Abstract
Hepatocellular carcinoma (HCC) is a common malignancy that affects a large number of patients worldwide, with an increasing incidence in the United States and Europe. The therapies that are currently available for patients with inoperable HCC have limited benefits. Although molecular targeted therapies against selected cell signaling pathways have shown some promising results, their impact has been minimal. There is a need to identify and explore other targets for the development of novel therapeutics. Several non-protein coding RNAs (ncRNA) have recently been implicated in hepatocarcinogenesis and tumor progression. These ncRNA genes represent promising targets for cancer. However, therapeutic targeting of ncRNA genes has not been employed for HCC. The use of antisense oligonucleotides and viral vector delivery approaches has been shown to be feasible approaches to modulate ncRNA expression. HCC is an optimal cancer to evaluate novel RNA based therapeutic approaches because of the potential of effective delivery and uptake of therapeutic agents to the liver. In this review, we discuss selected ncRNA that could function as potential targets in HCC treatment and outline approaches to target ncRNA expression. Future challenges include the need to achieve site-specific targeting with acceptable safety and efficacy.
Keywords: Antagomirs, HCC, LNA-antimiR, miRNA, non-coding RNA, ultraconserved
INTRODUCTION
Hepatocellular carcinoma (HCC) is a deadly cancer that affects a large number of patients worldwide, with an increasing incidence in the United States and Europe [1]. The therapies that are currently available for patients with inoperable HCC have limited benefits. There has been much interest in molecular targeted therapies that can target selected cell signaling pathways and protein mediators of tumor growth and progression. One such agent, sorafenib, has been evaluated and entered clinical practice for HCC [2]. Although such molecular targeted therapies have shown some promising results, their impact has been minimal. There is a need to identify and explore other targets for the development of novel therapeutics. A number of transcribed RNA genes that do not encode for protein, non-coding RNAs (ncRNA), have recently been implicated in hepatocarcino-genesis and tumor progression. As a result, these ncRNA genes represent promising targets for cancer and offer promise of novel therapeutic approaches for liver cancers. In this review, we discuss selected ncRNAs that can function as potential targets in HCC treatment and outline approaches to target ncRNA expression.
ncRNAs IN HCC BIOLOGY
Over the last decade there has been growing evidence to support the existence and a functional role of ncRNAs in human cancers [3–5]. Broadly, ncRNAs can be classified in two classes based on their size: small RNAs of less than 200 nucleotides (nt) and long RNAs > 200 nt. Both these groups comprise of functionally heterogeneous types of RNA. Although several groups of ncRNAs with functional similarity have been recognized, a distinct functional or structural basis for further classification is not available. Several ncRNAs have been identified and characterized and their involvement in cellular physiology or tumor biology has been elucidated. However, a large number of ncRNA transcripts remain uncharacterized and their role in cancer biology remains undefined.
Amongst the small ncRNAs, the microRNAs (miRNA) have been the most extensively studied. miRNAs are small ncRNAs which are approximately 22nt in length that can modulate gene expression through mechanisms such as post-translational degradation of mRNAs (as extensively described in [6,7]). miRNAs have been shown to be altered in expression in HCC as well as in several other tumors [8, 9]. They have been shown to play a remarkable role in carcinogenesis through the modulation of expression of critical genes encoding tumor-suppressors or oncogenes. Thus, deregulated expression of miRNAs may contribute to the formation of cancer or its progression.
Several studies have identified alterations in miRNAs in HCC, with specific miRNAs consistently identified as either increased or decreased in HCC tissues and cell lines in comparison to normal counterparts (Table 1). Increased expression of selected miRNAs has been linked to alterations in critical biological processes. For example, miR-221/222 is frequently increased in HCC and can mediate specific oncogenic effects through mechanisms such as the suppression of the tumor suppressor gene phosphatase and tensin homolog (PTEN) [10, 11], blocking proliferation by inhibiting the p27 and p57 checkpoint proteins of the cell cycle [12, 13], activating the mammalian target of rapamycin pathway by suppressing the DNA damage-inducible transcript 4 (DDIT4) [13], and promoting apoptosis through the modulation of the B Cell lymphoma (Bcl-2) family members [14]. Similarly miR-21, a miRNA that is deregulated in many cancers and also frequently increased in HCC, can mediate specific effects through the inhibition of the PTEN and AKT pathway [15]. The combinatorial effect of a single miRNA on multiple genes, or of several miRNAs on a single gene emphasize the unique potential of these aberrantly expressed miRNAs as therapeutic targets if their primary cellular effect contributes to tumorigenesis or tumor behavior. Other miRNAs that are increased in HCC include miR-181, miR-106, miR-224, etc. [13].
