Abstract
PIK3CA, which encodes the catalytic p110α subunit of phosphatidylinositol 3-kinase α, is the most frequently mutated oncogene in human cancers. Targeting mutant p110α holds great promise for cancer therapy. However, it is challenging to develop p110α isoform-specific inhibitors. Most p110α mutations occur at two hotspot regions: an acidic cluster (E542, E545 and Q546) in the helical domain and a histidine residue (H1047) in the kinase domain. We recently discovered that p110α helical domain mutant proteins, but not the kinase domain mutant proteins, directly associate with IRS1. Moreover, we demonstrated that disruption of protein-protein interaction between p110α helical domain mutant and IRS1 inhibits the growth of tumors with such mutations. The direct protein interaction between IRS1 and p110α helical domain mutants may provide a more accessible target for developing novel precision cancer therapy.
Keywords: Cancer, Molecular biology, Genomics
Introduction
Targeted cancer therapy is transforming the way that doctors treat cancer patients. In contrast to standard chemotherapies that indiscriminately kill rapidly dividing cells in human body, targeted cancer therapies are designed to affect specific molecules that are part of the pathways and processes used by cancer cells to grow, divide, and metastasize (Gschwind et al., 2004). A major advantage of targeted therapies is that they elicit fewer toxic side effects than standard chemotherapies because they often cause little or no damage to normal cells. The best targets for cancer therapies are these molecules or pathways that are only present in cancer cells but not in normal cells (Gschwind et al., 2004). For example, genetically altered proteins resulting from somatic mutations and translocations are ideal targets. A paradigm of targeted cancer therapy is the successful treatment of chronic myelogenous leukemia (CML) with Imatinib (branded as Gleevec) (Druker et al., 2001). Over 90% of CML patients harbor a reciprocal translocation between chromosome 9 and 22, which results in an in-frame gene fusion between BCR and ABL1 gene. Imatinib inhibits the tyrosine kinase activity of BRC-Abl fusion protein, thereby killing only CML cells but not normal cells (Druker et al., 2001). Recently, the FDA approved Vemurafenib, a small molecule that inhibits mutant B-raf kinase activity, for treating metastatic melanoma patients with a BRAF V600E mutation. This success highlights the importance of targeting mutant oncoproteins for cancer therapy (Sosman et al., 2012). Given that recent cancer genome sequencing studies reveal that PIK3CA is the most frequently mutated oncogene in human cancers (Garraway and Lander, 2013), the mutant PIK3CA gene product, p110α, is an important target for cancer therapy. Here we review the promises and challenges of targeting mutant p110α.
The PI3K/AKT pathway is a central intracellular signaling pathway
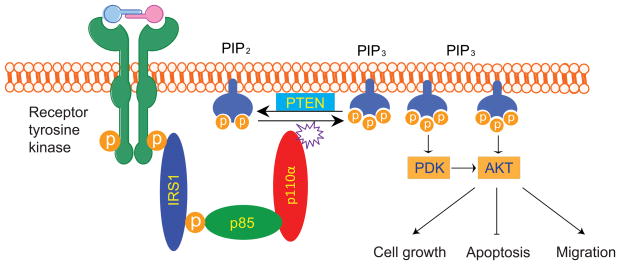
Phosphatidylinositol 3-kinases (PI3Ks) play key roles in regulating cell proliferation, survival and motility (Figure 1). The PI3K family proteins are classified into three groups (see more detailed discussion below). The class I PI3Ks consist of a catalytic p110 subunit and a regulatory p85 subunits. In the basal state, the regulatory p85 subunit stabilizes the catalytic p110 subunit and inhibits its enzymatic activity (Liu et al., 2009). Upon growth factor stimulation, the SH2 domains of p85 bind to the phospho-tyrosine residues on the receptor protein kinases or adaptor proteins such as insulin receptor substrate 1 (IRS1), thereby activating the lipid kinase activity of PI3Ks (Cantley, 2002). Activated PI3Ks convert phosphatidylinositol-4,5-bisphosphate (PIP2) to phosphatidylinositol-3,4,5-triphosphate (PIP3). PIP3 then recruits proteins that contain a pleckstrin homology domain (PH domain) including AKT serine/threonine kinase and 3-phosphoinositide-dependent kinase -1 (PDK1) to the cell membrane (Cantley, 2002). The active membrane associated AKT and PDK1 phosphorylate numerous protein targets including MDM2, GSK3β, IKK, tubrin and a subset of forkhead transcription factors (Cantley, 2002). The tumor suppressor gene Phosphatase and Tensin homolog deleted on chromosome Ten (PTEN) catalyzes the dephosphorylation of PIP3 to generate PIP2, thereby negatively regulating the PI3K signaling pathways (Li et al., 1997). Inactivating mutations of PTEN have been found to occur in several different tumor types (Li et al., 1997).
