Summary
Identification of cis-regulatory elements, such as enhancers and promoters, is very important not only for analysis of gene regulatory networks but also as a tool for targeted gene expression experiments. In this chapter, we introduce an easy but reliable approach to predict enhancers of a gene of interest by comparing mammalian and Xenopus genome sequences, and to examine their activity using a co-transgenesis technique in Xenopus embryos. Since the bioinformatics analysis utilizes publically available web-tools, bench biologists can easily perform it without any need for special computing capability. The co-transgenesis assay, which directly uses polymerase chain reaction (PCR) products, quickly screens for activity of the candidate elements in a cloning-free manner.
Keywords: cis-regulatory elements, enhancer, comparative genomics, multiple alignment, phylogenetic footprinting, transcription factor-binding motif, Xenopus transgenesis
1. Introduction
Identification and analysis of cis-regulatory elements with conventional approaches has generally been slow and laborious work. Although promoters with basal transcriptional activity may be easily identified as the short upstream sequences adjacent to transcription start sites, enhancers that control spatio-temporal specificity and strength level of gene expression are often located far from the basal promoter. Even if enhancers are found, it is not easy to identify transcription factors that bind to the enhancer by recognizing ambiguous sequences (1). However, recent progress in comparative genomics has markedly improved the situation. Comparison of orthologous genomic sequences from different vertebrate species identifies conserved non-coding elements (CNEs) as reliable candidates for enhancers, and their phylogenetic footprinting analysis predicts transcription factor-binding motifs (TFBMs) that are highly conserved through evolution (2). Analysis of the genome-wide distribution of CNEs showed that they are especially enriched around developmental regulatory genes where there are clearly conserved regulatory gene circuits involved (3). The power of the comparative genomics-based approach and biological significance of the conserved enhancers have already been demonstrated by a number of studies, including the analyses of a Shh limb enhancer and a Sox9 mandibular enhancer whose mutations are responsible for congenital polydactyly and cleft palate, respectively (4-6). There are also reports that show lack of evolutionary sequence conservation of vertebrate enhancers (7). However, if one focuses primarily on classic-style promoter bashing so as not to miss non-conserved elements, one may miss critical conserved enhancers located far away from basal promoters. In the above cases of Shh and Sox9, the enhancers are located more than 1 Mb away from their transcription start sites. If comprehensive identification of enhancers is necessary, it may be performed by more elaborate methods, for example, as with the ChIP-seq analysis using anti-p300 antibody followed by a large-scale transgenesis screening (8)(see Note 1).
In search of conserved enhancers, though fish genomic sequence is often used for comparison with mammalian sequences, the Xenopus tropicalis genome sequence may be more generally suitable for this purpose. This is because, while the Xenopus genome is evolutionarily distant enough from mammalian genomes for identifying functionally active CNEs, its structure is much closer to that of mammalian genomes than is the fish genome, where many duplications and rearrangements followed by subfunctionalization of cis-regulatory elements occurred after they had diverged from other vertebrates (9-11).
The power of combining the mammalian-Xenopus genome comparison with an efficient transgenesis technique in Xenopus was demonstrated by a cis-regulatory analysis of foxe3, a transcription factor gene essential for lens development (12). This study demonstrated that the classic-style promoter analysis and the comparative genomics-based approach gave consistent results by identifying the same enhancer. The phylogenetic footprinting analysis of this enhancer identified nine conserved TFBMs in its 0.4 kb length, and their mutational analyses in founder transgenics led to the discovery of Notch signaling as a new lens-inducing signal.
The approach described in this chapter includes the following steps: (a) retrieving large orthologous genome sequences of a gene of interest from comparative genome browsers; (b) aligning the retrieved multiple sequences; (c) extracting orthologous CNE sequences; (d) generating a phylogenetic footprint of these CNEs; (e) searching for conserved TFBMs in the CNEs; and (f) performing the co-transgenesis assay to assess possible enhancer activity of the CNEs in vivo (12). The comparative genome browsers, such as the ECR Browser (13), show pre-made, multiple pair-wise alignments of vertebrate genome sequences. While such alignments are quite useful for quickly looking at sequence conservation associated with a gene of interest, their parameter settings are typically adjusted for whole genome comparisons and are often too stringent for precise analysis of a specific gene locus. Thus, we use the comparative genome browsers as a source of large orthologous genome sequences, then re-align them using a sensitive genome aligner, PipMaker, for identifying CNEs (14). PipMaker often discovers CNEs that are not found by the comparative genome browsers due to their relatively low sequence conservation or to local inversions in compared genomes. Furthermore, while the comparative genome browsers show just pair-wise alignments at the nucleotide level view, PipMaker can generate a multiple alignment that contains more than three sequences. After the PipMaker analysis, the CNE sequences can be extracted from the output files and further re-aligned using a more sensitive, short sequence aligner, ClustalW, for phylogenetic footprinting (15). In the co-transgenesis assay, a PCR-amplified CNE fragment is injected into Xenopus laevis eggs along with a basal promoter-GFP-polyA cassette. Since these two DNA fragments are integrated together into the host chromosome at the same site at the one-cell stage, GFP exhibits non-mosaic expression in the embryos if the injected CNE has enhancer activity. Since the co-transgenesis assay does not involve any cloning, one can finish enhancer screening of ~10 CNEs within two weeks, which appears to be a typical number of CNEs associated with one gene.