Table 1.
Examples of non-coding RNAs that are aberrantly expressed in HCC.
| UP Regulated |
DOWN Regulated |
|
|---|---|---|
| microRNAs | ||
| miR-221/222 | miR-199a | |
| miR-21 | miR-26 | |
| miR-106 | miR-29 | |
| miR-181 | miR-122 | |
| miR-18 | Let7 | |
| miR-182 | miR-101 | |
| miR-224 | miR-148 | |
| miR-34a | miR-125 | |
| miR-20 | miR-214 | |
| Long ncRNAs | TUC338 HULC | MEG3 |
A reduction of some miRNAs has also been reported in HCC. Loss of miR-26 can result in multiple cellular effects [16, 17]. Along with the over-expression of some miRNAs, the loss of other miRNAs is responsible for the promotion of HCC. The reduction of miR-199a-3p in HCC results in hyper-activation of the MET signaling and the extracellular-signal related kinase pathway [18–20]. miR-122 is usually lost in HCC and induces hepatocarcinogenesis by promoting aberrant proliferation, modulation of cell adhesion and induction of an hepatoblastic signature [21–23]. Lack of miR-29 promotes apoptosis via a mitochondrial pathway involving Bcl-2 and Mcl-1 [24] but can also have more general effects by acting on the methylation machinery and repressing tumor suppressor genes [25].
Recent evidence supports the involvement of miRNAs in the early phases of hepatocarcinogenesis. miRNA expression profiling of non-tumorous liver was distinct from primary HCCs, although there were no discernable differerences between primary HCC and HCC with venous metastases [26]. Aberrant expression of miRNAs in the early phases was confirmed by other studies that revealed aberration of selected miRNAs, such as miR-216a and miR-100 in pre-neoplastic lesions [27, 28].
Although the involvement of long ncRNAs (size greater than 200nt) is less well established, selected long ncRNAs have been shown to be aberrantly expressed in human HCC. The literature regarding the functional roles of long ncRNA transcripts is sparse, and their mechanisms of action are poorly understood. It has been postulated that they can act as enhancers of transcription of protein-coding gene expression. Although their precise functional effects are not established, it is likely that long ncRNAs can modulate gene expression in HCC cells. Other postulated mechanisms by which long ncRNAs may exert effects on gene expression is by regulation of transcription, by regulation of mRNA dynamics at a post-transcriptional level, or mediate epigenetic changes [4, 5, 29]. Genome wide expression profiling approach has identified individual long ncRNAs that are abundantly expressed in HCC cell lines and tissues [30–32]. These include HULC and TUC338. The involvement of these long ncRNAs in HCC is supported by studies showing reduction of cellular growth when ncRNA expression is inhibited.
POTENTIAL ncRNA TARGETS IN HCC
There are several ncRNAs that are potential targets for therapeutic intervention, and of these some promising targets are briefly discussed here as examples of the utility of this approach.
miR-26
miR-26 is an example of a miRNA with promise for developing novel ncRNA based therapies for HCC. This miRNA has been shown to have a multitude of effects in the liver such as modulation of hepatocyte growth and inflammatory responses that involve several key mediators of hepatocarcinogenesis such as interleukin-6 [16, 17]. miR-26 can modulate HCC growth by triggering the cyclin-dependent cell arrest in resting phases of the cell cycle and can also reduce development of liver tumors in animal models [17]. Consistently, miR-26 was found to be reduced in human HCC in comparison to adjacent normal liver [16]. Thus, reintroduction of miR-26 may be an effective strategy to counteract the promotion and progression of HCC. In addition, tumors with low miR-26 levels have a poorer prognosis with a reduction in overall survival. Moreover, miR-26 expression can predict response to adjuvant interferon suggesting a potential role for miR-26 targeted therapies in combination regimens.
miR-221/222
Another example of miRNAs that can be further studied for therapeutic targeting are miR-221/222. miR-221 and miR-222 belong to a cluster on chromosome X and have been implicated in several malignancies in addition to HCC. In HCC, miR221 and miR222 were found significantly up-regulated when compared with adjacent liver tissues, and their expression is higher in poorly differentiated HCC [13]. For this reason miR-221/222 represents an ideal candidate for therapies aiming at miRNA silencing. As discussed above, there are several mechanisms through which miR-221/222 can promote liver growth such as modulation of cell cycle progression and proliferation through effects on key regulatory molecules such as p27Kip1, and cyclin-dependent kinase inhibitor 1C p57 (CDKN1C/p57) [12, 13, 33]. At the same time miR-221/222 can inhibit apoptosis by modulating the expression of the pro-apototic Bcl2-modifying factor (Bmf) [14]. miR-221/222 can also control the survival pathways related to Akt-mTOR and thereby enhance effects on cell survial and invasion [10, 11, 13]. In line with these in vitro data, human HCCs with high expression of miR-221/222 are correlated with worse outcome [12, 14, 34]. Although the effects downstream of miR-221 have been more and more elucidated, the mechanisms responsible for the increased expression of miR-221/222 in HCC are less clear. Recent evidence suggests that miR-221/222 are induced by MET through the activator protein 1 (AP-1) transcription factor, but further studies are warranted [10].
