Abstract
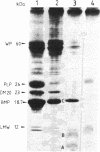
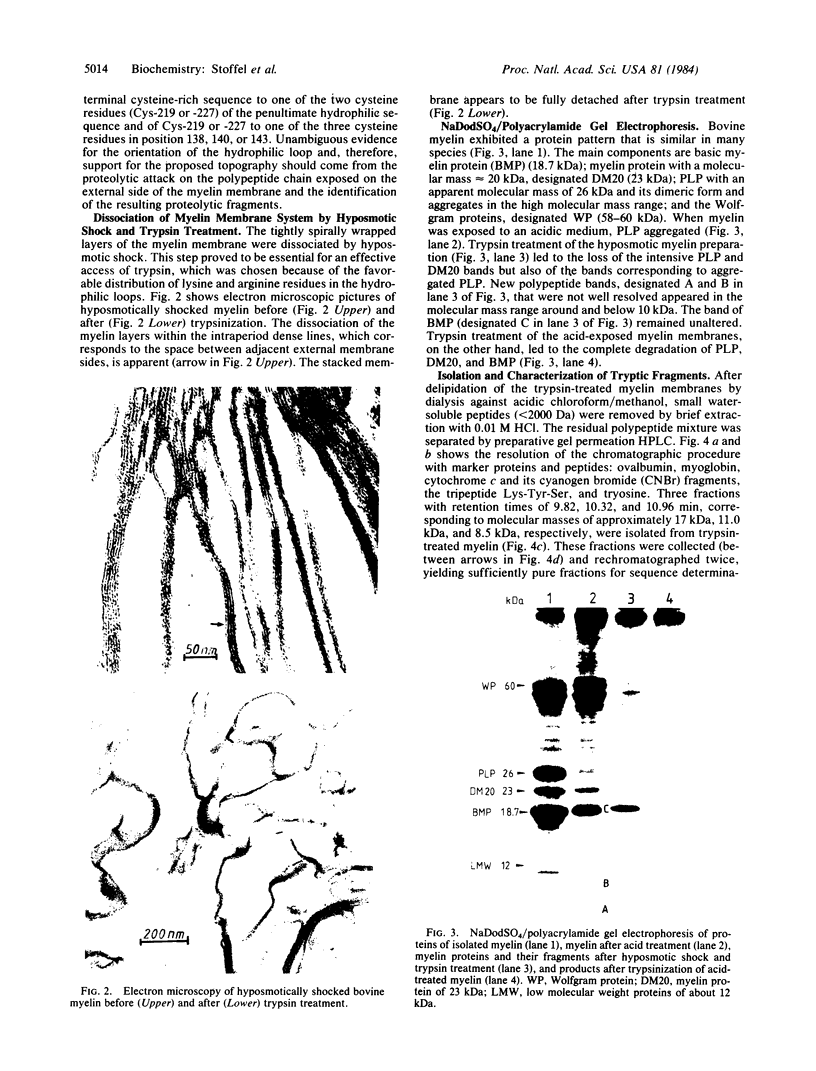
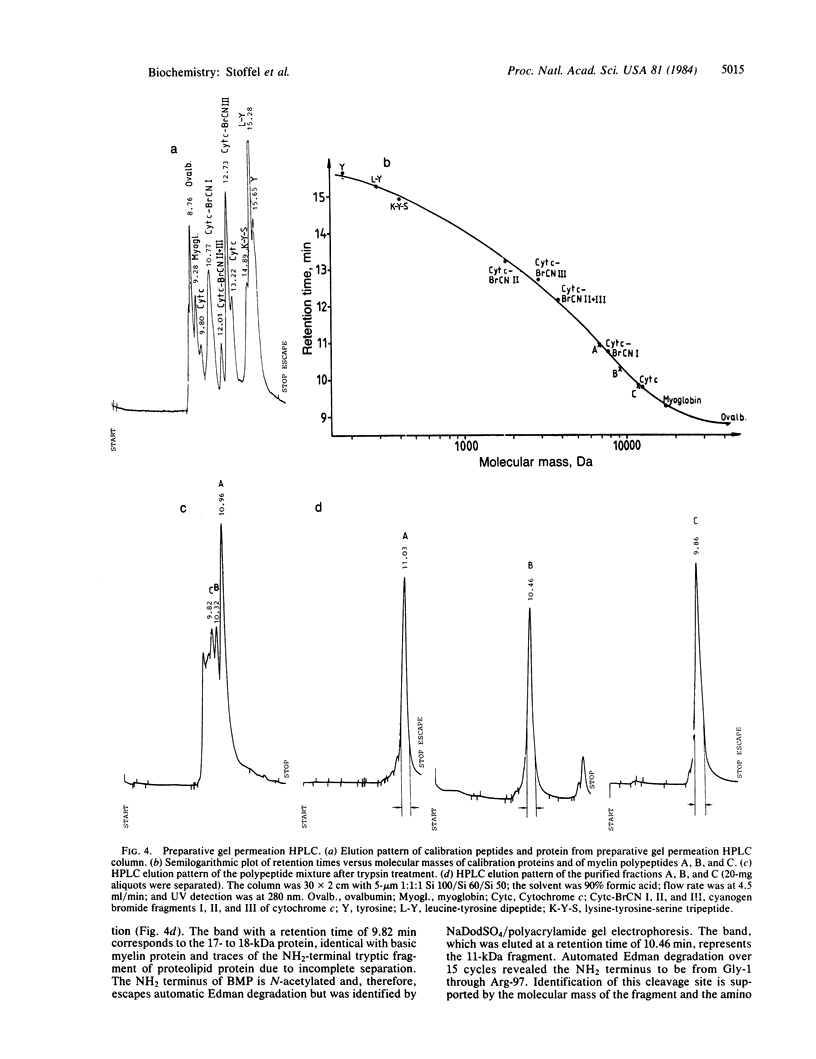
Proteolipid protein (PLP) of central nervous system myelin is one of the most hydrophobic integral membrane proteins. It consists of a 276-residue-long polypeptide chain with five strongly hydrophobic sequences of 26, 30, 39, 12, and 36 residues, respectively, linked by highly charged hydrophilic sequences. Hyposmotically dissociated bovine myelin membranes were treated with trypsin. PLP was completely cleaved into smaller fragments, whereas basic myelin protein remained essentially unaltered. The proteins and tryptic peptides of myelin were separated after the removal of the short, water-soluble peptides into three large fragments of 11, 7.3, and 9.0 kDA, respectively. They were characterized by their molecular mass and NH2-terminal amino acid sequences, which proved that trypsin cleaved predominantly at Arg-97 yielding the 11-kDa fragment from Gly-1 through Arg-97, at Arg-126 releasing the 7.3-kDa fragment from Gly-127 through Lys-191, and at Lys-191 releasing the 9-kDa fragment from Thr-192 through Phe-276. We propose that PLP is integrated into the lipid bilayer of myelin with the NH2 terminus and three positively charged hydrophilic loops oriented toward the extracytosolic side of the membrane, whereas one strongly negative hydrophilic loop and the positively charged COOH terminus cover the cytosolic side of the lipid bilayer. Basic myelin protein remains protected against tryptic cleavage, which indicates its apposition to the cytosolic side of the membrane. These cleavage sites of trypsin support the suggested orientation of PLP in the myelin membrane and thereby extend our knowledge about the molecular arrangement of the components of this membrane. In demyelinating processes membrane desintegration could be initiated by proteolysis at the external surfaces of proteolipid protein in a similar way as described here.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boggs J. M., Moscarello M. A. Structural organization of the human myelin membrane. Biochim Biophys Acta. 1978 Apr 10;515(1):1–21. doi: 10.1016/0304-4157(78)90006-0. [DOI] [PubMed] [Google Scholar]