In the postgenomic era, one of the central aims of developmental biology is, undoubtedly, the global understanding of the gene regulatory networks (GRNs) that are hardwired in the genome. While pioneering GRN studies have achieved remarkable success in sea urchin, fruit fly and nematode, this type of study has been very limited in vertebrates (16). This is mostly because that the vertebrates have much larger genomes than these invertebrates, which makes it difficult to perform the genome-wide cis-regulatory analysis that is essential for GRN studies. However, the approach described in this chapter effectively combines the comparative genomics tools and the high-throughput transgenesis assay in Xenopus to overcome this problem. The resulting collection of cis-regulatory elements will be a powerful starting point for a genome-wide analysis of regulatory interactions between transcription factors and their target genes, and provide us a framework for elucidating the GRNs that are conserved in vertebrates.
2. Materials
2.1. Bioinformatics tools
Comparative genome browser: ECR Browser (http://ecrbrowser.dcode.org/)
- Genome aligner software: (basic) PipMaker, Advanced PipMaker, and MultiPipMaker (http://pipmaker.bx.psu.edu/pipmaker/)
- PipMaker and Advanced PipMaker generate pairwise alignments, and MultiPipMaker is used to align three or more sequences. MultiPipMaker is generally used because of the utility of multiple genome alignments. Links to detailed instructions of these programs are found in the website.
Supporting software tool for PipMaker: PipHelper (http://pipmaker.bx.psu.edu/piphelper/)
Simple sequence manipulation software tool: Range Extractor DNA (http://www.bioinformatics.org/sms2/range_extract_dna.html)
Short sequence aligner software: ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/)
TFBM search programs: rVista (http://rvista.dcode.org/) (17), ConSite (http://asp.ii.uib.no:8090/cgi-bin/CONSITE/consite) (18)
TFBM database: Transfac (http://www.gene-regulation.com/index.html, a free version is available from http://www.gene-regulation.com/cgi-bin/pub/databases/transfac/search.cgi), JASPAR (http://jaspar.cgb.ki.se/cgi-bin/jaspar_db.pl) (19)
It is helpful if one has a standard DNA analysis software tool, such as VectorNTI (Invitrogen) or equivalent.
2.2. Reagents and equipment for preparation of DNA fragments for Xenopus co-transgenesis assay
Other reagents and equipment for obtaining Xenopus eggs and transgenesis are found in Chapter 11 of this book, and that for in situ hybridization analysis are found in references (20).
・ X. tropicalis genomic DNA: 50 ng/μL (see Note 2)
・ pBSSK+βGFP (12): a pBluescript plasmid carrying the chicken β-actin basal promoter (21) adjacent to a GFP-polyA cassette. This plasmid is available from H. Ogino. A plasmid carrying the gata2 basal promoter linked to the GFP-polyA (22), which is available from R. M. Grainger, may provide more sensitivity for the co-transgenesis assay since it appears to impart higher levels of transcriptional activity to enhancers without causing background expression on its own.
・ QIAquick PCR Purification Kit (Qiagen)
・ QIAquick Gel Extraction kit (Qiagen)
・ Primer pairs for amplification of CNEs: 10 pmol/μL in TE (see Methods for details), stored at −20°C.
・ Platinum Pfx DNA polymerase: 2.5 U/μL (Invitrogen), or an equivalent proof-reading DNA polymerase for high fidelity PCR.
For the following, generic reagents and equipment are likely to suffice.
・ 50 mM MgSO4 (included in a set of the Platinum Pfx DNA polymerase)
・ dNTP mixture, 2.5 mM each (Invitrogen)
・ BamHI: 10-20 U/μL (New England Biolabs)
・ XhoI: 10-20 U/μL (New England Biolabs)
・ 10× NEB3 buffer (for double-digestion with BamHI and XhoI, New England Biolabs)
・ 1 × TAE buffer: 40 mM Tris-acetate, 1mM EDTA, pH 8.0
・ Agarose (molecular biology grade)
・ Ethidium bromide solution: 10 μg/mL (Sigma)
・ TE buffer: 10 mM Tris-HCl, 1mM EDTA, pH 8.0
・ 10 mM KCl
・ Ethanol (96-100%, molecular biology grade)
・ 70% Ethanol (prepare by mixing 96-100% ethanol and distilled water)
・ Distilled water
・ Heating block or water bath (for restriction enzyme digestion at 37°C and for melting agarose gel at 50°C)
・ GeneAmp PCR System 2400 (PerkinElmer) or an equivalent standard thermal cycler
・ UV transilluminator (for visualizing ethidium bromide-stained DNA after gel electrophoresis)
・ Microcentrifuge
3. Methods
The following sections describe how to identify and analyze enhancers in Xenopus, using as an example our analysis of the 5’ flanking region of a paired domain homeobox gene, pax6 (23). This is only a small part of the large cis-regulatory region surrounding pax6, which spans at least 180 kb in mouse genome and contains more than nine enhancers (23) (24). The “C” region identified in the following sections was originally reported to be an enhancer for expression in the photoreceptors of the developing mouse retina (24). However, our analysis in Xenopus revealed its early embryonic activity in the developing optic vesicle and overlying presumptive lens ectoderm (H. Ogino and R. M. Grainger, unpublished).
