuclear foci to restore splicing. A similar effect was also observed in another DM1 cell lines with both WT and mutated ASRE-(U) (Supplementary Figure S4b).
As the cardiac isoform of TNT, cTNT, is the subunit of troponin complex with an important role in the regulation of muscle contraction. There are two major cTNT isforms generated by alternative splicing of exon 5 with different calcium sensitivity to the myofilament, and this alternative splicing event is developmentally regulated.9,14 Exon 5 of cTNT is included in the mature mRNAs during early development of the heart and skeletal muscle, but in adult heart, the exon is skipped. In DM1 patients, cTNT is misspliced to cause inappropriate inclusion of exon 5 in adult cardiac muscle. As expected, exon 5 was predominately excluded in normal cells but was mainly included in DM1 cells (Figure 5c). The expression of ASRE-(U) induced the skipping of exon 5 in mature cTNT mRNAs to partially restore normal splicing pattern. The mutated ASRE-(U) again has a residual activity in changing splicing, whereas the ASRE-(G) had no effect (Figure 5c). A similar effect was observed in GM04033 with both WT and mutated ASRE-(U) (Supplementary Figure S4c).
The Nfix gene encodes NF-I/X CAAT box-binding transcription factor that functions as a switch in the development of skeletal muscle from embryonic to fetal stage. A recent study showed that the Nfix exon 9 was aberrantly included in mouse models of DM.12 Again we found that the expression of ASRE-(U) caused a significant decrease of exon 9 inclusion to reverse the DM1-specific splicing (Figure 5d). A similar effect was also observed in GM04033 with ASRE-(U) (Supplementary Figure S4d).
SET and MYND domain containing protein 1 (SMYD1) is another DM1-affected gene that is specifically expressed in skeletal and cardiac muscles. SMYD1 functions as a histone methyltransferase to regulate gene transcription and is essential for myogenic differentiation.41 Exon 5 of Smyd1 mRNAs is aberrantly included in human DM1 sample and HSALR and Mbnl1ΔE3/ΔE3 mice model.12 ASRE-(U) expression decreased the inclusion of exon 5 in mature mRNAs, and a partial restoration was again obtained with the mutated ASRE-(U) (Figure 5e).
The last gene we tested, SERCA1, functions as a regulator of intracellular Ca2+ homeostasis in skeletal muscle cells. This gene has two major splice variants (SERCA1a and SERCA1b) resulting from the alternative splicing of its exon 22.42,43 Exon 22 is included in adult muscle but excluded in skeletal muscle of DM1 patients, which is regulated by MBNL1.16,17 The expression of ASRE-(U), and to a lesser extent mutated ASRE-(U), induced the inclusion of exon 22 in mature SERCA1 mRNAs to partially restore normal splicing pattern (Figure 5f), whereas the ASRE-(G) has no obvious effect on its splicing. As controls, we also examined the effect of ASREs on the WT cells in a parallel experiment and found that the splicing of the above genes were unaffected (Supplementary Figure S5).
Discussion
Here, we reported the first attempt to develop a treatment of DM1 with an artificial enzyme. This new gene therapy approach used a custom-designed RNA endonuclease to specifically bind and cleave (CUG)n repeats. Compared with conventional gene therapy that used MBNL1 to restore normal adult-splicing patterns and reduce muscle myotonia,19 our method directly targets the toxic RNA repeats. Because multiple muscleblind proteins are sequestered and other RNA-binding factors (e.g., CUGBP1 and staufen1) are also affected in DM1 patients, conventional gene therapy approach cannot restore the normal level of all the factors affected. On the other hand, this protein-based approach will take advantage of available gene therapy tools (e.g., AAV vectors) to achieve efficient delivery in muscle and heart, thus circumventing the difficulties of in vivo delivery associated with antisense oligonucleotide–based methods.