- Crang A. J., Rumsby M. G. The labelling of lipid and protein components in isolated central-nervous-system myelin with dansyl chloride. Biochem Soc Trans. 1977;5(1):110–112. doi: 10.1042/bst0050110. [DOI] [PubMed] [Google Scholar]
- Edman P., Begg G. A protein sequenator. Eur J Biochem. 1967 Mar;1(1):80–91. doi: 10.1007/978-3-662-25813-2_14. [DOI] [PubMed] [Google Scholar]
- FOLCH J., LEES M. Proteolipides, a new type of tissue lipoproteins; their isolation from brain. J Biol Chem. 1951 Aug;191(2):807–817. [PubMed] [Google Scholar]
- Laursen R. A., Samiullah M., Lees M. B. Gene duplication in bovine brain myelin proteolipid and homology with related proteins. FEBS Lett. 1983 Sep 5;161(1):71–74. doi: 10.1016/0014-5793(83)80732-7. [DOI] [PubMed] [Google Scholar]
- Lee A. G. Functional properties of biological membranes: a physical-chemical approach. Prog Biophys Mol Biol. 1975;29(1):3–56. doi: 10.1016/0079-6107(76)90019-5. [DOI] [PubMed] [Google Scholar]
- Lees M. B., Chao B. H., Laursen R. A., L'Italien J. J. A hydrophobic tryptic peptide from bovine white matter proteolipid. Biochim Biophys Acta. 1982 Mar 18;702(1):117–124. doi: 10.1016/0167-4838(82)90034-6. [DOI] [PubMed] [Google Scholar]
- Lees M. B., Chao B. H., Lin L. F., Samiullah M., Laursen R. A. Amino acid sequence of bovine white matter proteolipid. Arch Biochem Biophys. 1983 Oct 15;226(2):643–656. doi: 10.1016/0003-9861(83)90334-x. [DOI] [PubMed] [Google Scholar]
- McIntosh T. J., Robertson J. D. Observations on the effect of hypotonic solutions on the myelin sheath in the central nervous system. J Mol Biol. 1976 Jan 15;100(2):213–217. doi: 10.1016/s0022-2836(76)80149-0. [DOI] [PubMed] [Google Scholar]
- Mehl E., Halaris A. Stoichiometric relation of protein components in cerebral myelin from different species. J Neurochem. 1970 May;17(5):659–668. doi: 10.1111/j.1471-4159.1970.tb00545.x. [DOI] [PubMed] [Google Scholar]