3.1. Identification of CNEs associated with a gene of interest
1. Extract genomic sequences from genome assembly data
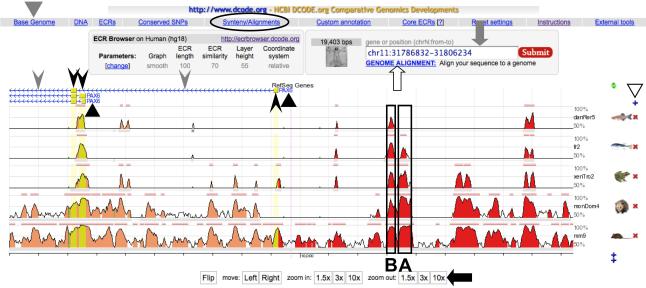
The first step in generating an alignment is identifying orthologous genomic regions of a gene of interest in multiple species and creating their text files in FASTA format (see Note 3). This can be done easily with the ECR Browser, where one can choose a base genome for the alignment from 13 vertebrate species including X. tropicalis, and then view a stacked pair-wise genome alignment of any gene or location that was generated between the base genome and other vertebrate genomes (see Note 4). Figure 1 is a screen shot of the ECR Browser window, where the 5’ flanking region of human PAX6 (hg18 chr11: 31786832 - 31806234; position 31786832 - 31806234 of chromosome 11 in human genome assembly 18) is chosen as the base genome sequence and is aligned with orthologous regions from mouse, opossum, Xenopus, Fugu, and zebrafish genome sequences. The regions with high sequence similarity are graphically displayed as peaks, whose height and width represent their sequence conservation level and length, respectively. One can change the base genome species, add or remove genomes from the alignment, and adjust the length of the alignment by clicking the buttons on the browser window (see the legend for Fig. 1). To retrieve the sequences aligned in the window, click the “Synteny/Alignments” button (circled in Fig. 1), which will open a new window showing a list of the sequences (see Note 5). In this window, click check boxes of the sequences that are needed for the subsequent PipMaker analysis, and then click the “Mulan” button at the left bottom corner (see Note 6). A link list will be visible for the sequences in FASTA format (ex. seq1.fa, seq2.fa). Download all the necessary sequences.
Fig. 1.
A screen shot of the ECR Browser window. Above the schematic representation of the alignment, the gene names (PAX6, indicated by black triangles), exons (boxes indicated by black arrowheads) and introns (horizontal lines indicated by gray arrowheads) in the base genome are shown. Arrows associated with the horizontal line representing introns indicate transcriptional orientation of the gene. The conserved regions shown as peaks include not only exons but also CNEs located in introns and intergenic regions. Among these CNEs, those indicated as A and B (boxed) were shown to have enhancer activity in the pancreas and lens/cornea in transgenic mouse analyses, respectively (32). One can change the base genome species by clicking the “Base Genome” button (indicated by a gray triangle) at the upper left, but it is not necessary to choose Xenopus at this step, even if it will be used as the base sequence in a subsequent PipMaker analysis. On the right can be seen a + or an × next to the genomes shown (white triangle), permitting addition or removal of genomes from the alignment. The length of the alignment is adjusted using buttons such as “10 × ” at the bottom of the window (black arrow), or by directly inputting a desired genomic position of the base genome into the rectangular box at the upper right (gray arrow). One may also input just a gene name into the rectangular box to move to another gene locus. If one needs to align user-identified sequence to the genomes in this browser, click the “GENOME ALIGNMENT” button below the rectangular box (white arrow), and input the sequence as a FASTA format. For details, go to the instruction page of ECR Browser (http://ecrbrowser.dcode.org/ecrInstructions/ecrInstructions.html).
2. Build accompanying files
The second step is to build Exons, Mask, and Underlay files for the sequence that is chosen to be the base sequence in the PipMaker analysis. The Exons and Mask files indicate the location of exons and repetitive sequences in the base sequence, respectively. The Underlay file instructs the program to shade specific gene structures (ex. exons and introns) with colors in the output. To build these files, open the FASTA file of the sequence that will be used as the base sequence in the PipMaker analysis below (ex. the FASTA file of human PAX6 downloaded in the previous step, using TextEdit on a Macintosh or Notepad on a Windows computer to open it), and find the name of genome assembly and position of the sequence in the assembly in the header line of the text (ex. hg18 chr11:31786832-31806234). Next, go to the PipHelper website (http://pipmaker.bx.psu.edu/piphelper/), choose the genome assembly (ex. Mammal, Human, Mar. 2006 (NCBI36/hg18)), enter the position (ex. chr11:31786832-31806234), choose an annotation source for exons (ex. RefSeq Genes), and click the “submit” button. This will generate a link list for downloading the Exons, Mask, and Underlay files. If these files for the chosen base sequence are not available from PipHelper, one has to manually create them (see Note 7).
3. Generate a multiple genomic sequence alignment
At the PipMaker website, click on MultiPipMaker and enter the number of sequences to be aligned. On the next page, select “Generate nucleotide level view (PDF)” option to receive a raw sequence alignment data in addition to a schematic “Pip (Percent identity plot)” view as the output files. In the “First sequence” section, give a desired label for the base sequence, upload the base sequence in FASTA format, the Mask, Exons, and Underlay files for the base sequence, and check the “use as default in pip” box. An annotation file is not necessary for the analysis. Below the “First sequence” section, enter labels and upload sequence files for the additional orthologous sequences in FASTA format, and check the “Search both strands”, “Show all matches” and “High sensitivity and low time limit” options (see Note 8). Underlay files are not necessary for these sequences. After entering the necessary information, click the “submit” button.