Using cultured cells derived from DM1 patients, we showed that the designer RNA endonucleases decreased the level of toxic RNAs (Figure 3), rescued the number of nuclear foci (Figure 4), and restored the normal splicing patterns of multiple pre-mRNAs affected in DM1 (Figure 5). Interestingly, we have found that the abnormal splicing of some genes (such as BIN1, SMYD1, and SERCA1) can be partially restored by the mutated ASRE-(U) with an inactive PIN domain, whereas the others can only be reversed by active ASRE-(U). Such results could be caused by two possible reasons. First, it was reported that the D1353A mutation of PIN did not completely abolish its RNA endonuclease activity inside cells, although this RNase activity is undetectable in vitro with purified recombinant PIN.29,37 Second, the mutated ASRE-(U) could also function as a competitor to disperse the sequestered MBNL proteins from nuclear foci and thus caused splicing restoration. Consistently, MBNL1 was shown to be the primary regulator of alternative splicing of BIN1,15 SMYD1,12 and SERCA117 in DM1 patients. For the genes whose splicing was reversed by WT ASRE-(U) but not by inactive ASRE-(U), it is possible that their splicing is mainly regulated by splicing factors other than MBNL in DM1 cells. For example, the splicing of cTNT and IR was mainly controlled by CUGBP9,13 but also can be regulated by MBNL in an opposite way.14 Therefore, dispersing of MBNL proteins by inactive ASRE-(U) had less effect on their splicing, and an active ASRE-(U) was required for splicing restoration.
We found that the degradation of (CUG)n repeat is the most efficient for long repeats (Figure 2b), presumably because the long repeats have more binding sites of the enzyme. The binding between PUFs and RNA was also affected by RNA structure. We speculate that PUF(G) preferably bound its targets in single-stranded RNA rather than in the hairpin structure of a long repeat, whereas PUF(U) and PUF(C) recognized 8-nt binding site in both structures (Figure 1c). Since (CUG)12 is long enough to form a hairpin structure that is similar to (CUG)20 or (CUG)480, ASRE(G) was found to slightly degrade (CUG)480 but not (CUG)12 (Figure 2b). This explanation assumes that (CUG)n repeat adopts a hairpin structure inside cells similar to its in vitro structure.34,35,44 Since there is no direct evidence of (CUG)n structure inside cells, other explanations are possible.
Compared with native RNA endonucleases (e.g., RNase A or H), we found that ASREs have a fairly slow catalytic rate with kcat ranging from 10 to 50 per minute.29 However, the catalytic efficiency (kcat/KM) of ASREs is reasonably high for typical RNA endonucleases (kcat/KM >107 (mol/l)−1 minute−1) due to the high binding affinity between the enzyme and its substrate (i.e., a low KM). This finding suggested that the catalytic efficiency of ASREs are mainly provided by the tight binding between the enzyme and its cognate RNA substrate, consistent with our finding that the short RNA repeats with less binding sites are poor substrates for ASRE-catalyzed degradation (Figure 2b). Such catalytic property provided a favorable feature for the application of ASRE as DM1 treatment, as the WT alleles of DMPK in DM1 patients are much less efficiently being cleaved by ASRE. Consistently, we found that ASRE-(U) can only decrease the level of DMPK mRNAs by ~40% (Figure 3d) even after a treatment for 10 days, suggesting that ASRE indeed has low activity toward WT DMPK mRNAs.
As the proof of concept for a new therapeutic approach, there are remaining questions need to be addressed in the future. For example, the ASRE-(U) recognize its targets by 8-nt, suggesting that there are possible off-target effect in other genes. However, the low catalytic rate of ASRE limited its off-target effect toward low-repetitive substrate, which is supported by the finding that the mRNA levels of the two genes with low number of CUG repeats (CASK: 16 repeats, MAP3K4: 11 repeats) are barely affected by ASRE-(U) (Supplementary Figure S6). Nevertheless, a thorough assessment of the levels for all other RNAs by mRNA-seq is needed in DM1 animal model treated with ASREs. Our study paves the way for the future test of in vivo efficacy of ASREs in DM1 animals, which can be carried out with standard gene delivery vectors for muscle and heart (e.g., AAV). Because the applications of AAV vectors have been extensively tested for muscular dystrophy and neurodegenerative diseases,45,46 our approach can benefit from these studies in the next stage of therapy development.