Figure 1. The PI3K/AKT signaling pathway.
Growth factor stimulation leads to activation of receptor protein tyrosine kinases, which in turn phosphorylate adaptor proteins such as IRS1. Phospho-IRS1 then recruits the PI3K heterodimers (p85 regulatory and p110 catalytic subunits) and activates the lipid kinase activity. The activated PI3Ks convert PIP2 to PIP3. The second messenger PIP3 recruits PDK and AKT to cytoplasmic membrane, where PDK phosphorylates and activates AKT. Activated AKT phosphorylates an array of downstream targets that control cell proliferation, apoptosis and cell migration.
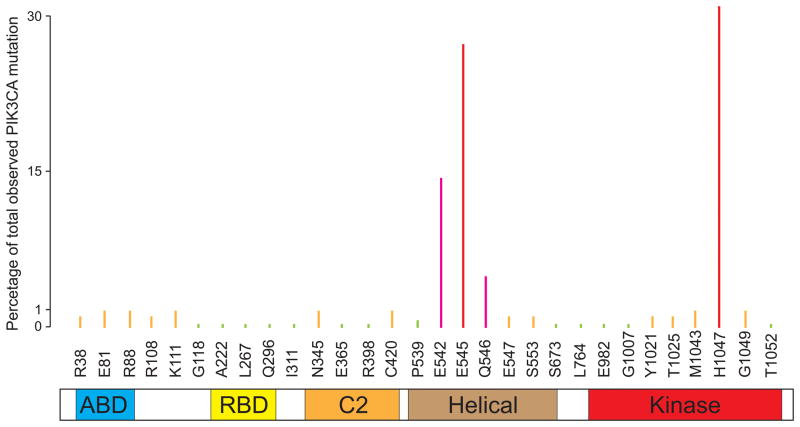
The discovery of PIK3CA/p110α mutation in human cancers provides an excellent target for cancer therapy
In the mutational analysis of PI3K gene family, Samuel et al. discovered almost a decade ago that PIK3CA (encoding p110α) was mutated in a variety of human cancers including brain, breast, colon, gastric and lung cancers (Samuels et al., 2004). Follow-up studies by many laboratories and recent comprehensive cancer genome sequencing of a large collection of various human cancers by The Cancer Genome Atlas (TCGA) project revealed that PIK3CA is the most frequently mutated oncogene in human cancers (Table 1) (Bachman et al., 2004, Broderick et al., 2004, Campbell et al., 2004, Lee et al., 2005, Levine et al., 2005, Li et al., 2005, Oda et al., 2005, Garraway and Lander, 2013). Interestingly, nearly all of the mutations are heterozygous and the majority are clustered in at two hotspot regions: an acidic cluster (E542, E545 and Q546) in the helical domain and a histidine residue (H1047) in the kinase domain (Figure 2). Results from in vitro lipid kinase assays indicated that the hotspot mutations, H1047R and E545K, resulted in increased lipid kinase activity, suggesting that the mutant p110α indeed acts as an oncogene (Samuels et al., 2004). Targeted inactivation of the PIK3CA oncogenic mutations in human colorectal cancer cell lines showed that oncogenic alleles of PIK3CA render cancer cells growth advantage in low serum condition, attenuation of apoptosis and facilitating tumor invasion (Samuels et al., 2005). Interestingly, mutant p110α appears to selectively activate AKT1 and the forkhead transcription factors FKHR and FKHRL1 (Samuels et al., 2005). Given that the oncogenic mutant p110α is a lipid kinase, it is a very promising drug target for cancer therapy as it is actionable, but a potential limitation is specificity.
Table 1.
Mutation frequency of PIK3CA in major human cancers.
| Cancer type | Mutation frequency | References |
|---|---|---|
| Brain | 5–20% | (Samuels et al., 2004, Hartmann et al., 2005) |
| Breast | 21–28% | (Bachman et al., 2004, Saal et al., 2005) |
| Cervix | 36% | (Janku et al., 2011) |
| Colorectal | 30% | (Samuels et al., 2004) |
| Endometrium | 23% | (Garcia-Dios et al., 2013) |
| Esophagus | 6–11% | (Dulak et al., 2013, Phillips et al., 2006) |
| Gastric | 9–25% | (Li et al., 2005, Samuels et al., 2004) |
| Liver | 28–35% | (Lee et al., 2005, Colombino et al., 2012) |
| Lung | 5% | (Yamamoto et al., 2008) |
| Ovary | 25% | (Levine et al., 2005) |
| Pancreas | 11% | (Schonleben et al., 2006) |
| Thyroid | 11% | (Garcia-Rostan et al., 2005) |
Figure 2. Somatic mutation of PIK3CA/p110α in human cancers.