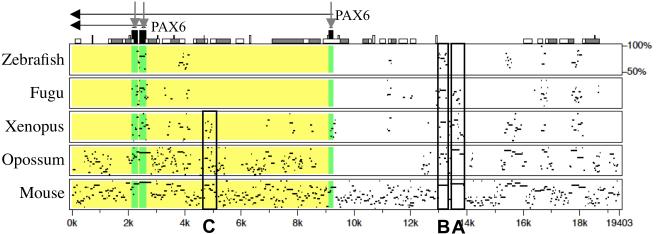
One will receive by e-mail a “Pip view” file that schematically shows the alignment (Fig. 2), and a “nucleotide level view” file that shows an actual multiple alignment of the genomic sequences (Fig. 3). In the Pip view, the height and width of the Pip represent the sequence conservation level and length, respectively. In our example, comparison of Figures 1 and 2 reveals that PipMaker discovered not only the CNEs shown in the ECR Browser window (including the previously identified enhancers shown as A and B in Fig. 1), but also some additional CNEs. One of them, which is marked as “C” in Figure 2, is conserved in tetrapods, but not in fish. In the nucleotide level view of the C region (Fig. 3), the Xenopus, opossum, and mouse nucleotides that are identical to the human nucleotides are shown as dots in the multiple alignment. This multiple alignment is useful for cloning of the sequences and for preliminary search for conserved TFBMs.
Fig. 2.
A pip view output of MultiPipMaker analysis that was generated with sequences downloaded from the ECR Browser window shown in Figure 1. The alignments between the base sequence (human) and zebrafish, Fugu, Xenopus, opossum, and mouse sequences are shown from the top to the bottom. Above the alignment, the gene name (PAX6) and its transcriptional orientation (horizontal black arrows) are shown. The black boxes on the top of the Pip view (indicated by vertical gray arrows) indicate the exons, and other boxes indicate repeats, transposons and other sequence features in the base sequence (go to PipMaker Instructions for details, http://pipmaker.bx.psu.edu/pipmaker/pip-instr.html). The regions of the zebrafish, Fugu, Xenopus, opossum, and mouse sequences that correspond to exons and introns of the base sequence are also shaded in the alignments with dark gray and light gray, respectively. In addition to the pancreas enhancer (A) and lens/cornea enhancer (B), the C region, which is conserved only in tetrapods, is boxed.
Fig. 3.
A nucleotide level view of MultiPipMaker analysis. Only part of the alignment that corresponds to the C region is shown.
3.2. Phylogenetic footprinting analysis of a CNE
1. Re-align orthologous CNE sequences using ClustalW
It is very helpful to re-align the CNE sequences with a more accurate aligner for short sequences, ClustalW, and then shade conserved nucleotides with a multiple alignment editor, Jalview (25), for better visualization of the alignment and more efficient search of TFBMs. First, open the genome sequence files that were used for the PipMaker analysis, find the orthologous CNE sequences identified by PipMaker (in our example, the human, mouse, opossum and Xenopus sequences for the C region of Pax6), and generate new text files in FASTA format for just the CNE sequences. This step can be easily done if you use Range Extractor DNA or a standard DNA analysis tool, such as VectorNTI. Next, go to ClustalW website, select the “DNA” mode, enter a set of the orthologous CNE sequences in a Multi-FASTA format (see Note 3), and click the “submit” button. One sees a raw ClustalW alignment of the input sequences in the next window.
2. Shade conserved nucleotides in a ClustalW alignment using Jalview
In the ClustalW alignment window, click “Result Summary”, and then click the “Start Jalview” button. A new window appears, showing the sequence alignment. In this Jalview window, the alignment format can be modified as desired. For example, first click the “Format” menu, and then click “Wrap” to change the default “single column view” to a “multi-column view” so that the alignment length fits to the window width. Next, click “Color”, and then click “Percent identity” to shade conserved nucleotides. Although the default setting shows a schematic representation of the sequence conservation under the alignment column, it can be removed by clicking “View” menu and then deselecting “Show Annotations”. After the modification, the alignment image can be saved using the “screen shot” function (push “cmd” + “shift” + “4” + “space” keys, or use the packaged program “Grab” on a Macintosh, and “Print Screen” key on a Windows computer). Unfortunately, at this ClustalW website Jalview does not have a menu to export the alignment image to one's computer (see Note 9).
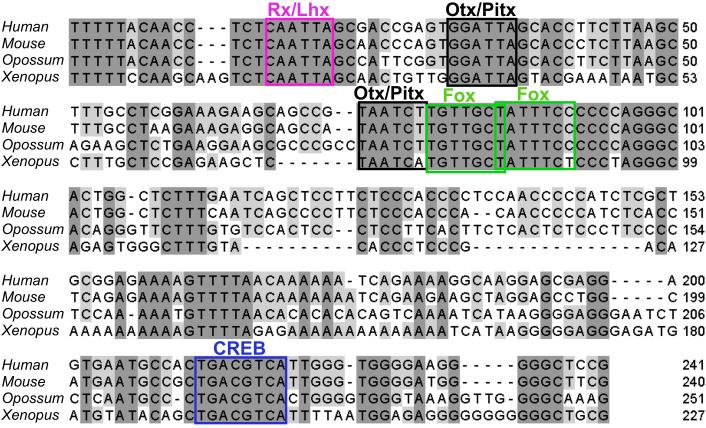
Figure 4 is an example of the Jalview output that shows a multiple alignment of the C region identified by the PipMaker analysis (compare with Fig. 3). Methods used for identifying the TFBMs shown in this figure are described in the next section.