Materials and Methods
Yeast three-hybrid assays. Different PUF domains were cloned into the pACT2 plasmid and modified for distinct targets using QuikChange kits (Agilent Technologies, La Jolla, CA ). The target sequences were inserted into pIIIA-MS2-2 vector using SmaI and SphI.32 Yeast three-hybrid assays were performed in YBZ-1 strain. The activity of β-galactosidase was measured using LacZ reporter assay as described previously.32 Briefly, the yeast colonies were randomly chosen and inoculated into individual wells of a 96-well plate and incubated overnight at 30 °C with shaking to reach mid-log phase. The reaction was set up by transferring 20 µl of yeast culture into a new plate containing 180 µl of Z-buffer.32 After incubation at 37 °C for 2 hours, 80 µl of 1 mol/l carbonate solution was added into each well to stop the reaction. The A405 was measured using a spectrophotometer to quantify the yellow-colored product (nitrophenol). The β-galactosidase units were calculated according to the difference of A405 between the samples and the background calibrated by culture densities.
Cell culture and transfection. COS7 cells were grown in Dulbecco's modified Eagle's medium (Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum. These cells were cotransfected with 0.5 µg of DT12 or DT480 reporters and 1 µg of ASREs expression plasmid using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Forty-eight hours after transfection, the cells were collected for purification of total proteins and RNA. The ASRE levels were determined by western blot with anti-FLAG M2 (Sigma, St Louis, MO), and the levels of mRNA were measured by real-time RT-PCR.
DM1 broblasts (GM04602 or GM04033) were purchased from Coriell Cell Repositories (Camden, NJ). The fibroblasts were maintained in minimal essential medium (Gibco) supplemented with 15% fetal bovine serum and nonessential amino acids at 37 °C under 5% CO2 as recommended by Coriell.
Conversion from fibroblasts to skeletal muscle. MyoD cDNA was cloned into lentiviral vector pCDH-CMV-MCS-EF1-Puro (System Biosciences, Mountain View, CA). The lentivirus production was carried out according to the manufacturer's instructions. The supernatants containing the viral vector were used to infect DM1 fibroblasts. After 24 hours of incubation, the infected cells were selected with normal culture media containing 5 µg/ml puromycin for 7 days. The MyoD-transduced fibroblasts (myoblasts) were reinfected with ASREs lentiviral supernatant following the manufacturer's protocols. Infected broblasts were incubated in differentiation medium (Dulbecco's modified Eagle's medium supplemented with 1% heat-inactivated horse serum, 10 µg/ml insulin, and 10 µg/ml transferrin) for 10 days to differentiate to myocytes. The expression of ASREs and desmin were determined by western blot.