The p110α subunit consist of several different domains: ABD: adapter binding domain (AKA: p85 binding domain); RBD: Ras-binding domain; C2: C2 domain; helical: helical domain; kinase: kinase domain.
Challenges of targeting mutant p110α
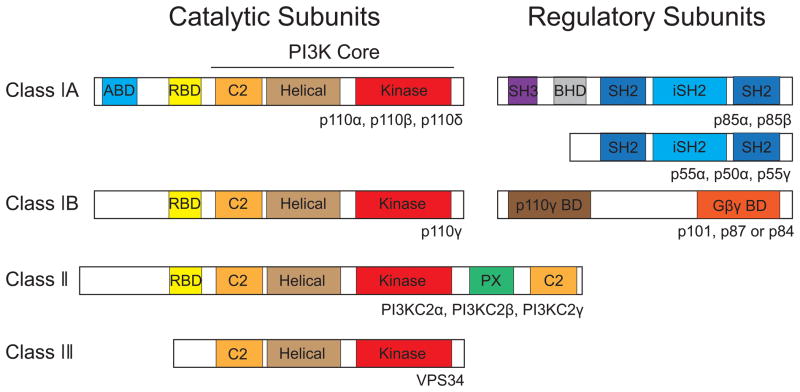
Although scientists at both pharmaceutical companies and academic institutions have made enormous effort to develop drugs to inhibit mutant p110α enzymatic activity, it turns out that it is difficult to find small molecules that specifically inhibit p110 isoforms (Liu et al., 2009). Based on the substrate specificities and structural features, PI3K family members are divided into three classes (Figure 3). Class I PI3Ks are heterodimers consisting of a p110 catalytic subunit and a p85 regulatory subunit. The p85 regulatory subunit modulates the activity, stability and localization of the enzyme. Three genes PIK3R1, PIK3R2 and PIK3R3 encode p85α (and its splice variants p55α and p50α), p85β and p55γ respectively. All these regulatory subunits are collectively referred as p85. The class I PI3Ks convert PIP2 to PIP3. Both Class II and class III PI3K family members have only a single catalytic subunit. The class II PI3Ks (PIKC2α, PIKC2β and PIKC2γ) use phosphatidylinositol or phosphatidylinositol-4-phosphate as substrates; whereas the single class III PI3K member, VPS34, converts phosphatidylinositol to phosphatidylinositol-3-phosphate.
Figure 3. Structures of PI3Ks.
PI3Ks are classified into three groups: class I, class II and class III. Class I PI3Ks are heterodimers consisting of a catalytic p110 subunit and a p85 regulatory subunit. The class I PI3Ks are further divided into class IA and class IB subgroups. Class IA PI3Ks consist of PI3Kα, PI3Kβ and PI3Kδ with p110α, p110β, and p110δ as their catalytic subunit respectively. The catalytic subunit can form a heterodimer with any of the p85 subunit (p85α, p85β, p55α, p50α and p55γ). The class IB PI3K subgroup consists of a p110γ catalytic subunit and one of the regulatory subunits (p101, p87 and p84). The class II and III PI3Ks consist of only a catalytic polypeptide. Several different domains are present including SH3: Src homology 3 domain; BHD: BCR homology domain; SH2: Src homology 2 domain; iSH2: Inter-Src homology 2 domain; p110γ BD: p110γ binding domain; Gβγ BD: G beta gamma protein complex binding domain; PX: phosphoinositide-binding structural domain.
The class I PI3Ks have been intensively characterized. This group of PI3Ks is further divided into class IA and class IB subgroups. The Class IA PI3Ks have three members: PI3Kα, PI3Kβ and PI3Kδ. The catalytic subunits of these three enzymes share similar domain structures: an N-terminal adaptor-binding domain (ABD, also called p85 binding domain), a Ras-binding domain (RBD), a C2 domain, a helical domain and a catalytic domain (Figure 3). The p110 catalytic subunit of the class IA PI3Ks form a heterodimer with one of the p85 regulatory subunits (p85α, p85β, p55α, p55γ and p50α). The class IB PI3K consists of the catalytic subunit p110γ and regulatory subunit p101 (Figure 3). The overall domain structure of p110γ is similar to these of PI3Kα, PI3Kβ and PI3Kδ, but it lacks the ABD domain (Figure 3). While p110α and p110β are ubiquitously expressed, p110δ and p110γ are mainly expressed in leukocytes (Liu et al., 2009). Interestingly, only p110α is mutated in human cancers (Zhao and Vogt, 2008a). Recent crystal structure analyses indicate that the structures of class I PI3K catalytic domains are quite similar and that the ATP binding pockets of these enzymes are almost identical (Vadas et al., 2011), which may explain why it is difficult to develop p110α isoform-specific inhibitors.