TUC338
TUC338 is a long ncRNA that contains a sequence which is very highly conserved across species. TUC338 is increased in HCC compared to non-malignant hepatocytes [30]. Moreover, TUC338 expression progressively increases from normal liver to cirrhosis, a preneoplastic state, to HCC. Thus, the expression correlates with disease progression, and raises the possibility that it may be involved in malignant transformation. In vitro studies showed the abilities of TUC338 to stimulate anchorage dependent and anchorage independent growth of human and murine HCC cells. These effects are mediated by the control of the G1/S checkpoint of the cell cycle through the inhibition of p16INK4a and the activation of the cyclin D1/ cyclin dependent kinases (CDK) 4–6. Thus, targeting long ncRNA such as TUC338 may be a promising strategy to modulate HCC growth.
APPROACHES TO TARGET miRNA EXPRESSION
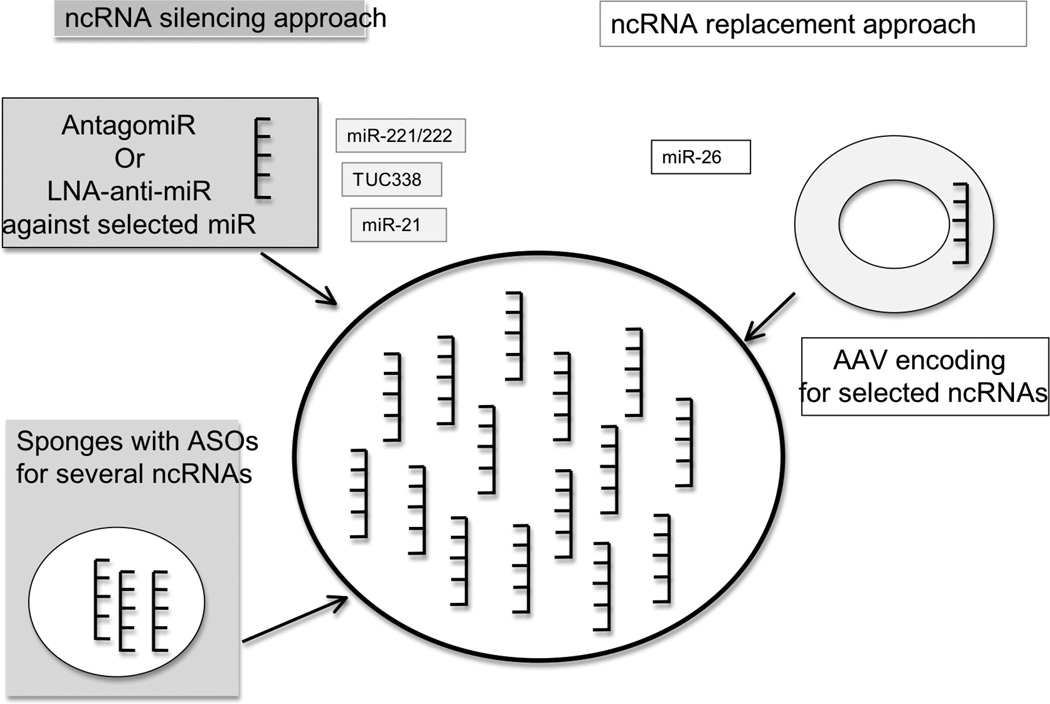
For ncRNA such as selected miRNAs that are deregulated in expression in cancer and that have been shown to have relevant cancer enhancing effects, a therapeutic approach would aim to normalize their expression. For miRNAs that are decreased in expression, restoration can be achieved by the use of miR-replacement therapies comprising of mature functional miRNA or miRNA mimetics. Therapeutic strategies to replace miRNAs have been recently attempted [35]. For ncRNAs such as miRNAs that are over-expressed in HCC, ncRNA targeting strategies have focused on approaches to silence genes and inhibiting miRNA expression by antisense approaches (Fig. 1).
Fig. (1).