Fig. 4.
Phylogenetic footprinting analysis of Pax6 C region. The ClustalW alignment was shaded with Jalview, in which conserved TFBMs that were identified by rVista analysis are mapped.
3. Searching for TFBMs conserved in CNEs
Program tools for searching TFBMs are classified into three groups: pattern search programs, weight matrix search programs, and Hidden Markov Model (HMM)-based programs (1). Because HMM-based programs require some special mathematical knowledge, pattern and weight matrix search programs are more widely used for conventional analysis. We often use rVista for pattern search, and ConSite for a weight matrix search. This is because these two programs search TFBMs conserved in a pair of orthologous sequences, whereas most other programs work only on a single sequence (see Note 10). Although rVista and ConSite have default sets of TFBMs, they are limited in number and poorly defined TFBMs that may not be relevant to in vivo binding. Hence we strongly recommend creating a user-defined set by collecting TFBMs that are likely involved in regulation of a CNE of interest from the literature and TFBM databases for use in these programs (see Note 11).
Go to the rVista website, input two orthologous CNE sequences in FASTA format (in our example, the human and Xenopus sequences of the C region used for the ClustalW analysis), and click “Submit”. Next, select “User-defined consensus sequences”, enter the TFBMs (in our example, CREB/ATF tgacgtca, Otx/Pitx taakcy, Rx/Lhx taatkr, Fox trttkvy), and click “Submit”. The “Results” window will then show TFBMs conserved between the human and Xenopus sequences. Since rVista shows the TFBMs only one by one in the resulting pair-wise alignment, one has to draw the identified TFBMs in the Jalview alignment for making a visually effective, final output, as shown in Figure 4. Conservation of the TFBMs in other aligned sequences (the mouse and opossum sequences in Fig. 4) is also checked at this step.
3.3. Analysis of enhancer activity of CNEs by co-transgenesis assay in Xenopus
1. Amplify CNEs by genomic PCR
First, amplify a Xenopus CNE sequence from X. tropicalis genomic DNA by PCR. PCR primers should have BamHI sites and an extra 3-base overhang at the 5’ ends for efficient digestion of the resulting PCR product with BamHI. The PCR product with BamHI-protruding ends will be efficiently ligated to the reporter cassette with a compatible end, after their injection into Xenopus eggs (see below). If the CNE sequence under study has BamHI sites internally, BglII sites may be added to the primer ends in place of the BamHI sites so that the PCR product has the protruding ends compatible with the BamHI end of the reporter cassette (see Note 12). In our example, the primers used for amplification of the C region of Xenopus pax6 are as follows (BamHI sites are underlined with the extra 3 bases): C-1, GACGGATCCTTCCAAGCCTTGAATTGACCATCTG; C-2, GACGGATCCACTGATCGCTTCCAAAAACCCAG. They were designed to bind to the 5’ and 3’ flanking regions of the C element in Xenopus genome (see Note 13). The PCR condition used with these primers is shown in Table 1.
Table 1.
The reaction mixture and cycle setting used for amplification of the C region of Xenopus Pax6
| Reagents | Volume (μL) |
|---|---|
| Distilled water | 57 |
| 10 × Pfx buffer | 20 |
| 50 mM MgSO4 | 2 |
| dNTP mix (2.5 mM each) | 12 |
| C-1 Primer (10 pmol/μL) | 3 |
| C-2 Primer (10 pmol/μL) | 3 |
| X. tropicalis genomic DNA (50 ng/μL) | 2 |
| Platinum Pfx DNA polymerase (2.5 U/μL) | 1 |
The cycle setting: 94°C 5 min. → (94°C 30 sec. → 58°C 1 min. → 68°C 1 min.) × 38 cycles → 68°C 7 min.
2. Purify DNA fragments (CNEs and a reporter cassette) for the co-transgenesis assay
After the PCR reaction, take 1/10 volume of the resulting reaction for evaluation by agarose gel electrophoresis (0.7-1.5% agarose in 1 × TAE buffer) and stain the gel with ethidium bromide (0.5 μg/mL in 1 × TAE buffer) to check if the expected product is amplified as a single band (see Note 14). In our example, the primer sets for the Xenopus C element are expected to amplify a fragment of about 0.4 kb that contains a conserved 227 base sequence in the middle of the fragment. If the product has the expected size, purify the remaining product using the QIAquick PCR purification kit, and then digest with BamHI at 37°C overnight to expose their BamHI protruding ends. In parallel, digest pBSSK+βGFP with BamHI and XhoI to cut out a reporter cassette (β-actin basal promoter-GFP-polyA) at 37°C overnight (see Note 15).
