Abstract
Background
The rapid identification of acid-fast bacilli recovered from patient specimens as Mycobacterium tuberculosis complex (MTC) is critically important for accurate diagnosis and treatment. A thin-layer immunochromatographic (TLC) assay using anti-MPB64 or anti-MPT64 monoclonal antibodies was developed to discriminate between MTC and non-tuberculosis mycobacteria (NTM). Capilia TB-Neo, which is the improved version of Capilia TB, is recently developed and needs to be evaluated.
Methods
Capilia TB-Neo was evaluated by using reference strains including 96 Mycobacterium species (4 MTC and 92 NTM) and 3 other bacterial genera, and clinical isolates (500 MTC and 90 NTM isolates). M. tuberculosis isolates tested negative by Capilia TB-Neo were sequenced for mpt64 gene.
Results
Capilia TB-Neo showed 100% agreement to a subset of reference strains. Non-specific reaction to M. marinum was not observed. The sensitivity and specificity of Capilia TB-Neo to the clinical isolates were 99.4% (99.6% for M. tuberculosis, excluding M. bovis BCG) for clinical MTC isolates and 100% for NTM isolates tested, respectively. Two M. tuberculosis isolates tested negative by Capilia TB-Neo: one harbored a 63-bp deletion in the mpt64 gene and the other possessed a 3,659-bp deletion from Rv1977 to Rv1981c, a region including the entire mpt64 gene.
Conclusions
Capilia TB-Neo is a simple, rapid and highly sensitive test for identifying MTC, and showed better specificity than Capilia TB. However, Capilia TB-Neo still showed false-negative results with mpt64 mutations. The limitation should be recognized for clinical use.
Keywords: Capilia TB-Neo, Mycobacterium tuberculosis complex identification, mpt64 gene
Background
Tuberculosis remains a major threat to global health, and therefore, rapid identification of the causative M. tuberculosis complex (MTC) is critical. Liquid culture detection is now widely used for managing HIV-co-infected and drug-resistant tuberculosis, and liquid culture can improve the recovery of acid-fast bacilli and decreases the time to detection. However, because other non-tuberculosis mycobacterium (NTM) species may also grow, it is important to identify MTC from positive culture for rapid and appropriate management of tuberculosis.
Several methods are available to identify mycobacteria. Conventional biochemical tests are generally time-consuming [1,2] and not surely reproducible, while more recently developed techniques involving molecular biology [3-7] or high-performance liquid chromatographic analysis of mycolic acid [8] are accurate and rapid, but these require expensive devices. In contrast, immunochromatographic species identification tests, Capilia TB (TAUNS, Izunokuni, Japan), SD BIOLINE TB Ag MPT64 rapid (Standard Diagnostics, Inc., Korea) and BD MGIT™ TBc Identification Test (Becton, Dickinson and Company, USA) have been adopted as a cheap, rapid, and accurate alternative in clinical laboratories around the world [9-11]. However, false positives to Mycobacterium marinum, Staphylococcus aureus[9,12], and false negatives in MPB64 mutants [10,13] have occasionally been reported. Capilia TB-Neo (TAUNS, Izunokuni, Japan), an improved version of Capilia TB, has recently been developed to overcome these problems. In this study, we evaluated the performance of Capilia TB-Neo with reference strains and clinical isolates. Any false-negative MTC clinical isolate detected by Capilia TB-Neo were further investigated relative genes.
Methods
Reference strains and clinical isolates
Reference strains of 96 Mycobacterium species and subspecies (4 MTC and 92 NTM) and 3 other genera with acid-fastness (Nocardia asteroids, Rhodococcus equi and Rhodococcus aichiense) were used for the evaluation (Table 1). A total of 500 MTC and 90 NTM clinical isolates (10 M. abscessus, 4 M. chelonae, 13 M. fortuitum, 8 M. gordonae, 15 M. avium complex, 7 M. intracellulare, 3 M. nonchromogenicum, 5 M. scrofulaceum, 4 M. xenopi, 15 M. kansasii, 1 M. gastri, 2 M. peregrinum, 1 M. intermedium, 1 M. szulgai, and 1 M. marinum) were selected to provide a representative sample of the isolates available from Miroku Medical Laboratory Co., Ltd. (Saku, Japan) from 2009 to 2010, and the collection from the Ryoken survey in 2002 and 2007. The clinical isolates were collected from patients as a part of routine examination. No ethical approval was required for this type of laboratory based study only using isolates. Reference strains and clinical isolates were cultured with OADC-supplemented Middlebrook 7H9 broth (Becton, Dickinson and Company, USA) and 2% Ogawa medium (Kyokuto Pharmaceutical Industrial Co., Japan) at 37°C or 30°C.