Therapeutic approaches to target non coding RNAs. For ncRNA that are increased in expression in HCC, such as miR-221/222, miR-21, or TUC-338, therapeutic strategies to reduce their expression are considered. AntagomiRs are chemically modified, cholesterol conjugated, single strand RNA analogues complementary to selected miRNAs (or other ncRNAs). The stability and specificity of antisense miRNA could be increased by conjugation to locked nucleic acid (LNA). Several antisense oligonucleotides could potentially be combined in the so-called sponges. For ncRNA that are decreased in expression in HCC such as miR-26, potential replacement strategies using adenovirus-associated virus may be feasible.
Antisense Oligonucleotides (ASO)
Antisense oligonucleotides (ASO) are single-strand DNA molecules that complement target RNA through sequence-specific base pairing. Although they were discovered more than 30 years ago [36], the only ASO therapy approved by the US Food and Drug Administration to date is fomivirsen sodium (Vitravene, Isis, Carlsbad, CA), to target cytomegalovirus mRNA in retinitis. The major limitation of ASO is the delivery and their poor stability. Intravenously administered encapsulated ASOs were shown to be effectively incorporated in the liver [37] suggesting that the promise of the use of ASOs to target diseases in the liver. ASOs have also been developed to target miRNAs. Krutzfeldt et al. [38] designed chemically modified, cholesterol conjugated, single-strand RNA analogues complementary to miRNAs, termed antagomirs. The intravenous administration of antagomir-122 in mice resulted in a marked decrease of endogeneous levels of miR-122 by inducing specific miRNA degradation. Levels of miR-122 were undetectable for 23 days after injections. Concomitant reduction in expression of miR-122 target genes involved in cholesterol metabolism was accompanied with a decrease in plasma cholesterol levels for two weeks. Experiments with other miRNAs that are expressed in several tissues outside the liver (such as miR-16) showed that antagomirs may achieve broad distribution and have therapeutic silencing effects in most tissues. Antagomirs were well tolerated with no side effects on mice body weight or liver necrosis. Anti-miRNA oligonucleotides have also been proved effective for targeting metastatic liver tumors [39]. Current clinical trials in HCC for ASO therapy include antisense approaches to target XIAP mRNA, but no antagomirs have reached this phase of clinical investigation (Table 2). However, there are promising preclinical data that have been recently reported. Park et al reported an 80–90% of reduction of miR-221 in the liver of HCC-bearing mice treated with cholesterol-tagged anti-miR-221. Tumor growth was stabilized and markers of proliferation inhibited. These data would support further clinical studies on the use of anti-sense oligonucleotides targeted towards critical HCC specific miRNA such as miR-221 [40, 41].
Table 2.
Clinical trials involving antisense therapies in liver diseases.
| Test Compound | Phase | Disease | Combination Therapy |
Line of Theray | Primary Endpoint |
Secondary Endpoint |
Status | |
|---|---|---|---|---|---|---|---|---|
| ASO | XIAP Antisense (AEG35156) | I–II | advanced HCC | sorafenib | first (prior local therapy allowed) | safety and tolerability | response rate | recruiting |
| HIF-1α Antisense (EZN-2968) | I | solid tumors with liver metastases | alone | - | effect on HIF-1α mRNA | safety; effects on HIF1α protein; resposne rate | recruiting | |
| LNA-antimiR | Miravirsen (LNA-antimiR-122) SPC2649 | I | healthy volunteers (men) | alone | NA | safety and tolerability | pharmacokinetics; pharmacodynamics | completed |
| Miravirsen (LNA-antimiR-122) SPC2650 | I | healthy volunteers | alone | NA | safety and tolerability | pharmacokinetics; pharmacodynamics | active but not recruiting | |
| Miravirsen (LNA-antimiR-122) SPC2651 | II | hepatitis C | alone | first | safety and tolerability | pharmacokinetics; effect on viral titer | recruiting |
Locked Nuclei Acid-antimiR (LNA-antimiR)
In order to increase the stability and the specificity of antisense miRNAs, LNA-modified oligonucleotides have been synthesized as unconjugated LNA/DNA mixmers with a complete phosphorothioate backbone [42]. LNA-antimiRs show improved potency of inhibition in vitro compared to ASO. In mice, intraperitoneal injections of LNA-antimiR-122 were shown to be more efficient in antagonizing miR-122 compared to antagomirs. Following LNA-antimiR injection miR-122 sequesters in a heteroduplex with LNA-antimiR. Primate studies with LNA-antimiR have also been performed in African green monkeys. Systemic administration of phosphate buffered saline-formulated LNA-antimiR-122 resulted in dose-dependent and sustained decrease in total plasma cholesterol for seven weeks in