After the digestion, separate the DNA fragments by agarose gel electrophoresis, stain with ethidium bromide, cut out the desired DNA bands from the gel, and then purify the DNA using the QIAquick Gel Extraction Kit. The expected length of the reporter cassette is about 1.0 kb. After the purification with the QIAquick Gel Extraction Kit, precipitate the DNA with ethanol once, and dissolve in 10 mM KCl solution so that the final concentration will be 50-100 ng/μL (see Note 16). The purified DNA solution is stored at −20°C until use in transgenesis.
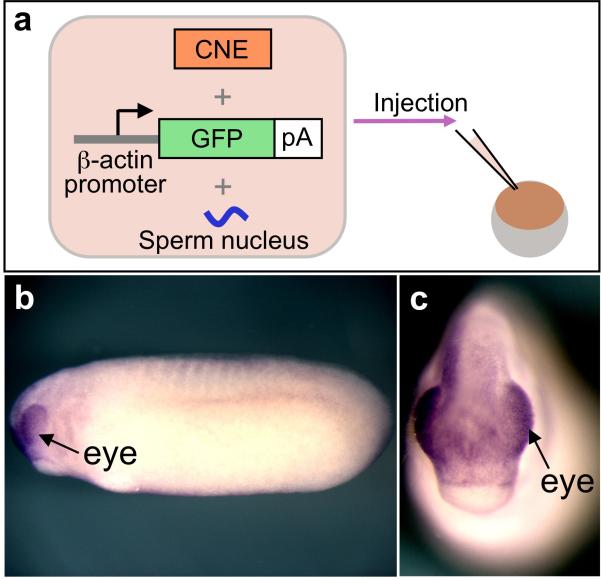
3. Co-inject a purified CNE fragment together with the reporter cassette into X. laevis eggs
A standard protocol for the sperm nuclear transplantation method of X. laevis transgenesis is found in Chapter 11 (see Note 17). For the co-transgenesis, mix the purified CNE fragment and the reporter cassette in a molar ratio of 2 : 1, and inject this mixture with the sperm nuclei into the eggs (Fig. 5A). After the injection and subsequent development, fix the resulting embryos at desired developmental stages, and subject them to in situ hybridization with antisense GFP probe. The in situ hybridization analysis is necessary for detecting GFP expression in embryos using this transgenesis protocol, because the expression from a β-actin basal promoter of the reporter cassette is not strong enough for epifluorescence microscopy in most cases. However, because the background expression from this promoter is very weak, it makes it possible to clearly distinguish the enhancer-driven expression from the promoter background in an in situ hybridization analysis. The GFP probe can be generated by transcribing the HindIII-digested pBSSK+βGFP with T7 RNA polymerase. For in situ hybridization and probe synthesis, follow the standard protocol for Xenopus (20). The gata2 promoter discussed in the Materials section may elicit more robust expression of CNE's by co-transgenesis and therefore direct detection of expression by GFP fluorescence, though there may be a significant delay between the transcription of GFP mRNA and the accumulation and/or maturation of GFP protein for fluorescence during development.
Fig. 5.
The co-transgenesis assay and GFP expression in the resulting embryo detected by in situ hybridization. (A) A schematic representation of the co-injection of a CNE fragment, the reporter cassette and the sperm nucleus into Xenopus eggs. (B, C) GFP expression in the embryo co-injected with the C element of Xenopus Pax6 and the GFP reporter cassette. Arrows indicate eye-specific expression of GFP (B, lateral view; C, frontal view).
If the injected CNE has enhancer activity, approximately 5-20% of the developing embryos typically express GFP, though the fraction of expressing embryos depends on a number of factors, including the strength of the enhancer. If the enhancer activity is relatively weak, the color reaction of the in situ hybridization experiment may take 1-2 days. Figures 5B and C show the example of GFP expression in the transgenic embryo generated with the C region and the reporter cassette. GFP expression was detected in this case in the eye (see Note 18).
Acknowledgements
This work was supported by the Global COE Program in NAIST (Frontier Biosciences: strategies for survival and adaptation in a changing global environment), Grant-in-Aid for Scientific Research (C) (20579002 and 23570256) from JSPS and Grant-in-Aid for Scientific Research on Innovative Areas (21200064) from MEXT, Japan, to H. Ogino, by Grant-in-Aid for Young Scientists (B) (21770234) from JSPS and Research for Promoting Technological Seeds (A) (10-099) from JST, Japan, to H. Ochi, and by CREST (JST). This work was also supported by NIH grants EY00675, EY017400, EY018000 and RR013221 to R. Grainger.
Footnotes
p300 is a transcriptional coactivator that is a near-ubiquitously expressed component of enhancer-associated protein assemblies. Since ChIP-seq analysis showed that p300 is significantly enriched within 10 kb up- and downstream of promoters, it may be effective to search non-conserved enhancers in the −10 kb to +10 kb region in addition to searching for enhancer activity in CNEs (8). It is also noteworthy that another ChIP-seq analysis revealed that enhancers of early developmental genes display three overlapping chromatin signatures, monomethylation of histone H3 at lysine 4 (H3K4me1), trimethylation of histone H3 at lysine 27, and p300-binding, in embryonic stem cells (26). Incorporation of such information about genome-wide epigenetic modifications may give the comparative genomics approach even more promise.
One may follow the Xenopus standard protocols for collecting blood cells from the heart, and purification of the genomic DNA from the blood cells or embryos (20). The DNeasy Blood & Tissue Kit (Qiagen) also works well.