Table 1.
List of reference strains and the results of identification of MTC by using Capilia TB-Neo, SD MPT64, and TBc ID
| Species | Strain | Capilia TB-Neo | SD MPT64 | TBc ID | Species | Strain | Capilia TB-Neo | SD MPT64 | TBc ID |
|---|---|---|---|---|---|---|---|---|---|
|
M. tuberculosis H37Rv |
ATCC27294 |
+ |
+ |
+ |
M. interjectum |
ATCC51457 |
- |
- |
- |
|
M. africanum |
ATCC25420 |
+ |
+ |
+ |
M. intermedium |
ATCC51848 |
- |
- |
- |
|
M. bovis |
ATCC19210 |
+ |
+ |
+ |
M. intracellulare |
ATCC13950 |
- |
- |
- |
|
M. microti |
ATCC19422 |
+ |
+ |
+ |
M. kansasii |
ATCC12478 |
- |
- |
- |
|
M. abscessus |
ATCC19977 |
- |
- |
- |
M. kubicae |
ATCC700732 |
- |
- |
- |
|
M. acapulcensis |
ATCC14473 |
- |
- |
- |
M. lactis |
ATCC27356 |
- |
- |
- |
|
M. agri |
ATCC27406 |
- |
- |
- |
M. lentiflavum |
ATCC51985 |
- |
- |
- |
|
M. aichiense |
ATCC27280 |
- |
- |
+ |
M. madagascariense |
ATCC49865 |
- |
- |
- |
|
M. alvei |
ATCC51304 |
- |
- |
- |
M. malmoense |
ATCC29571 |
- |
- |
- |
|
M. asiaticum |
ATCC25276 |
- |
- |
- |
M. marinum |
ATCC00927 |
- |
- |
+ |
|
M. aurum |
ATCC23366 |
- |
- |
- |
M. moriokaense |
ATCC43059 |
- |
- |
- |
|
M. austroafricanum |
ATCC33464 |
- |
- |
- |
M. mucogenicum |
ATCC49650 |
- |
- |
- |
|
M. avium subsp. avium |
ATCC25291 |
- |
- |
- |
M. neoaurum |
ATCC25795 |
- |
- |
- |
|
M. avium subsp. paratuberculosis |
ATCC19698 |
- |
- |
- |
M. nonchromogenicum |
ATCC19530 |
- |
- |
- |
|
M. avium subsp. “suis” |
ATCC19978 |
- |
- |
- |
M. novum |
ATCC19619 |
- |
- |
- |
|
M. avium subsp. silvaticum |
ATCC49884 |
- |
- |
- |
M. obuense |
ATCC27023 |
- |
- |
- |
|
M. branderi |
ATCC51789 |
- |
- |
- |
M. paraffinicum |
ATCC12670 |
- |
- |
- |
|
M. brumae |
ATCC51384 |
- |
+ |
- |
M. parafortuitum |
ATCC19686 |
- |
- |
- |
|
M. celatum |
ATCC51131 |
- |
- |
- |
M. peregrinum |
ATCC14467 |
- |
- |
- |
|
M. celatum II |
ATCC51130 |
- |
- |
- |
M. petroleophilum |
ATCC21497 |
- |
- |
- |
|
M. chelonae chemovar niacinogenes |
ATCC35750 |
- |
- |
- |
M. phlei |
ATCC11758 |
- |
- |
- |
|
M. chelonae subsp. chelonae |
ATCC35752 |
- |
- |
- |
M. porcinum |
ATCC33776 |
- |
- |
- |
|
M. chitae |
ATCC19627 |
- |
- |
+ |
M. poriferae |
ATCC35087 |
- |
- |
- |
|
M. chlorophenolicum |
ATCC49826 |
- |
- |
- |
M. pulveris |
ATCC35154 |
- |
- |
- |
|
M. chubuense |
ATCC27278 |
- |
- |
- |
M. rhodesiae |
ATCC27024 |
- |
- |
- |
|
M. confluentis |
ATCC49920 |
- |
- |
- |
M. scrofulaceum |
ATCC19981 |
- |
- |
- |
|
M. conspicuum |
ATCC700090 |
- |
- |
- |
M. senegalense |
ATCC35796 |
- |
- |
- |
|
M. cookii |
ATCC49103 |
- |
- |
- |
M. septicum |
ATCC700731 |
- |
- |
- |
|
M. diernhoferi |
ATCC19340 |
- |
- |
- |
M. shimoidei |
ATCC27962 |
- |
- |
- |
|
M. duvalii |
ATCC43910 |
- |
- |
- |
M. shinshuense |
ATCC33728 |
- |
- |
- |
|
M. engbaekii |
ATCC27353 |
- |
- |
- |
M. simiae |
ATCC25275 |
- |
- |
- |
|
M. flavescens |
ATCC14474 |
- |
- |
- |
M. smegmatis |
ATCC19420 |
- |
- |
- |
|
M. fortuitum subsp. acetamidolyticum |
ATCC35931 |
- |
- |
- |
M. smegmatis |
ATCC700084 |