Currently, several pan-class I PI3K inhibitors (GDC-0941, BKM120, PX866 and BAY 80-6946) and dual PI3K/mTOR inhibitors (e. g. BEZ235) are in early-stage clinical trials (Rodon et al., 2013). There are also some PI3Kβ-specific and PI3Kδ-specific inhibitors under clinical evaluation for their efficacy against various tumor types (Rodon et al., 2013). It is worth noting that Genentech has recently developed a so-called β-spare PI3K inhibitor GDC-0032, which selectively inhibits p110α, p110δ and p110γ, but sparing the p110β isoform. Early-stage clinical trials in breast cancer patients has shown great promise. Patients harboring PIK3CA/p110α had at least partial clinical responses to the β-spare PI3K inhibitor (Juric D, personal communication), suggesting that a p110α isoform-specific inhibitor should be more potent and less toxic. Nonetheless, drugs that specifically target the mutant p110α should be highly advantageous.
Mutant-specific protein interaction between IRS1 and helical domain mutations of p110α may provide a more accessible target for cancer therapy
Given the challenges encountered with development of p110α-specific inhibitors, alternative approaches are needed to target mutant p110α for cancer therapy. Our recent discovery that the helical domain p110α mutants directly interact with IRS1 may provide a more accessible target for developing drugs to treat cancer patients harboring those p110α mutations (Hao et al., 2013). We discuss below the relevant background and therapeutic implications of our findings.
-
The p110α helical domain and kinase domain mutations exert their oncogenic functions through distinct mechanisms.
As mentioned above, most of p110α mutations occur at two hotspot regions: an acidic cluster (E542, E545 and Q546) in the helical domain and a histidine residue (H1047) in the kinase domain. The E545K and H1047R are the two most frequently observed p110α somatic mutations in human cancers. Interestingly, several recent studies indicate that the E545K and H1047R mutations exert their oncogenic functions through distinct mechanisms: (1) Zhao and Vogt have shown that the helical domain and the kinase domain mutant proteins could synergistically transform chicken embryonic fibroblasts (Zhao and Vogt, 2008b), providing the first clue that the two mutations may induce oncogenic transformation through different pathways. Consistently, they further demonstrated that the p110α helical domain mutants require the Ras-binding domain for their transformation activities, whereas the oncogenic activity of the H1047R mutant depends on the ABD domain. (2) Pang et al. showed that expression of the p110α E545K mutant produce a more severe metastatic phenotype than that of the H1047R mutant in a breast cancer cell line (Pang et al., 2009). (3) In contrast, the p110α H1047R mutant, but not the E545K mutant, is found to enhance HER2-mediated transformation of immortalized mammary epithelial cells (Chakrabarty et al., 2010). Structural analysis indicates that the p110α H1047R mutation alters the interaction between PI3Ka and the cell membrane, thereby activating its kinase activity (Mandelker et al., 2009). It has been suggested that the helical domain mutations activate their enzymatic activities by disrupting the inhibitory effect of the p85 subunits (Huang et al., 2007, Miled et al., 2007).