Definitions of FASTA and Multi-FASTA format are found in Oxford University CBRG website (http://www.compbio.ox.ac.uk/bioinformatics_faq/format_examples.shtml).
The UCSC Genome Browser (http://genome.ucsc.edu/) and Vista Browser (http://genome.lbl.gov/vista/index.shtml) provide alternate options for retrieval of orthologous genomic sequences of a gene of interest. In the genome alignment window of the UCSC Genome Browser, click the “Convert” menu, and follow the directions. In the alignment window of the Vista Browser, click “Show Alignment(s)”, and then click “Sequence (softmasked)” in the newly opened page.
The “Synteny/Alignments” window sometimes show two or more sequences as orthologs for a single base sequence. In such a case, one has to check whether the genome browser identified true multiple orthologs generated by a species-specific genome duplication (this often occurs in a mammalian-fish comparison), or misidentified closely related genes as the orthologs. For an example of the latter case, the genome browsers often misidentify both mouse Sox2 and Sox3 as the orthologs of human SOX2. A very useful database, Metazome, can help one to check orthologous relationships between genes from different species by showing their synteny conservation (http://www.metazome.net/).
This webpage is designed to forward the sequences to Mulan, a genome alignment tool that searches sequence conservation with higher sensitivity than ECR Browser in a limited length of orthologous genomic sequences. Since the sensitivity of Mulan is still lower than that of PipMaker, we usually choose to use Pipmaker for identifying CNEs. In our experience, Mulan often misses small CNEs that are found by PipMaker in mammalan-Xenopus genome comparison. Furthermore the nucleotide level output of Mulan is not a typical multiple sequence alignment and less accessible than that of PipMaker for bench biologists to extract CNE sequences for subsequent phylogenetic footprinting analysis, though the schematic view of the alignment generated by Mulan is better than that of PipMaker.
One can easily generate a Mask file using the RepeatMasker tool at the ISB website (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker) from a genome sequence of interest. For Exons and Underlay files, one has to generate these by analyzing gene structures using a standard DNA analysis software tool, such as Blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi) or Vector NTI.
The “Show all matches” setting makes it possible for one region of the base sequence to align with several regions of the second sequence because of partial sequence duplications, or because of incomplete masking of low-complexity regions. The “Chaining” setting allows identifying and plotting only matches that appear in the same relative order in the base and second sequences. With the “Single coverage” setting, PipMaker selects a highest-scoring set of alignments such that any position in the base sequence can appear in at most one alignment. We often use “Show all matches”, which leads to “multi-hits” of low complexity regions, such as repetitive elements, of the base sequence to the aligned sequences and distinguishes them from CNEs with enhancer activity, which are generally unique in the genome and show a “single-hit” between the base and aligned sequences. The “High sensitivity and low time limit” option increases the MultiPipMaker sensitivity so that it can identify weak conservation between a sequence pair at a great evolutionary distance, such as human and Fugu sequences. The default setting without this option is tuned for comparison of two mammalian sequences. Further detailed instructions for these settings are found on the Advanced PipMaker instructions website (http://pipmaker.bx.psu.edu/pipmaker/pip-instr3.html).
Although Jalview can be easily used at this ClustalW website, its function is in part restricted. If its full function is needed, go to the Jalview website of University of Dundee (http://www.jalview.org/index.html), where one can use ClustalW, fully functional Jalview, and easily save the resulting alignment image to a local computer.
For searching TFBMs in a single sequence, Patch, Match, and P-Match are available for public use (http://www.gene-regulation.com/pub/programs.html). Patch and Match are pattern search and weight matrix search programs, respectively. P-Match combines pattern matching and weight matrix approaches thus providing higher accuracy of recognition than either of the methods alone. One can choose Transfac or a user-defined set of TFBMs for running these programs.
One can develop a custom set by selecting TFBMs from a TFBM database, such as Transfac or JASPAR (http://jaspar.cgb.ki.se/cgi-bin/jaspar_db.pl) (19). A collection of binding motifs of homeodomain proteins is also useful (27). It is also strongly recommended to search the literature for specific TFBMs that are likely involved in regulation of a CNE of interest and validated by stringent tests defining them as sites used in vivo. Many of the TFBMs in databases such as Transfac were generated by in vitro binding experiments and may include weak sites that do not work or are not relevant in vivo. Thus, one may easily be misled by the multitude of false positives if the TFBM collection from the database alone is used for the search.
In the co-transgenesis assay, the addition of compatible protruding ends to both an enhancer fragment and the reporter cassette increases the number of transgenic embryos with enhancer-driven reporter expression by approximately two-fold (C. Uchiyama, H. Ochi and H. Ogino, unpublished observation). The ligation reaction between these co-injected DNA fragments may be mediated by the homologous end-joining activity of Xenopus eggs (28).
A web-tool, Primer3 (http://primer3.sourceforge.net/) is convenient for designing PCR primers.
One may follow standard molecular cloning protocols for the restriction enzyme digestion, agarose gel epectrophoresis, and ethanol precipitation of DNA (29).
Digestion of 10 μg of pBSSK+βGFP would be sufficient for preparing the reporter cassette for 10-20 rounds of co-transgenesis.