- |
- |
- |
|
M. fortuitum subsp. fortuitum |
ATCC06841 |
- |
- |
- |
M. sphagni |
ATCC33027 |
- |
- |
- |
|
M. fortuitum subsp. fortuitum |
ATCC49403 |
- |
- |
- |
M. szulgai |
ATCC35799 |
- |
- |
- |
|
M. gadium |
ATCC27726 |
- |
- |
+ |
M. terrae |
ATCC15755 |
- |
- |
- |
|
M. gallinarum |
ATCC19710 |
- |
- |
- |
M. terrae |
DSMZ43540 |
- |
- |
- |
|
M. genavense |
ATCC51234 |
- |
- |
- |
M. terrae |
DSMZ43541 |
- |
- |
- |
|
M. gilvum |
ATCC43909 |
- |
- |
- |
M. terrae |
DSMZ43542 |
- |
- |
- |
|
M. goodii |
ATCC700504 |
- |
- |
- |
M. thermoresistibile |
ATCC19527 |
- |
- |
- |
|
M. gordonae |
ATCC14470 |
- |
- |
- |
M. tokaiense |
ATCC27282 |
- |
- |
- |
|
M. gordonae group B
19
|
KK33-08 |
- |
- |
- |
M. triplex |
ATCC700071 |
- |
- |
- |
|
M. gordonae group C
19
|
KK33-53 |
- |
- |
- |
M. triviale |
ATCC23292 |
- |
- |
- |
|
M. gordonae group D
19
|
KK33-46 |
- |
- |
- |
M. vaccae |
ATCC15483 |
- |
- |
- |
|
M. haemophilum |
ATCC29548 |
- |
- |
- |
M. valentiae |
ATCC29356 |
- |
- |
- |
|
M. hassiacum |
ATCC700660 |
- |
- |
- |
M. wolinskyi |
ATCC700010 |
- |
- |
- |
|
M. heckeshornense |
DSMZ44428 |
- |
- |
- |
M. xenopi |
ATCC19250 |
- |
- |
- |
|
M. heidelbergense |
ATCC51253 |
- |
- |
- |
Nocardia asteroides |
ATCC19247 |
- |
- |
- |
|
M. hiberniae |
ATCC49874 |
- |
- |
- |
Rhodococcus equi |
ATCC6939 |
- |
- |
- |
| Rhodococcus aichiense | ATCC33611 | - | - | - |
Identification of mycobacteria
Mycobacterium species of the clinical isolates were identified using one or more of the following approaches: (i) the DNA or RNA amplification kits Cobas Amplicor PCR (Roche Diagnostics, Japan) and TRC Rapid (Tosoh Bioscience, Japan); (ii) the DNA-DNA hybridization DDH Mycobacteria Kit (Kyokuto Pharmaceutical Industrial Co., Japan); and (iii) 16S rRNA gene sequencing, supplementary [7]. The isolates identified as MTC were further examined by multiplex PCR analysis of cfp32, the region of difference (RD) 9, and RD12 according to the method of Nakajima et al. [14]. When MTC species other than M. tuberculosis sensu stricto were detected, they were further characterized with respect to RD1, RD4, RD7, and MiD3 [15]. If M. bovis Bacillus Calmette-Guerin (BCG) was identified, additional multiplex PCR analyses were performed to test for RD2, RD14, RD15, RD16, and SenX3-RegX3 to distinguish sub-strains of BCG [16,17]. The multiplex PCR amplification was performed using a Type-it Microsatellite PCR Kit (QIAGEN, Japan). Each PCR reaction contained 1.0 μl of DNA template, 6.25 μl of Type-it multiplex PCR Master mix, 1.25 μl of Q-solution, 0.25 μl of each primer (10 pmol/μl) and an appropriate amount of molecular grade water for a total reaction volume of 13 μl. The thermal profile was as follows: (i) 95°C (5 min); (ii) 28 cycles of 95°C (0.5 min), 58 or 55°C (1.5 min), 72°C (0.5 min); and (iii) a final extension step at 68 or 60°C (10 or 30 min). The amplified products were analyzed by 3% agarose gel electrophoresis. The expected RD loci for each MTC species are summarized in Table 2.
Table 2.