primates, reduced expression of miR-122, and accumulation of the LNA-antimiR in the liver. Clearance of LNA-antimiR compound from the liver occurred after around 3 months with decrement of effects on miRNA and target mRNA expression. No collateral effects were noted in primates, with the exception of transient alteration of creatinine phosphokinase, aminotransferases and bilirubin in those animals who underwent liver biopsies [42]. miR-122 can bind to the HCV genome and act as a co-factor for the accumulation of viral RNA in cultured liver cells [43]. LNA-antimiR-122 (SPC3649, Santaris Pharma, Hørsholm, Denmark) has been assessed in HCV chronically infected chimpanzees as a new anti-HCV therapy [44]. Administration of 5 mg/kg of SPC3649 resulted in reduction of HCV RNA in the serum after 3 weeks of therapy. Concomitantly miR-122 levels in the liver were remarkably reduced up to 8 weeks after the last dose as a result of the sequestration of miR-122 in a heteroduplex with SPC3649. Interestingly, viral resistance to therapy did not occur and no rebound in viremia during treatment or occurrence of adaptive mutations in the miR-122 seed sites was noted. Histological improvement was also noted in addition to prolonged suppression of viremia. The tolerability profile was safe and no abnormalities in liver function tests were noted. On the bases of these results phase I trials on healthy volunteers have been conducted to delineate the safety profile of SPC3649 in humans and a phase II trial is ongoing in hepatitis C patients (Table 2).
Other Strategies for miRNA Targeting
miRNA sponges are transcripts that are expressed from strong promoters and can function as competitive inhibitors. They contain binding sites for miRNAs either in a noncoding transcript or in the 3’-untranslated region (UTR) of a reporter gene. They can contain sites for one miRNA family or for a combination of miRNAs. When vectors encoding these sponges are transiently transfected into culture cells, sponges repress miRNA targets with effects similar to those with ASO. The use of viral vectors enabled continuous miRNA inhibition over long duration [45]. The effects of sponges remain to be determined in vivo. The use of short hairpin RNAs is another strategy of note. A short hairpin RNA contains a tight hairpin turn that can silence gene expression via RNA interference. shRNA use a vector with a U6 promoter to ensure constant expression when introduced in a cell. This vector is usually passed on to daughter cells, allowing the gene silencing to be inherited. The shRNA hairpin structure is cleaved by the cellular machinery into siRNA, which is then bound to the RNA-induced silencing complex (RISC) that in turn binds to and cleaves mRNAs which match the siRNA. The intravenous injection of a pool of shRNA-deliver AAV vectors into mice can achieve robust shRNA expression in hepatocytes and result in dose-dependent liver injury and death [46]. Adenovirus-mediated delivery of shRNA has been extensively used to target specific and selected liver genes but concerns remain about their safety profile in humans. However, the favorable safety profile of adenovirus associated virus (AAV) mediated delivery of shRNA anti microRNA makes this a promising approach.
EFFECTIVE DELIVERY OF NCRNA DIRECTED THERAPIES TO THE LIVER
HCC is an optimal cancer to evaluate novel RNA based targeting approaches because of the potential of effective delivery and uptake of therapeutic agents to the liver. The likelihood of progress in targeting ncRNA for the treatment of liver cancer is increased by demonstrations of the availability and feasibility of both viral and non-viral systems for effective delivery of ncRNA targeting molecules to the liver [47].
Viral Delivery Methods
Gene-therapy vectors based on delivery of a miRNA or miRNA-mimetic by AAV appears promising. Kota et al. [17] proposed an AAV-based miRNA delivery in which miR-26a is delivered to the liver of mice with MYC-induced HCC to replace this miRNA which is usually reduced in HCC. A single tail vein injection of scAAV8.miR-26a resulted in high miR-26a expression level in the liver after 3 weeks with no signs of hepatotoxicity or modulation of other miRNA pathways. Moreover, cAAV8.miR-26a protected mice from developing liver tumors by reducing proliferative potential and inducing apoptotic activation in hepatocytes. The advantage of this technique is that miRNAs delivered in viral vectors are continually transcribed and allows sustained high level of expression in target tissues. The use of tissue-specific promoters could restrict this expression to selected cell types. AAV-based gene therapy has been extensively studied, and several clinical studies are now ongoing in cancer patients. However no AAV gene therapy protocols are under investigation in HCC, neither AAV-miRNA delivery in humans.