-
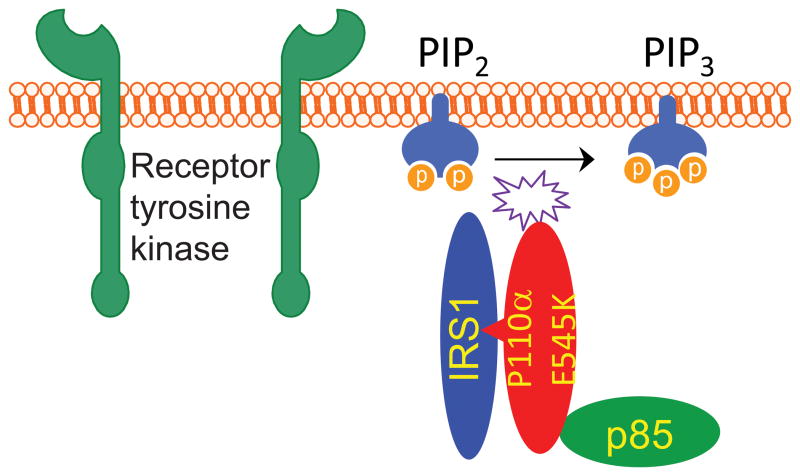
Direct interaction between p110α helical domain mutants and IRS1 drives tumorigenesis. Our recent studies however indicate that the weakened p110α-p85 interaction caused by the p110α helical domain mutations is not sufficient for these mutant proteins to exert their oncogenic functions. In an attempt to determine how p110α E545K and p110α H1047R differentially activate oncogenic signaling pathways, we tried to identify proteins that might differentially bind to wild-type (WT) and mutant p110α. To this end, we discovered that IRS1 binds to p110α E545K mutant but not to WT or H1047R mutant under serum-starvation conditions (Hao et al., 2013). In addition, other hotspot p110α mutations in the helical domain including E542K, E545A, E545G and Q546K as well as relatively rare mutations including K111N in the ABD domain and N345K in the C2 domain also gain interaction with IRS1 under serum-starvation conditions (Hao et al., 2013). We provided compelling evidence that the p110α E545K mutant-IRS1 interaction plays a critical role in tumorigenesis and demonstrated how this mutant-specific protein interaction may be exploited for cancer therapy. Firstly, in the IRS1 KO DLD1 cells harboring the p110α E545K mutation, reconstitution of an IRS1 deletion construct that does not interact with p110α E545K mutant protein produces smaller xenograft tumors compared to the full-length IRS1 reconstituted cells (Hao et al., 2013). Secondly, a p110α E545K mutant stapled peptide, but not the WT counterpart, specifically inhibits xenograft tumor growth of CRC cells with a p110α E545K mutation (Hao et al., 2013). Mechanistically, our data suggest that p110α E545K mutant-IRS1 interaction activates the mutant p110α lipid kinase in two ways: (1) it brings the mutant enzyme complexes from cytosol to plasma membrane; (2) it stabilizes the mutant p110α proteins (Hao et al., 2013). We propose that the helical domain mutations of p110α, as well as some of the mutations in the ABD and C2 domain, induce conformational changes that enable p110α to directly interact with IRS1, which does not require IRS1 tyrosine phosphorylation and p85 proteins (Figure 4). In support, a recent study shows that oncogenic mutations of p110α in various domains indeed induce conformational changes of its protein structure that may affect its structural interactions (Burke et al., 2012).
Our studies may provide an explanation for observed different phenotypes produced by the helical and kinase domain mutations. Using isogenic breast cancer cell lines, Pang and colleagues demonstrated that expression of p110α helical domain mutations increase the sensitivity of cancer cells to chemo-attractants and induce much stronger metastatic phenotypes than the H1047R kinase domain mutation (Pang et al., 2009). These phenotypic differences cannot be simply explained by the increased enzymatic activities of the helical and kinase domain mutations. In fact, Vasudevan et al. observed that the basal levels of phospho-AKT are lower in cancer cell lines expressing p110α helical domain mutations compared to these cell lines expressing the H1047R kinase domain mutation (Vasudevan et al., 2009). Consistently, when complexed with p85 regulatory subunit, the p110α H1047R kinase domain mutant protein displays higher lipid kinase activity in vitro than the E545K helical domain mutant (Samuels et al., 2005). We showed that the p110α helical domain mutants, but not the kinase domain mutant, directly associate with IRS1 without growth factor stimulation (Hao et al., 2013). In this regard, we suggest that the p110α helical domain mutant-IRS1 protein complexes may contain additional proteins that do not exist in the p110α kinase domain mutant protein complexes, thereby producing distinct phenotypes.
-
The protein interaction between IRS1 and p110α helical domain mutants is independent of the p85 regulatory subunit.
Given that p110α is brought to the IRS1 complex through interaction between phospho-IRS1 and p85 regulatory subunit when cells are stimulated with growth factor, it is important to demonstrate that the p110α helical domain mutant-IRS1 interaction is independent of p85. Our studies provided several lines of evidence indicating that interaction between the p110α helical domain mutant proteins and IRS1 is not mediated by binding of p85 SH2 domain with pY residues on IRS1. Firstly, p110α helical domain mutant-IRS1 interaction is independent of IRS1 tyrosine phosphorylation (Hao et al., 2013). Secondly, a p110α E545K mutant protein (p110α E545KΔABD) devoid of the ability to bind with p85 proteins still interacts with IRS1 (Hao et al., 2013). This result also provides an explanation for the observation made by Vogt and colleagues that the helical domain mutants of p110α do not require the binding of p85 to transform chicken embryonic fibroblasts (Zhao and Vogt, 2008b). Thirdly, we show here that ablation of p85 proteins does not reduce the p110α E545K mutant-IRS1 interaction (Hao et al., 2013). Interestingly, diminishment of p85 proteins actually enhances the p110α E545K mutant-IRS1 interaction, suggesting that p85 may compete with IRS1 to bind to the p110α helical mutants (Hao et al., 2013). Fourthly, in the absence of p85 proteins, recombinant IRS1 proteins bind in vitro to p110α E545K mutant but not WT p110α proteins (Hao et al., 2013). Fifthly, an IRS1 deletion that abrogates IRS1 interaction with p110α E545K mutant still binds well with p85 proteins (Hao et al., 2013). Sixthly, a p110α E545K mutant peptide, which disrupts the p110α E545K mutant-IRS1 interaction, does not affect the binding of the mutant p110α to p85 (Hao et al., 2013). However, we point out that even though p85 proteins are not required for direct interaction between the p110α helical domain mutants and IRS1, they may participate in an interaction that provides stability to the mutant because ablation of p85 proteins results in significantly reduction of the levels of p110α protein (Hao et al., 2013).