Oligonucleotide primers used in PCR and direct sequencing
| Target gene | Primer ID | Nucleotide sequence (5'-3') | Size (bp) | Ref. no. |
|---|---|---|---|---|
| MTC identification |
|
|
|
|
| 16S rRNA |
285 |
GAGAGTTTGATCCTGGCTCAG |
1028 |
7 |
| |
264 |
TGCACACAGGCCACAAGGGA |
|
|
| |
259 |
TTTCACGAACAACG GACAA |
591 |
|
| cfp32 |
Rv0577F |
ATGCCCAAGAGAAGCGAATACAGGCAA |
786 |
14 |
| |
Rv0577R |
CTATTGCTGCGGTGCGGGCTTCAA |
|
|
| RD9 |
Rv2073cF |
TCGCCGCTGCCAGATGAGTC |
600 |
14 |
| |
Rv2073cR |
TTTGGGAGCCGCCGGTGGTGATGA |
|
|
| RD12 |
Rv3120F |
GTCGGCGATAGACCATGAGTCCGTCTCCAT |
404 |
14 |
| |
Rv3120R |
GCGAAAAGTGGGCGGATGCCAG |
|
|
| RD1 |
ET1 |
AAGCGGTTGCCGCCGACCGACC |
|
15 |
| |
ET2 |
CTGGCTATATTCCTGGGCCCGG |
|
|
| |
ET3 |
GAGGCGATCTGGCGGTTTGGGG |
|
|
| RD4 |
Rv1510F |
GTGCGCTCCACCCAAATAGTTGC |
1033 |
15 |
| |
Rv1510R |
TGTCGACCTGGGGCACAAATCAGTC |
|
|
| RD7 |
Rv1970F |
GCGCAGCTGCCGGATGTCAAC |
1116 |
15 |
| |
Rv1970R |
CGCCGGCAGCCTCACGAAATG |
|
|
| MiD3 |
IS1561F |
GCTGGGTGGGCCCTGGAATACGTGAACTCT |
530 |
15 |
| |
IS1561R |
AACTGCTCACCCTGGCCACCACCATGGACT |
|
|
| Distinguish sub-strains of BCG |
|
|
|
|
| RD2 |
RD2l |
CCAGATTCAAATGTCCGACC |
|
16 |
| |
RD2r |
GTGTCATAGGTGATTGGCTT |
|
|
| RD14 |
RD14l |
CAGGGTTGAAGGAATGCGTGTC |
|
16 |
| |
RD14r |
CTGGTACACCTGGGGAATCTGG |
|
|
| RD15 |
RD8l |
ACTCCTAGCTTTGCTGTGCGCT |
|
16 |
| |
RD8r |
GTACTGCGGGATTTGCAGGTTC |
|
|
| RD16 |
RD16nf |
ACATTGGGAAATCGCTGCTGTTG |
|
17 |
| |
RD16nr |
GGCTGGTGTTTCGTCACTTC |
|
|
| SenX3-RegX3 |
C3 |
GCGCGAGAGCCCGAACTGC |
|
16 |
| |
C5 |
GCGCAGCAGAAACGTCAGC |
|
|
| Sequencing |
|
|
|
|
| mpt64 (Rv1980c) |
mpb64W-F |
ACTCAGATATCGCGGCAATC |
1061 |
this study |
| |
mpb64W-R |
CGATCACCTCACCTGGAGTT |
|
|
| Rv1977 |
Rv1977F |
GTTTCCCGAGATCAGCTCAA |
348 |
this study |
| |
Rv1977R |
ATCTCGTCGTGTGTCACCAG |
|
|
| Rv1981c |
Rv1981F |
GATCGAATGCAGGCTGGTAT |
399 |
this study |
| Rv1981R | ACTACTACCGCGGTGACGAC | |||
Capilia TB-Neo, SD MPT64, and TBc ID
The validation of Capilia TB-Neo (TAUNS, Izunokuni, Japan) was conducted using the aforementioned reference strains as well as MTC and NTM clinical isolates. In addition, SD BIOLINE TB Ag MPT64 rapid (SD MPT64: Standard Diagnostics, Inc. Korea) and BD MGIT™ TBc Identification Test (TBc ID: Becton, Dickinson and Company, USA), detect MPT64 which is the same as MPB64, were tested using reference strains and NTM clinical isolates. Each test was performed according to the manufacturer’s instructions. Briefly, clinical isolates growing on Ogawa medium were suspended in 1 ml of sterile distilled water, and the suspension subjected to the test. Similarly, positive liquid cultures of reference strains (McFarland No. 1 to 2) were directly subjected to each test. Positive test results were indicated by a red line in the test area after 15 min.