The last ethanol precipitation step is for removal of residual carryover of the gel extraction buffer and the DNA elution buffer from the kit. The DNA elution buffer in the kit contains Tris buffer, which is toxic for Xenopus embryos and sometimes causes ectopic transgene expression if introduced into the embryos with the DNA.
The co-transgenesis assay uses the sperm nuclear transplantation method. Because X. laevis eggs are larger and more tolerant to the sperm nuclear transplantation than X. tropicalis eggs, we usually choose X. laevis for this experiment. The sperm nuclear transplantation method may be somewhat technically demanding for those who are not familiar with the Xenopus system. In this case, one may clone a CNE fragment into a basal promoter-GFP reporter plasmid and perform the Xenopus transgenic assay using a more convenient technique, such as the I-SceI meganuclease method or others (Chapter 12-14) (30, 31). It is slower than the co-transgenesis assay, but still faster and more reliable than any enhancer assay in other vertebrates, and can be performed with X. tropicalis if necessary.
One may also use another eye enhancer of pax6, which is located in intron 7, as a positive control for the co-transgenesis assay (30). This enhancer may exhibit stronger activity than that of the C region in tailbud embryos. The primers for amplification of this intronic enhancer of Xenopus tropicalis pax6 are as follows (BamHI sites are underlined with the extra 3 bases): int7-1, GACGGATCCACTGGGTGGGGGTAATTCCT; int7-2, GACGGATCC GGGAGATAAATACAGGGGGTC. The PCR condition for this primer pair is nearly the same as that shown in Table 1, except that the annealing temperature should be 54°C. The expected length of the amplicon is about 0.6 kb.
References
- 1.Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 2.Pennacchio LA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 3.Woolfe A, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lettice LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 5.Sagai T, et al. Phylogenetic conservation of a limb-specific, cis-acting regulator of Sonic hedgehog (Shh) Mamm Genome. 2004;15:23–34. doi: 10.1007/s00335-033-2317-5. [DOI] [PubMed] [Google Scholar]
- 6.Benko S, et al. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat Genet. 2009;41:359–364. doi: 10.1038/ng.329. [DOI] [PubMed] [Google Scholar]
- 7.Fisher S, et al. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006;312:276–279. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- 8.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hellsten U, et al. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasahara M, et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- 11.Canestro C, Yokoi H, Postlethwait JH. Evolutionary developmental biology and genomics. Nat Rev Genet. 2007;8:932–942. doi: 10.1038/nrg2226. [DOI] [PubMed] [Google Scholar]
- 12.Ogino H, Fisher M, Grainger RM. Convergence of a head-field selector Otx2 and Notch signaling: a mechanism for lens specification. Development. 2008;135:249–258. doi: 10.1242/dev.009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ovcharenko I, et al. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz S, et al. MultiPipMaker and supporting tools: Alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res. 2003;31:3518–3524. doi: 10.1093/nar/gkg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 16.Stathopoulos A, Levine M. Genomic regulatory networks and animal development. Dev Cell. 2005;9:449–462. doi: 10.1016/j.devcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Loots GG, Ovcharenko I. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 2004;32:W217–221. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandelin A, Wasserman WW, Lenhard B. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004;32:W249–252. doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Portales-Casamar E, et al. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sive H, Grainger R, Harland R. Early Development of Xenopus laevis - A LABORATORY MANUAL. Cold Spring Harbor Laboratory Press; New York: 2000. [Google Scholar]
- 21.Kost TA, Theodorakis N, Hughes SH. The nucleotide sequence of the chick cytoplasmic β-actin gene. Nucleic Acids Res. 1983;11:8287–8301. doi: 10.1093/nar/11.23.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navratilova P, et al. Systematic human/zebrafish comparative identification of cis-regulatory activity around vertebrate developmental transcription factor genes. Dev Biol. 2009;327:526–540. doi: 10.1016/j.ydbio.2008.10.044. [DOI] [PubMed] [Google Scholar]
- 23.Kleinjan DA, et al. Long-range downstream enhancers are essential for Pax6 expression. Dev Biol. 2006;299:563–581. doi: 10.1016/j.ydbio.2006.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu PX, et al. Regulation of Pax6 expression is conserved between mice and flies. Development. 1999;126:383–395. doi: 10.1242/dev.126.2.383. [DOI] [PubMed] [Google Scholar]
- 25.Waterhouse AM, et al. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noyes MB, et al. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehman CW, Trautman JK, Carroll D. Illegitimate recombination in Xenopus: characterization of end-joined junctions. Nucleic Acids Res. 1994;22:434–442. doi: 10.1093/nar/22.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press; New York: 2001. [Google Scholar]
- 30.Ogino H, Ochi H. Resources and transgenesis techniques for functional genomics in Xenopus. Dev Growth Differ. 2009;51:387–401. doi: 10.1111/j.1440-169X.2009.01098.x. [DOI] [PubMed] [Google Scholar]
- 31.Ogino H, McConnell WB, Grainger RM. High-throughput transgenesis in Xenopus using I-SceI meganuclease. Nat Protoc. 2006;1:1703–1710. doi: 10.1038/nprot.2006.208. [DOI] [PubMed] [Google Scholar]
- 32.Kammandel B, et al. Distinct cis-essential modules direct the time-space pattern of the Pax6 gene activity. Dev Biol. 1999;205:79–97. doi: 10.1006/dbio.1998.9128. [DOI] [PubMed] [Google Scholar]