-
Disrupting protein-protein interaction between p110α helical domain mutant and IRS1 provide a novel approach for development of drugs targeting cancers with these mutations.
Increasing evidence suggests that protein-protein interactions are druggable targets (White et al., 2008). In fact, small molecules that inhibit the MDM2-p53 interaction are being tested in clinical trials to treat cancer patients (Essmann and Schulze-Osthoff, 2011). Moreover, ABT-737, a small-molecule BH3-mimetic that inhibits protein-protein interaction of BCL2 family proteins, is also being evaluated in clinical trials for its efficacy in haematologic malignancies (Khaw et al., 2011). Our observation that a p110α E545K mutant staple peptide, but not the WT counterpart, specifically inhibits xenograft tumor growth of CRC cells with a p110α E545K mutation provides a proof-of-principle for targeting this mutant specific protein interaction for cancer therapy. The discovery of frequent mutations of PIK3CA in human cancer provides a strong rationale for inhibition of mutated p110α activities for targeted cancer therapy (Samuels et al., 2005). However, as discussed above, it has remained a challenge to develop p110α isoform-specific inhibitors. Our data suggest that disruption of the interactions between helical domain p110α mutants and IRS1 may be exploited as a more accessible targeted therapy approach for cancer patients harboring such mutations. Given that this mutant p110α specific interaction exists only in tumor cells that harbor these particular mutations, but not in normal tissues, drugs targeting this cancer-specific interaction should have no or minimal side effects because they do not perturb normal cellular functions. It is worth noting that our p110α E545K mutant stapled peptide is not stable in blood stream, which prevents it being delivered systematically. A mean to improve the pharmacokinetics of the peptide is to substitute the peptide bonds with chemical bonds to generate a functional peptidomimetics. Alternatively, small molecules that can disrupt protein-protein interaction between IRS1 and p110α helical domain mutant could be identified through high-throughput screening.
Figure 4. Protein interaction between helical domain mutation of p110α and IRS1 drives tumorigenesis.
In cancer cells harboring p110α helical domain mutations, IRS1 directly binds to mutant p110α and brings it the cell membrane, thereby activating the PI3K-AKT signaling pathway independent of any stimulation by growth factor.
Prospective
Mutant p110α is excellent target for cancer therapy. Much of current effort has been devoted to develop p110α-isoform specific inhibitors, which turns out to be a challenge task. Our recent finding that p110α helical domain mutants gain direct interaction with IRS1 raises new hope of targeting this mutant specific protein interaction for cancer therapy. The key will be to screen or design small molecules with favorable pharmacokinetics that disrupt the protein-protein interaction between p110α helical domain mutant and IRS1. In this regard, a co-crystal structure of p110α helical domain mutant and IRS1 would be very helpful for both in silico design and optimization of compounds that disrupts the p110α mutant-specific protein-protein interaction. Lastly, drugs targeting mutant p110α may also be used to treat hemimegalencephaly, because recent next-gen sequencing analyses revealed that p110α is also mutated in patients with this disease (Lee et al., 2012).
Acknowledgments
This work is supported by NIH grants R21CA160060, R01CA127590, R01HG004722, P50CA150964 and P30 CA043703.