Sequencing of the mpt64 gene
Any false-negative M. tuberculosis isolate detected by Capilia TB-Neo was further analyzed by sequencing mpt64 and surrounding genes by using the primers listed in Table 2. Each PCR reaction contained 1.0 μl of DNA template, 12.5 μl of Type-it multiplex PCR Master mix, 2.5 μl of Q-solution, 0.5 μl of each primer (10 pmol/μl) and an appropriate amount of molecular grade water for a total reaction volume of 25 μl. The thermal profile was as follows: (i) 95°C (5 min); (ii) 30 cycles of 95°C (0.5 min), 62°C (1.5 min), 72°C (1 min); and (iii) final extension at 60°C (10 min). The amplified product was analyzed by 3% agarose gel electrophoresis and was purified using Mag Extractor (TOYOBO, Japan). The purified DNA products were subjected to direct sequencing using an ABI 377 automatic sequencer (Applied Biosystems, USA) and BigDye Terminator Cycle Sequencing v 3.1 (Applied Biosystems, USA), according to the manufacturer’s instructions. DNA sequences of mpt64 from each isolate were compared with M. tuberculosis H37Rv by using Genetyx-win ver. 5.2 (Genetyx Co., Japan).
Results
Each of the three kits (Capilia TB-Neo, SD MPT64, and TBc ID) was tested using the 99 reference strains. Capilia TB-Neo correctly produced positive results for four MTC (M. tuberculosis, M. africanum, M. bovis, and M. microti) and negative results for 92 NTM and 3 non-mycobacterial species (other genera) with acid-fastness, while SD MPT64 and TBc ID generated several false positives (Table 1). The sensitivity and specificity of Capilia TB-Neo to reference strains were 100%.
Of the 500 MTC clinical isolates tested, 497 were identified as MTC by Capilia TB-Neo. The other 3 isolates that tested negative by using Capilia TB-Neo also tested negative by using SD MPT64 and TBc ID. All three kits produced negative results for all 90 NTM clinical isolates examined. Thus, The sensitivity and specificity of Capilia TB-Neo to the clinical isolates were 99.4% and 100%, respectively.
The multiplex PCR system identified 492 M. tuberculosis isolates out of 500. Five isolates, which were cfp32-, RD9-, RD4-, RD7-, and MiD3-positive, but RD12-negative, were initially identified as M. canettii. However, colonies of these isolates showed a consistent rough surface on solid medium, and subsequent sequencing of hsp65 indicated that the isolates had the genotype of M. tuberculosis sensu stricto (data not shown). These isolates were collected from different areas of Japan. Consequently, 497 isolates were identified as M. tuberculosis. The remaining 3 isolates were deficient in RD1, RD4, RD7, RD9, and RD12, and therefore were identified as M. bovis BCG. Two of these isolates were confirmed as M. bovis BCG Tokyo based on the unique size of RD16, and the third isolate had the same RD pattern as BCG Connaught and BCG Montreal, as for RD2, RD14, RD15, RD16 and SenX3-RegX3 (Figure 1). Among the 3 MTC isolates that tested negative by Capilia TB-Neo, 2 isolates were M. tuberculosis and the other was M. bovis BCG Connaught or BCG Montreal (Table 3).
Figure 1.
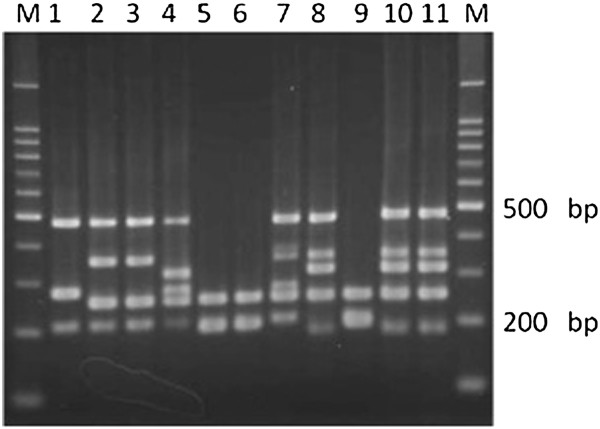
Multiplex PCR analysis of Mycobacterium bovis BCG sub-strains and clinical isolates. 1: BCG Pasteur (ATCC35734), 2: BCG Glaxo (ATCC35741), 3: BCG Copenhagen (ATCC27290), 4: BCG Russian (ATCC35740), 5: BCG Montreal (ATCC35735), 6: BCG Connaught (ATCC35745), 7: BCG Danish, 8: BCG Tokyo, 9: Sample 421, 10: Sample 467, 11: Sample 475, M: Size marker.
Table 3.