RNA extraction and RT-PCR. Total RNA was extracted from cultured cells using TRIzol (Invitrogen) according to the manufacturer's protocol. The RNA was treated with RNase-free DNase I (Promega, Madison, WI) at 37 °C for 1 hour and then incubated at 70 °C for 15 minutes to inactivate DNase. First-strand cDNA was synthesized using high capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA) with random primers. DMPK and GAPDH mRNA levels were determined by real-time PCR using Applied Biosystem 7500 fast Real-Time PCR System and Thermo Scientific Maxima SYBR Green/ROX qPCR Master Mix. For the analysis of alternative splicing of IR, BIN1, cTNT, Nfix, SMYD1, and SERCA1, 2 µl cDNA was used as the template for semi-quantitative PCR as described previously (primers are shown in Supplementary Table S1).47
Fluorescence in situ hybridization and immunofluorescence. Fluorescence in situ hybridization was performed to detect the (CUG)n repeats in DM1 myocytes using Cy5-labeled (CAG)10 probe. The cells were inoculated and cultured in cover slide and were washed with phosphate-buffered saline (PBS) and then with cytoskeleton buffer (300 mmol/l sucrose, 100 mmol/l NaCl, 3 mmol/l MgCl2, 1.2 mmol/l phenylmethylsulfonyl fluoride, and 10 mmol/l piperazine-N,N'-bis(ethanesulfonic acid), pH 6.8) before the experiments. The slides were incubated for 5 minutes in cytoskeleton buffer with 0.5% Triton X-100 at 4 °C, fixed in 4% paraformaldehyde in PBS for 15 minutes, and then rinsed in 70% ethanol at 4 °C. The cells were dehydrated through a series of ethanol incubation for 3 minutes each in 70, 80, 95, and 100% ethanol. The cells were incubated with probe mixture (50% formamide, 2× saline sodium citrate (SSC), 0.2% bovine serum albumin, 10% dextran sulfate, 2 mmol/l vanadyl sulfate, 0.5 mg/ml yeast tRNA, and 50 ng probe) at 37 °C for 3 hours in a humidified chamber. The slides were washed three times with 2×SSC/50% formamide for 10 minutes at room temperature and once at 37 °C followed by three times wash with 2×SSC at room temperature. The slides were transferred to a jar with 4×SSC/0.1% Tween. For immunofluorescence after fluorescence in situ hybridization, the slides were then incubated with 3% bovine serum albumin/PBS 30 minutes at 37 °C, incubated with primary anti-MBNL1 antibody 3A4 (1:1,000, Santa Cruz Biotechnology, Dallas, TX) in 3% bovine serum albumin/PBS at 37 °C for 90 minutes, washed three times with 4×SSC/0.1% Tween, and incubated with the secondary antibody (Alexa Fluor 488-labeled goat anti-mouse IgG, Invitrogen) in 3% bovine serum albumin/PBS for 30 minutes. Following the incubation, cells were washed three times with 4×SSC/0.1% Tween. The slides were mounted with mounting media with 4′,6-diamidino-2-phenylindole (Vector laboratories, Burlingame, CA).
SUPPLEMENTARY MATERIAL Figure S1. Design of ASREs. Figure S2. The expression of ASREs have no effect on DMPK mRNA in differentiated normal human fibroblasts. Figure S3. Cellular effects of ASREs in GM04033 cells. Figure S4. The reverse of aberrant alternative splicing by ASREs expression in GM04033 cells. Figure S5. The expression of ASREs have no effect on alternative splicing in normal cells. Figure S6. Effect of ASRE-(U) on other mRNA containing short CUG repeat. Table S1. Primers used in detection of different mRNA isoforms.
Acknowledgments
We thank Thomas Cooper from Baylor to generously provide reporters with CUG repeats. This work is supported by the NIH grants (R21AR061640) to Z.W. and partially supported by research grants from the National Natural Science Foundation of China (31125011, 31071148, and 31270844) to Y.J. W.Z. is supported by a student scholarship from the Chinese Scholarship Council (No. 2011632095). The authors have a patent application involving the ASRE (application number PCT/US2011/040933) and also seek to apply this technique through Macro Science Solutions LLP for therapeutic use.