References
- Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, Konishi H, Karakas B, Blair BG, Lin C, Peters BA, Velculescu VE, Park BH. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3:772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- Broderick DK, Di C, Parrett TJ, Samuels YR, Cummins JM, McLendon RE, Fults DW, Velculescu VE, Bigner DD, Yan H. Mutations of PIK3CA in anaplastic oligodendrogliomas, high-grade astrocytomas, and medulloblastomas. Cancer Res. 2004;64:5048–5050. doi: 10.1158/0008-5472.CAN-04-1170. [DOI] [PubMed] [Google Scholar]
- Burke JE, Perisic O, Masson GR, Vadas O, Williams RL. Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110alpha (PIK3CA) Proc Natl Acad Sci U S A. 2012;109:15259–15264. doi: 10.1073/pnas.1205508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell IG, Russell SE, Choong DY, Montgomery KG, Ciavarella ML, Hooi CS, Cristiano BE, Pearson RB, Phillips WA. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–7681. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- Chakrabarty A, Rexer BN, Wang SE, Cook RS, Engelman JA, Arteaga CL. H1047R phosphatidylinositol 3-kinase mutant enhances HER2-mediated transformation by heregulin production and activation of HER3. Oncogene. 2010;29:5193–5203. doi: 10.1038/onc.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombino M, Sperlongano P, Izzo F, Tatangelo F, Botti G, Lombardi A, Accardo M, Tarantino L, Sordelli I, Agresti M, Abbruzzese A, Caraglia M, Palmieri G. BRAF and PIK3CA genes are somatically mutated in hepatocellular carcinoma among patients from South Italy. Cell death & disease. 2012;3:e259. doi: 10.1038/cddis.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R, Ohno-Jones S, Sawyers CL. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, Bandla S, Imamura Y, Schumacher SE, Shefler E, McKenna A, Carter SL, Cibulskis K, Sivachenko A, Saksena G, Voet D, Ramos AH, Auclair D, Thompson K, Sougnez C, Onofrio RC, Guiducci C, Beroukhim R, Zhou Z, Lin L, Lin J, Reddy R, Chang A, Landrenau R, Pennathur A, Ogino S, Luketich JD, Golub TR, Gabriel SB, Lander ES, Beer DG, Godfrey TE, Getz G, Bass AJ. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45:478–486. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann F, Schulze-Osthoff K. Translational approaches targeting the p53 pathway for anticancer therapy. Br J Pharmacol. 2011 doi: 10.1111/j.1476-5381.2011.01570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Dios DA, Lambrechts D, Coenegrachts L, Vandenput I, Capoen A, Webb PM, Ferguson K, Anecs, Akslen LA, Claes B, Vergote I, Moerman P, Van Robays J, Marcickiewicz J, Salvesen HB, Spurdle AB, Amant F. High-throughput interrogation of PIK3CA, PTEN, KRAS, FBXW7 and TP53 mutations in primary endometrial carcinoma. Gynecologic oncology. 2013;128:327–334. doi: 10.1016/j.ygyno.2012.11.037. [DOI] [PubMed] [Google Scholar]
- Garcia-Rostan G, Costa AM, Pereira-Castro I, Salvatore G, Hernandez R, Hermsem MJ, Herrero A, Fusco A, Cameselle-Teijeiro J, Santoro M. Mutation of the PIK3CA gene in anaplastic thyroid cancer. Cancer Res. 2005;65:10199–10207. doi: 10.1158/0008-5472.CAN-04-4259. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Gschwind A, Fischer OM, Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat Rev Cancer. 2004;4:361–370. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- Hao Y, Wang C, Cao B, Hirsch BM, Song J, Markowitz SD, Ewing RM, Sedwick D, Liu L, Zheng W, Wang Z. Gain of interaction with IRS1 by p110alpha-helical domain mutants is crucial for their oncogenic functions. Cancer Cell. 2013;23:583–593. doi: 10.1016/j.ccr.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C, Bartels G, Gehlhaar C, Holtkamp N, von Deimling A. PIK3CA mutations in glioblastoma multiforme. Acta neuropathologica. 2005;109:639–642. doi: 10.1007/s00401-005-1000-1. [DOI] [PubMed] [Google Scholar]
- Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, Vogelstein B, Gabelli SB, Amzel LM. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- Janku F, Lee JJ, Tsimberidou AM, Hong DS, Naing A, Falchook GS, Fu S, Luthra R, Garrido-Laguna I, Kurzrock R. PIK3CA mutations frequently coexist with RAS and BRAF mutations in patients with advanced cancers. PLoS One. 2011;6:e22769. doi: 10.1371/journal.pone.0022769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaw SL, Huang DC, Roberts AW. Overcoming blocks in apoptosis with BH3-mimetic therapy in haematological malignancies. Pathology. 2011;43:525–535. doi: 10.1097/PAT.0b013e32834b1b34. [DOI] [PubMed] [Google Scholar]