Results of PCR detection and Capilia TB-Neo of MTC with clinical isolates
|
Species interpretation (Number of isolates) |
Banding pattern |
Capilia TB-Neo |
% |
||||||
|---|---|---|---|---|---|---|---|---|---|
| cfp32 | RD9 | RD12 | RD4 | RD7 | MiD3 | RD1 | |||
|
M. tuberculosis (490) |
+ |
+ |
+ |
NT |
NT |
NT |
NT |
+ |
98.0 |
|
M. tuberculosis (2) |
+ |
+ |
+ |
NT |
NT |
NT |
NT |
- |
0.4 |
|
"M. canettii" (5)a |
+ |
+ |
- |
+ |
+ |
+ |
+ |
+ |
1.0 |
|
M. bovis BCG Tokyo (2)b |
+ |
- |
- |
- |
- |
+ |
- |
+ |
0.4 |
| M. bovis BCG Connaught (1)c | + | - | - | - | - | + | - | - | 0.2 |
aConfirmed to be M. tuberculosis by hsp65 sequencing and morphology, bConfirmed by contracted RD16, cConfirmed by absence of RD2 and RD15, and contracted SenX3-RegX3, NT: Not tested.
Mutations in the mpt64 gene were detected by sequencing two M. tuberculosis isolates with negative results by Capilia TB-Neo. One isolate had a deletion of 63 bp from nucleotides 196 to 258 (amino acids position 43 to 63), and the other had a deletion of 3,659 bp from nucleotide 874 in Rv1977 to nucleotide 905 in Rv1981c, which included the whole mpt64 gene.
Discussion
In many industrialized countries, the ability to rapidly distinguish between MTC and NTM is critical in clinical practice. Indeed, the anti-tuberculosis drug resistance survey in Japan revealed that 19.3% of all clinical mycobacterial isolates are NTM [18], underscoring the importance of rapid and accurate detection of MTC from acid-fast bacillus-positive culture. The immunochromatographic assay kit for the identification of MTC is now widely used in many countries. Capilia TB-Neo is the improved version of Capilia TB, and has been subjected to few clinical evaluations. Here, we report good overall performance of the kit but with several limitations.
In this study, the sensitivity of Capilia TB-Neo was 99.4% to clinical MTC isolates or 99.6% excluding M. bovis BCG, while the specificity of the kit tested to clinical NTM isolates was 100%. However, the isolation of BCG could present a practical problem. The M. bovis BCG Tokyo strain is sporadically isolated in Japan as a complex of vaccination or bladder cancer therapy, and is identified as MTC with the kit [19]. Some BCG strains such as Connaught, Pasteur, and Tice lack RD2 including the mpt64 gene, but RD2 is conserved in others such as Tokyo, Moreau, and Russia [16]. This issue should be properly addressed to avoid confusion. Although it is difficult to discriminate BCG Tokyo from MTC with mpt64/mpb64, their differentiation would be an important advance in the development of a future TLC product. The weak false-positive reaction to M. marinum that was reported using Capilia TB [12] was not observed in this study, and resulted in better specificity. The minimum detection concentration of M. tuberculosis for Capilia TB-Neo was 105 CFU/ml (data not shown), which was one-tenth than that for the previous kit. There was a report that Cpilia TB-Neo was higher sensitivity than Capilia TB [20]. In summary, the overall performance of Capilia TB-Neo was better than Capilia TB in both sensitivity and specificity.
SD MPT64 and TBc ID were also tested with reference strains. Both SD MPT64 and TBc ID showed false-positive results against several NTM strains in this study. Kodama et al. [12] reported that no M. marinum strains grown on 2% Ogawa medium tested positive by using the Capilia TB, while all strains grown on 3 kinds of liquid medium, MGIT (Becton Dickinson, Japan), KRD medium (Japan BCG Laboratory, Japan) and Myco Acid (Kyokuto Pharmaceutical Industrial Co. Ltd., Japan), eventually displayed a positive reaction that intensified with time. Kodama et al. speculated that nonspecific antigen which could make complex with anti-MPB64 antibody may be produced in liquid mediums, but not on solid medium. Considering the effect of liquid culture, the original bacterial suspensions giving false-positive results, that were prepared from liquid and solid culture, were then re-tested before and after 10-fold dilution. Interestingly, none of these diluted strains tested positive in these kits, but bacterial concentrations were high enough for positive results in case of MTC. These results implied that a high concentration of bacterial antigens could induce non-specific reactions in SD MPT64 and TBc ID. The manufacturer’s instructions for the TBc ID indicate that this kit may be used up to 10 days after a positive MGIT alarm. This non-specific reaction should be properly addressed in clinical practice, and the users should perform morphological characterization with a microscope to identify cord formation.