Unwanted immune responses are a major concern with gene therapy. Antigen presentation by professional antigen-presenting cells (APCs) results in the induction of a cellular immune response. To prevent immune-mediated clearance of transduced cells, Annoni et al. [48] recently developed an interesting strategy to avoid expression of a gene-encoded construct in APCs. They incorporated sequences for the APC-specific microRNA miR-142 into an antigen-encoding transgene, so that the expression of miR-142 in APC resulted in suppression of the transgene selectively in the APC. This approach has been extended to the liver, given that the liver is known to be a site of tolerance induction, particularly in the context of organ transplantation. Thus, miR-122 has been used to de-target the expression of a transgene from hepatocytes [48].
Non-viral Delivery Methods
Liposomal or nanoparticle delivery systems are being evaluated for delivery of synthetic anti-ncRNA therapies. One approach involves the use of stable nucleic-acid lipid particles incorporated with poly ethylene glycol polymer chains [49]. The delivery of siRNAs by this system to the livers of non-human primates showed increased efficacy but reduced toxicity [45]. Another approach has been the use of ligands such as N-acetylgalactosamine, galactose, apolipoprotein or alpha-tocopherol that bind to cell surface receptors expressed on hepatocytes and can thereby to achieve targeting to the liver [50–54]. These delivery approaches enable the delivery of ncRNA therapeutics such as anti-miRNA to liver cells.
CHALLENGES
Despite the considerable promise of ncRNAs as therapeutic targets, there are several challenges that will need to be addressed prior to the therapeutic application of these approaches. First, the safety of the approach will need to be established with certainty. Therapeutic agents comprised of double stranded RNA, or the use of viral delivery systems may potentially lead to activation of innate immune responses. Targeting ncRNAs that are involved in regulation of gene expression may result in un-expected off target gene effects. Modulation of miRNA expression and competition with endogeneous miRNAs may have potential effects on normal cellular physiology. As an example, saturation of exportin 5, a miRNA nucleus membrane transporter has been hypothesized to result in adverse effects within the liver [46]. Next, the effectiveness of approaches to target ncRNAs to stop or reduce tumor growth and spread, as well as improve survival in HCC will need to be demonstrated to establish therapeutic benefit in HCC. These issues will need to be systematically evaluated in preclinical studies but are neither insurmountable nor dissimilar to safety and effectiveness considerations for other molecular targeted therapies in development.
CONCLUSIONS
HCC is a major health problem worldwide, and moreover, the incidence is increasing in many regions such as the United States. The need for effective therapies is unmet, despite major advances in defining the molecular pathogenesis and identifying new approaches to treatment. An approach to systematically evaluate new molecular targets such as ncRNA is justified by this need. Several potential target ncRNA exist and the feasibility of effective targeting has been shown in several preclinical studies. It is hoped that these efforts will ultimately result in novel approaches to the therapy of liver cancer.
ACKNOWLEDGEMENTS
This work is supported by Grant DK069370 from the National Institutes of Health.
ABBREVIATIONS
- AAV
Adeno-associated Virus
- AP-1
Aactivator protein 1
- APCs
Antigen-presenting cells
- ASO
Antisense oligonucleotides
- Bcl-2
B cell lymphoma
- Bmf
Bcl2-modifying Factor
- CDK
Cyclin dependent Kinase
- CDKN1C/p57
Cyclin-dependent kinase inhibitor 1C p57
- DDIT4
DNA damage-inducible transcript 4
- DNA
Deoxyribonucleic acid
- HCC
Hepatocellular carcinoma
- LNA
Locked nucleic acid
- miRNA, miR
microRNA
- mTOR
Mammalianc target of rapamycin
- PTEN
Phosphatase and tensin homolog
- RISC
RNA-induced silencing complex
- RNA
Ribonucleic acid
- shRNA
Short hairpin RNA
- UCR
Ultraconserved region
- ucRNA
Ultraconserved non coding RNA
- UTR
Untranslated Region
Footnotes
CONFLICT OF INTEREST
None of the authors have anything to disclose.