Supplementary Material
References
- Turner C, Hilton-Jones D. The myotonic dystrophies: diagnosis and management. J Neurol Neurosurg Psychiatr. 2010;81:358–367. doi: 10.1136/jnnp.2008.158261. [DOI] [PubMed] [Google Scholar]
- Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3' end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- Mahadevan M, Tsilfidis C, Sabourin L, Shutler G, Amemiya C, Jansen G, et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3' untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- Wheeler TM, Thornton CA. Myotonic dystrophy: RNA-mediated muscle disease. Curr Opin Neurol. 2007;20:572–576. doi: 10.1097/WCO.0b013e3282ef6064. [DOI] [PubMed] [Google Scholar]
- Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem Soc Trans. 2009;37 Pt 6:1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, et al. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum Mol Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel-Chapuis A, Bélanger G, Yadava RS, Mahadevan MS, DesGroseillers L, Côté J, et al. The RNA-binding protein Staufen1 is increased in DM1 skeletal muscle and promotes alternative pre-mRNA splicing. J Cell Biol. 2012;196:699–712. doi: 10.1083/jcb.201108113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- Ho TH, Charlet-B N, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugier C, Klein AF, Hammer C, Vassilopoulos S, Ivarsson Y, Toussaint A, et al. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat Med. 2011;17:720–725. doi: 10.1038/nm.2374. [DOI] [PubMed] [Google Scholar]
- Kimura T, Nakamori M, Lueck JD, Pouliquin P, Aoike F, Fujimura H, et al. Altered mRNA splicing of the skeletal muscle ryanodine receptor and sarcoplasmic/endoplasmic reticulum Ca2+-ATPase in myotonic dystrophy type 1. Hum Mol Genet. 2005;14:2189–2200. doi: 10.1093/hmg/ddi223. [DOI] [PubMed] [Google Scholar]
- Hino S, Kondo S, Sekiya H, Saito A, Kanemoto S, Murakami T, et al. Molecular mechanisms responsible for aberrant splicing of SERCA1 in myotonic dystrophy type 1. Hum Mol Genet. 2007;16:2834–2843. doi: 10.1093/hmg/ddm239. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, et al. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci USA. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Shin J, Yuan Y, Beattie SG, Wheeler TM, Thornton CA, et al. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc Natl Acad Sci USA. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois MA, Lee NS, Rossi JJ, Puymirat J. Hammerhead ribozyme-mediated destruction of nuclear foci in myotonic dystrophy myoblasts. Mol Ther. 2003;7 5 Pt 1:670–680. doi: 10.1016/s1525-0016(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Lee JE, Bennett CF, Cooper TA. RNase H-mediated degradation of toxic RNA in myotonic dystrophy type 1. Proc Natl Acad Sci USA. 2012;109:4221–4226. doi: 10.1073/pnas.1117019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulders SA, van den Broek WJ, Wheeler TM, Croes HJ, van Kuik-Romeijn P, de Kimpe SJ, et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc Natl Acad Sci USA. 2009;106:13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, et al. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareiss PC, Sobczak K, McNaughton BR, Palde PB, Thornton CA, Miller BL. Dynamic combinatorial selection of molecules capable of inhibiting the (CUG) repeat RNA-MBNL1 interaction in vitro: discovery of lead compounds targeting myotonic dystrophy (DM1). J Am Chem Soc. 2008;130:16254–16261. doi: 10.1021/ja804398y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arambula JF, Ramisetty SR, Baranger AM, Zimmerman SC. A simple ligand that selectively targets CUG trinucleotide repeats and inhibits MBNL protein binding. Proc Natl Acad Sci USA. 2009;106:16068–16073. doi: 10.1073/pnas.0901824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf MB, Nakamori M, Matthys CM, Thornton CA, Berglund JA. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc Natl Acad Sci USA. 2009;106:18551–18556. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs-Disney JL, Parkesh R, Nakamori M, Thornton CA, Disney MD. Rational design of bioactive, modularly assembled aminoglycosides targeting the RNA that causes myotonic dystrophy type 1. ACS Chem Biol. 2012;7:1984–1993. doi: 10.1021/cb3001606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury R, Tsai YS, Dominguez D, Wang Y, Wang Z. Engineering RNA endonucleases with customized sequence specificities. Nat Commun. 2012;3:1147. doi: 10.1038/ncomms2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong CG, Hall TM. Engineering RNA sequence specificity of Pumilio repeats. Proc Natl Acad Sci USA. 2006;103:13635–13639. doi: 10.1073/pnas.0606294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- Dong S, Wang Y, Cassidy-Amstutz C, Lu G, Bigler R, Jezyk MR, et al. Specific and modular binding code for cytosine recognition in Pumilio/FBF (PUF) RNA-binding domains. J Biol Chem. 2011;286:26732–26742. doi: 10.1074/jbc.M111.244889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovska A, Razif MF, Nygård KK, Rackham O. A universal code for RNA recognition by PUF proteins. Nat Chem Biol. 2011;7:425–427. doi: 10.1038/nchembio.577. [DOI] [PubMed] [Google Scholar]
- Mooers BH, Logue JS, Berglund JA. The structural basis of myotonic dystrophy from the crystal structure of CUG repeats. Proc Natl Acad Sci USA. 2005;102:16626–16631. doi: 10.1073/pnas.0505873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalowski S, Miller JW, Urbinati CR, Paliouras M, Swanson MS, Griffith J. Visualization of double-stranded RNAs from the myotonic dystrophy protein kinase gene and interactions with CUG-binding protein. Nucleic Acids Res. 1999;27:3534–3542. doi: 10.1093/nar/27.17.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf CR, Opperman L, Wickens M. Chapter 14. Analysis of RNA-protein interactions using a yeast three-hybrid system. Meth Enzymol. 2008;449:295–315. doi: 10.1016/S0076-6879(08)02414-2. [DOI] [PubMed] [Google Scholar]
- Glavan F, Behm-Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 2006;25:5117–5125. doi: 10.1038/sj.emboj.7601377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita D, Zenno S, Lee WC, Saigo K, Tanokura M. Crystal structure of the PIN domain of human telomerase-associated protein EST1A. Proteins. 2007;68:980–989. doi: 10.1002/prot.21351. [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Santoro M, Masciullo M, Bonvissuto D, Bianchi ML, Michetti F, Silvestri G. Alternative splicing of human insulin receptor gene (INSR) in type I and type II skeletal muscle fibers of patients with myotonic dystrophy type 1 and type 2. Mol Cell Biochem. 2013;380:259–265. doi: 10.1007/s11010-013-1681-z. [DOI] [PubMed] [Google Scholar]
- Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, Harriss JV, et al. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet. 2002;31:25–32. doi: 10.1038/ng866. [DOI] [PubMed] [Google Scholar]
- Brandl CJ, Green NM, Korczak B, MacLennan DH. Two Ca2+ ATPase genes: homologies and mechanistic implications of deduced amino acid sequences. Cell. 1986;44:597–607. doi: 10.1016/0092-8674(86)90269-2. [DOI] [PubMed] [Google Scholar]
- MacLennan DH, Rice WJ, Green NM. The mechanism of Ca2+ transport by sarco(endo)plasmic reticulum Ca2+-ATPases. J Biol Chem. 1997;272:28815–28818. doi: 10.1074/jbc.272.46.28815. [DOI] [PubMed] [Google Scholar]
- Krzyzosiak WJ, Sobczak K, Wojciechowska M, Fiszer A, Mykowska A, Kozlowski P. Triplet repeat RNA structure and its role as pathogenic agent and therapeutic target. Nucleic Acids Res. 2012;40:11–26. doi: 10.1093/nar/gkr729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Cummins J, Huard J, Wang B. AAV-directed muscular dystrophy gene therapy. Expert Opin Biol Ther. 2010;10:395–408. doi: 10.1517/14712591003604690. [DOI] [PubMed] [Google Scholar]
- Terzi D, Zachariou V. Adeno-associated virus-mediated gene delivery approaches for the treatment of CNS disorders. Biotechnol J. 2008;3:1555–1563. doi: 10.1002/biot.200800284. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xiao X, Zhang J, Choudhury R, Robertson A, Li K, et al. A complex network of factors with overlapping affinities represses splicing through intronic elements. Nat Struct Mol Biol. 2013;20:36–45. doi: 10.1038/nsmb.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.