Several mutations in the mpt64 gene produce a negative test result for M. tuberculosis isolates in the TLC assay using anti-MPB64 monoclonal antibodies. To date, these include a 63-bp deletion from nucleotide 196, a 1-bp deletion from nucleotide 266, a point mutation at position 388 or 402, IS6110 insertion mutation at position 177 or 501, a 176-bp deletion from nucleotide 512, and a 1-bp insertion at position 287 [10,13,21]. In our study, 2 M. tuberculosis isolates gave false-negative results by using the Capilia TB-Neo, SD MPT64, and TBc ID. One isolate had a deletion of 63 bp from nucleotide 196 in the mpt64 gene as reported previously, and the other isolate possessed a 3,659-bp deletion from nucleotide 874 in Rv1977 to 905 in Rv1981c, including the whole mpt64 gene. To the best of our knowledge, this is the first report of a large deletion in mpt64. A transposon site hybridization (TraSH) study [22] indicated that mpt64 is not essential for infection or in vitro growth of M. tuberculosis. This large deletion mutant supported the finding.
In summary, the TLC assay detecting MPB64 or MPT64 can be applied to specimens prepared from liquid and solid culture. It does not need special reagents, instruments, or complex techniques. Capilia TB-Neo tested in this study showed excellent sensitivity with perfect specificity.
Conclusions
Capilia TB-Neo showed high sensitivity and specificity with clinical mycobacterial isolates, and 100% specificity to reference strains. However, 2 M. tuberculosis isolates were tested negative by Capilia TB-Neo because of mutations in the mpt64 gene, and positive to certain BCG sub-strain. This study, therefore, serves to emphasize the importance of careful use of the kit and the complementary techniques such as morphological identification.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KC carried out the TLC assays, molecular genetic studies, sequence alignment and drafted the manuscript. AA, HY, TS, and TK cultured clinical isolates. HY helped to draft the manuscript. YK prepared reference strains. KT and HY collected clinical isolates. SM was responsible for planning the study. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Kinuyo Chikamatsu, Email: chikamatsu@jata.or.jp.
Akio Aono, Email: aono@jata.or.jp.
Hiroyuki Yamada, Email: hyamada@jata.or.jp.
Tetsuhiro Sugamoto, Email: tsugamoto@jatahq.org.
Tomoko Kato, Email: dumbo22boko22@gmail.com.
Yuko Kazumi, Email: kazumi@jata.or.jp.
Kiyoko Tamai, Email: mml-idenshi@miroku-lab.co.jp.
Hideji Yanagisawa, Email: h-yanagisawa@miroku-lab.co.jp.
Satoshi Mitarai, Email: mitarai@jata.or.jp.
Acknowledgements
We thank TAUNS Co, Ltd (Izunokuni, Japan) for providing the Capilia TB-Neo, SD MPT64, and TBc ID.
References
- Public Health Mycobacteriology. Identification Test Techniques: Department of Health and Human Services. Atran: Public Health Service. CDC; 1985. A guide for the level III laboratory. [Google Scholar]
- Rastogi N, Goh KS, David HL. Selective inhibition of the Mycobacterium tuberculosis complex by p-nitro-a-acetylamino-b-hydroxypropiophenone (NAP) and p-nitrobenzoic acid (PNB) used in 7H11 agar medium. Res Microbiol. 1989;14:419–423. doi: 10.1016/0923-2508(89)90017-X. [DOI] [PubMed] [Google Scholar]
- Goto M, Oka S, Okuzumi K, Kimura S, Shimada K. Evaluation of acridinium ester-labeled DNA probes for identification of Mycobacterium tuberculosis and Mycobacterium avium-Mycobacterium intracellulare complex in culture. J Clin Microbiol. 1991;14:2473–2476. doi: 10.1128/jcm.29.11.2473-2476.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis P, Papaventsis D, Karabela S, Nikolaou S, Panagi M, Raftopoulou E, Konstantinidou E, Marinou I, Kanavaki S. Cepheid GeneXpert MTB/RIF Assay for Mycobacterium tuberculosis detection and rifampin resistance identification in patients with substantial clinical indications of tuberculosis and smear-negative microscopy results. J Clin Microbiol. 2011;14:3068–3070. doi: 10.1128/JCM.00718-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitarai S, Okumura M, Toyota E, Yoshiyama T, Aono A, Sejimo A, Azuma Y, Sugahara K, Nagasawa T, Nagayama N, Yamane A, Yano R, Kokuto H, Morimoto K, Ueyama M, Kubota M, Yi R, Ogata H, Kudoh S, Mori T. Evaluation of a simple loop-mediated isothermal amplification test kit for the diagnosis of tuberculosis. Int J Tuberc Lung Dis. 2011;14:1211–1217. doi: 10.5588/ijtld.10.0629. [DOI] [PubMed] [Google Scholar]