REFERENCES
- 1.Yang JD, Roberts LR. Hepatocellular carcinoma: A global view. Nat. Rev. Gastroenterol. Hepatol. 2010;7(8):448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, Schwartz M, Porta C, Zeuzem S, Bolondi L, Greten TF, Galle PR, Seitz JF, Borbath I, Haussinger D, Giannaris T, Shan M, Moscovici M, Voliotis D, Bruix J. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008;359(4):378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 3.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum. Mol. Genet. 2010;19(R2):R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Louro R, Smirnova AS, Verjovski-Almeida S. Long intronic noncoding RNA transcription: expression noise or expression choice? Genomics. 2009;93(4):291–298. doi: 10.1016/j.ygeno.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 5.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009;10(10):704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J. Pathol. 2011;223(2):102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heneghan HM, Miller N, Kerin MJ. MiRNAs as biomarkers and therapeutic targets in cancer. Curr. Opin. Pharmacol. 2010;10(5):543–550. doi: 10.1016/j.coph.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P, Gasparini P, Gonelli A, Costinean S, Acunzo M, Condorelli G, Croce CM. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16(6):498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Wong QW, Ching AK, Chan AW, Choy KW, To KF, Lai PB, Wong N. MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin. Cancer Res. 2010;16(3):867–875. doi: 10.1158/1078-0432.CCR-09-1840. [DOI] [PubMed] [Google Scholar]
- 12.Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Giovannini C, Croce CM, Bolondi L, Negrini M. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27(43):5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 13.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc. Natl. Acad. Sci. USA. 2010;107(1):264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gramantieri L, Fornari F, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Croce CM, Bolondi L, Negrini M. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin. Cancer Res. 2009;15(16):5073–5081. doi: 10.1158/1078-0432.CCR-09-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, Qin LX, Man K, Lo CM, Lee J, Ng IO, Fan J, Tang ZY, Sun HC, Wang XW. MicroRNA expression, survival, and response to interferon in liver cancer. N. Engl. J. Med. 2009;361(15):1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, Mendell JR, Mendell JT. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137(6):1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fornari F, Milazzo M, Chieco P, Negrini M, Calin GA, Grazi GL, Pollutri D, Croce CM, Bolondi L, Gramantieri L. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010;70(12):5184–5193. doi: 10.1158/0008-5472.CAN-10-0145. [DOI] [PubMed] [Google Scholar]
- 19.Henry JC, Park JK, Jiang J, Kim JH, Nagorney DM, Roberts LR, Banerjee S, Schmittgen TD. miR-199a-3p targets CD44 and reduces proliferation of CD44 positive hepatocellular carcinoma cell lines. Biochem. Biophys. Res. Commun. 2010;403(1):120–125. doi: 10.1016/j.bbrc.2010.10.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S, Lee UJ, Kim MN, Lee EJ, Kim JY, Lee MY, Choung S, Kim YJ, Choi YC. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2) J. Biol. Chem. 2008;283(26):18158–18166. doi: 10.1074/jbc.M800186200. [DOI] [PubMed] [Google Scholar]
- 21.Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, Yadav A, Nuovo G, Kumar P, Ghoshal K. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J. Biol. Chem. 2009;284(46):32015–32027. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28(40):3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E, Grazi GL, Croce CM, Bolondi L, Negrini M. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67(13):6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 24.Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH, Zhuang SM. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51(3):836–845. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- 25.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA. 2007;104(40):15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong CM, Wong CC, Lee JM, Fan DN, Au SL, Ng IO. Sequential alterations of miRNA expression in hepatocellular carcinoma development and venous metastasis. Hepatology. 2011 doi: 10.1002/hep.25512. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 27.Chen PJ, Yeh SH, Liu WH, Lin CC, Huang HC, Chen CL, Chen DS, Chen PJ. The androgen pathway stimulates microRNA-216a transcription to suppress the TSLC1 tumor suppressor gene in early hepatocarcinogenesis. Hepatology. 2012 doi: 10.1002/hep.25695. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 28.Petrelli A, Perra A, Schernhuber K, Cargnelutti M, Salvi A, Migliore C, Ghiso E, Benetti A, Barlati S, Ledda-Columbano GM, Portolani N, De Petro G, Columbano A. Sequential analysis of multistage hepatocarcinogenesis reveals that miR-100 and PLK1 dysregulation is an early event maintained along tumor progression. Oncogene. 2012 doi: 10.1038/onc.2011.631. J [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 29.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braconi C, Valeri N, Kogure T, Gasparini P, Huang N, Nuovo GJ, Terracciano L, Croce CM, Patel T. Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA. 2011;108(2):786–791. doi: 10.1073/pnas.1011098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, Shimizu M, Tili E, Rossi S, Taccioli C, Pichiorri F, Liu X, Zupo S, Herlea V, Gramantieri L, Lanza G, Alder H, Rassenti L, Volinia S, Schmittgen TD, Kipps TJ, Negrini M, Croce CM. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12(3):215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 32.Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M, Zatloukal K. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132(1):330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 33.Visone R, Russo L, Pallante P, De Martino I, Ferraro A, Leone V, Borbone E, Petrocca F, Alder H, Croce CM, Fusco A. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr. Relat. Cancer. 2007;14(3):791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Wang Y, Yu W, Chen J, Luo J. Expression of serum miR-221 in human hepatocellular carcinoma and its prognostic significance. Biochem. Biophys. Res. Commun. 2011;406(1):70–73. doi: 10.1016/j.bbrc.2011.01.111. [DOI] [PubMed] [Google Scholar]
- 35.Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7(6):759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 36.Stephenson ML, Zamecnik PC. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc. Natl. Acad. Sci. USA. 1978;75(1):285–288. doi: 10.1073/pnas.75.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soni PN, Brown D, Saffie R, Savage K, Moore D, Gregoriadis G, Dusheiko GM. Biodistribution, stability, and antiviral efficacy of liposome-entrapped phosphorothioate antisense oligodeoxynucleotides in ducks for the treatment of chronic duck hepatitis B virus infection. Hepatology. 1998;28(5):1402–1410. doi: 10.1002/hep.510280532. [DOI] [PubMed] [Google Scholar]
- 38.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 39.Huynh C, Segura MF, Gaziel-Sovran A, Menendez S, Darvishian F, Chiriboga L, Levin B, Meruelo D, Osman I, Zavadil J, Marcusson EG, Hernando E. Efficient in vivo microRNA targeting of liver metastasis. Oncogene. 2011;30(12):1481–1488. doi: 10.1038/onc.2010.523. [DOI] [PubMed] [Google Scholar]
- 40.Park JK, Kogure T, Nuovo GJ, Jiang J, He L, Kim JH, Phelps MA, Papenfuss TL, Croce CM, Patel T, Schmittgen TD. miR-221 silencing blocks hepatocellular carcinoma and promotes survival. Cancer Res. 2011;71(24):7608–7616. doi: 10.1158/0008-5472.CAN-11-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kasinski AL, Slack FJ. arresting the Culprit: targeted antagomir Delivery to Sequester oncogenic mir-221 in HCC. Molecular Therapy–Nucleic Acids. 2012 doi: 10.1038/mtna.2012.2. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 43.Jangra RK, Yi M, Lemon SM. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J. Virol. 2010;84(13):6615–6625. doi: 10.1128/JVI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4(9):721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441(7092):537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 47.Alexander IE, Cunningham SC, Logan GJ, Christodoulou J. Potential of AAV vectors in the treatment of metabolic disease. Gene Ther. 2008;15(11):831–839. doi: 10.1038/gt.2008.64. [DOI] [PubMed] [Google Scholar]
- 48.Annoni A, Brown BD, Cantore A, Sergi LS, Naldini L, Roncarolo MG. In vivo delivery of a microRNA-regulated transgene induces antigen-specific regulatory T cells and promotes immunologic tolerance. Blood. 2009;114(25):5152–5161. doi: 10.1182/blood-2009-04-214569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heyes J, Palmer L, Bremner K, MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control Release. 2005;107(2):276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 50.Rozema DB, Lewis DL, Wakefield DH, Wong SC, Klein JJ, Roesch PL, Bertin SL, Reppen TW, Chu Q, Blokhin AV, Hagstrom JE, Wolff JA. Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc. Natl. Acad. Sci. USA. 2007;104(32):12982–12987. doi: 10.1073/pnas.0703778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R, Borodovsky A, Borland T, Constien R, de Fougerolles A, Dorkin JR, Narayanannair Jayaprakash K, Jayaraman M, John M, Koteliansky V, Manoharan M, Nechev L, Qin J, Racie T, Raitcheva D, Rajeev KG, Sah DW, Soutschek J, Toudjarska I, Vornlocher HP, Zimmermann TS, Langer R, Anderson DG. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol. 2008;26(5):561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim SI, Shin D, Choi TH, Lee JC, Cheon GJ, Kim KY, Park M, Kim M. Systemic and specific delivery of small interfering RNAs to the liver mediated by apolipoprotein A-I. Mol. Ther. 2007;15(6):1145–1152. doi: 10.1038/sj.mt.6300168. [DOI] [PubMed] [Google Scholar]
- 53.Nishina K, Unno T, Uno Y, Kubodera T, Kanouchi T, Mizusawa H, Yokota T. Efficient in vivo delivery of siRNA to the liver by conjugation of alpha-tocopherol. Mol. Ther. 2008;16(4):734–740. doi: 10.1038/mt.2008.14. [DOI] [PubMed] [Google Scholar]
- 54.Sato A, Takagi M, Shimamoto A, Kawakami S, Hashida M. Small interfering RNA delivery to the liver by intravenous administration of galactosylated cationic liposomes in mice. Biomaterials. 2007;28(7):1434–1442. doi: 10.1016/j.biomaterials.2006.11.010. [DOI] [PubMed] [Google Scholar]