- Lee JH, Huynh M, Silhavy JL, Kim S, Dixon-Salazar T, Heiberg A, Scott E, Bafna V, Hill KJ, Collazo A, Funari V, Russ C, Gabriel SB, Mathern GW, Gleeson JG. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet. 2012;44:941–945. doi: 10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Soung YH, Kim SY, Lee HW, Park WS, Nam SW, Kim SH, Lee JY, Yoo NJ, Lee SH. PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene. 2005;24:1477–1480. doi: 10.1038/sj.onc.1208304. [DOI] [PubMed] [Google Scholar]
- Levine DA, Bogomolniy F, Yee CJ, Lash A, Barakat RR, Borgen PI, Boyd J. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin Cancer Res. 2005;11:2875–2878. doi: 10.1158/1078-0432.CCR-04-2142. [DOI] [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- Li VS, Wong CW, Chan TL, Chan AS, Zhao W, Chu KM, So S, Chen X, Yuen ST, Leung SY. Mutations of PIK3CA in gastric adenocarcinoma. BMC Cancer. 2005;5:29. doi: 10.1186/1471-2407-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelker D, Gabelli SB, Schmidt-Kittler O, Zhu J, Cheong I, Huang CH, Kinzler KW, Vogelstein B, Amzel LM. A frequent kinase domain mutation that changes the interaction between PI3Kalpha and the membrane. Proc Natl Acad Sci U S A. 2009;106:16996–17001. doi: 10.1073/pnas.0908444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, Inbar Y, Schneidman-Duhovny D, Wolfson HJ, Backer JM, Williams RL. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- Oda K, Stokoe D, Taketani Y, McCormick F. High frequency of coexistent mutations of PIK3CA and PTEN genes in endometrial carcinoma. Cancer Res. 2005;65:10669–10673. doi: 10.1158/0008-5472.CAN-05-2620. [DOI] [PubMed] [Google Scholar]
- Pang H, Flinn R, Patsialou A, Wyckoff J, Roussos ET, Wu H, Pozzuto M, Goswami S, Condeelis JS, Bresnick AR, Segall JE, Backer JM. Differential enhancement of breast cancer cell motility and metastasis by helical and kinase domain mutations of class IA phosphoinositide 3-kinase. Cancer Res. 2009;69:8868–8876. doi: 10.1158/0008-5472.CAN-09-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips WA, Russell SE, Ciavarella ML, Choong DY, Montgomery KG, Smith K, Pearson RB, Thomas RJ, Campbell IG. Mutation analysis of PIK3CA and PIK3CB in esophageal cancer and Barrett’s esophagus. Int J Cancer. 2006;118:2644–2646. doi: 10.1002/ijc.21706. [DOI] [PubMed] [Google Scholar]
- Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nat Rev Clin Oncol. 2013;10:143–153. doi: 10.1038/nrclinonc.2013.10. [DOI] [PubMed] [Google Scholar]
- Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmstrom PO, Mansukhani M, Enoksson J, Hibshoosh H, Borg A, Parsons R. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, Rago C, Huso DL, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Schonleben F, Qiu W, Ciau NT, Ho DJ, Li X, Allendorf JD, Remotti HE, Su GH. PIK3CA mutations in intraductal papillary mucinous neoplasm/carcinoma of the pancreas. Clin Cancer Res. 2006;12:3851–3855. doi: 10.1158/1078-0432.CCR-06-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, McArthur GA, Hutson TE, Moschos SJ, Flaherty KT, Hersey P, Kefford R, Lawrence D, Puzanov I, Lewis KD, Amaravadi RK, Chmielowski B, Lawrence HJ, Shyr Y, Ye F, Li J, Nolop KB, Lee RJ, Joe AK, Ribas A. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadas O, Burke JE, Zhang X, Berndt A, Williams RL. Structural basis for activation and inhibition of class I phosphoinositide 3-kinases. Sci Signal. 2011:4. doi: 10.1126/scisignal.2002165. [DOI] [PubMed] [Google Scholar]
- Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, Hennessy BT, Tseng H, Pochanard P, Kim SY, Dunn IF, Schinzel AC, Sandy P, Hoersch S, Sheng Q, Gupta PB, Boehm JS, Reiling JH, Silver S, Lu Y, Stemke-Hale K, Dutta B, Joy C, Sahin AA, Gonzalez-Angulo AM, Lluch A, Rameh LE, Jacks T, Root DE, Lander ES, Mills GB, Hahn WC, Sellers WR, Garraway LA. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AW, Westwell AD, Brahemi G. Protein-protein interactions as targets for small-molecule therapeutics in cancer. Expert Rev Mol Med. 2008;10:e8. doi: 10.1017/S1462399408000641. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Shigematsu H, Nomura M, Lockwood WW, Sato M, Okumura N, Soh J, Suzuki M, Wistuba II, Fong KM, Lee H, Toyooka S, Date H, Lam WL, Minna JD, Gazdar AF. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008;68:6913–6921. doi: 10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008a;27:5486–5496. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A. 2008b;105:2652–2657. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]