- Richter E, Rüsch-Gerdes S, Hillemann D. Evaluation of the genotype Mycobacterium assay for identification of Mycobacterial species from cultures. J Clin Microbiol. 2006;14:1769–1775. doi: 10.1128/JCM.44.5.1769-1775.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer B, Stockman L, Teschner KD, Roberts G, Bottger EC. Two-laboratory collaborative study on identification of Mycobacteria: molecular versus phenotypic methods. J Clin Microbiol. 1996;14:296–303. doi: 10.1128/jcm.34.2.296-303.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standardized Method for HPLC Identification of Mycobacteria. Department of Health and Human Services. Atranta: Public Health Service CDC; 1996. [Google Scholar]
- Abe C, Hirano K, Tomiyama T. Simple and rapid identification of Mycobacterium tuberculosis complex by immunochromatographic assay using anti-MPB64 monoclonal antibodies. J Clin Microbiol. 1999;14:3693–3697. doi: 10.1128/jcm.37.11.3693-3697.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillemann D, Rüsch-Gerdes S, Richter E. Application of the Capilia TB assay for culture confirmation of Mycobacterium tuberculosis complex isolates. Int J Tuberc Lung Dis. 2005;14:1409–1411. [PubMed] [Google Scholar]
- Muyoyeta M, de Haas PE, Mueller DH, van Helden PD, Mwenge L, Schaap A, Kruger C, Gey van Pittius NC, Lawrence K, Beyers N, Godfrey-Faussett P, Ayles H. Evaluation of the Capilia TB assay for culture confirmation of Mycobacterium tuberculosis infections in Zambia and South Africa. J Clin Microbiol. 2010;14:3773–3775. doi: 10.1128/JCM.01688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama A, Saito H. Evaluation of CapiliaTB for identification of Mycobacterium tuberculosis complex, with special reference to the culture medium. Jpn Society Clin Microbiol (in Japanese) 2007;14:109–118. [Google Scholar]
- Hirano K, Aono A, Takahashi M, Abe C. Mutations including IS6110 insertion in the gene encoding the MPB64 protein of Capilia TB negative Mycobacterium tuberculosis isolates. J Clin Microbiol. 2004;14:390–392. doi: 10.1128/JCM.42.1.390-392.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima C, Rahim Z, Fukushima Y, Sugawara I, Van der Zanden AGM, Tamaru A, Suzuki Y. Identification of Mycobacterium tuberculosis clinical isolates in Bangladesh by a species distinguishable multiplex PCR. BMC Infect Dis. 2010;14:118. doi: 10.1186/1471-2334-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huard RC, Fabre M, de Haas P, Lazzarini LC, van Soolingen D, Cousins D, Ho JL. Novel genetic polymorphisms that further delineate the phylogeny of the Mycobacterium tuberculosis complex. J Bacteriology. 2006;14:4271–4287. doi: 10.1128/JB.01783-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedwell J, Kario SK, Behr MA, Bygraves JA. Identification of substrains of BCG vaccine using multiplex PCR. Vaccine. 2001;14:2146–2151. doi: 10.1016/S0264-410X(00)00369-8. [DOI] [PubMed] [Google Scholar]
- Seki M, Sato A, Honda I, Yamazaki T, Yano I, Koyama A, Toida I. Modified multiplex PCR for identification of Bacillus Calmette-Guerin substrain Tokyo among clinical isolates. Vaccine. 2005;14:3099–3102. doi: 10.1016/j.vaccine.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Tuberculosis Research Committee (Ryoken) Drug-resistant Mycobacterium tuberculosis in Japan: a nationwide survey, 2002. Int J Tuberc Lung Dis. 2007;14:1129–1135. [PubMed] [Google Scholar]
- Morokuma Y, Uchida Y, Karashima T, Fujise M, Imamura S, Kayamori Y, Kang DC. Identification of Mycobacterium tuberculosis Complex Strains in Kyushu University Hospital. J J Clin Microbiol (in Japanese) 2008;14:177–182. [Google Scholar]
- Muyoyeta M, Mwanza WC, Kasese N, Cheeba-Lengwe M, Moyo M, Kaluba-Milimo D, Ayles H. Sensitivity, specificity, and reproducibility of the Capilia TB-Neo assay. J Clin Microbiol. 2013;14:4237–4239. doi: 10.1128/JCM.02441-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu MC, Chen HY, Wu MH, Huang WL, Kuo YM, Yu FL, Jou R. Evaluation of the rapid MGIT TBc identification test for culture confirmation of Mycobacterium tuberculosis complex strain detection. J Clin Microbiol. 2011;14:802–807. doi: 10.1128/JCM.02243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;14:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]