- Norton W. T. Isolation of myelin from nerve tissue. Methods Enzymol. 1974;31:435–444. doi: 10.1016/0076-6879(74)31049-x. [DOI] [PubMed] [Google Scholar]
- Nussbaum J. L., Jollès J., Jollès P. The apoprotein of the major rat brain myelin P7 proteolipid: alignment of the BNPS-skatole fragments and present state of the sequence. Biochimie. 1982 Jun;64(6):405–410. doi: 10.1016/s0300-9084(82)80578-6. [DOI] [PubMed] [Google Scholar]
- Poduslo J. F., Braun P. E. Topographical arrangement of membrane proteins in the intact myelin sheath. Lactoperoxidase incorproation of iodine into myelin surface proteins. J Biol Chem. 1975 Feb 10;250(3):1099–1105. [PubMed] [Google Scholar]
- Segrest J. P., Feldmann R. J. Membrane proteins: amino acid sequence and membrane penetration. J Mol Biol. 1974 Aug 25;87(4):853–858. doi: 10.1016/0022-2836(74)90090-4. [DOI] [PubMed] [Google Scholar]
- Stoffel W., Hillen H., Schroeder W., Deutzmann R. Primary structure of the C-terminal cyanogen bromide fragments II, III and IV from bovine brain proteolipid-apoprotein. Hoppe Seylers Z Physiol Chem. 1982 Aug;363(8):855–864. doi: 10.1515/bchm2.1982.363.2.855. [DOI] [PubMed] [Google Scholar]
- Stoffel W., Hillen H., Schröder W., Deutzmann R. Lipophilin (proteolipid apoprotein) of brain white matter. Purification and amino acid sequence studies of the four tryptophan fragments. Hoppe Seylers Z Physiol Chem. 1982 Nov;363(11):1397–1407. doi: 10.1515/bchm2.1982.363.2.1397. [DOI] [PubMed] [Google Scholar]
- Stoffel W., Hillen H., Schröder W., Deutzmann R. The primary structure of bovine brain myelin lipophilin (proteolipid apoprotein). Hoppe Seylers Z Physiol Chem. 1983 Oct;364(10):1455–1466. doi: 10.1515/bchm2.1983.364.2.1455. [DOI] [PubMed] [Google Scholar]
- Stoffel W., Schröder W., Hillen H., Deutzmann R. Analysis of the primary structure of the strongly hydrophobic brain myelin proteolipid apoprotein (lipophilin). Isolation and amino acid sequence determination of proteolytic fragments. Hoppe Seylers Z Physiol Chem. 1982 Sep;363(9):1117–1131. doi: 10.1515/bchm2.1982.363.2.1117. [DOI] [PubMed] [Google Scholar]
- Tanford C. The hydrophobic effect and the organization of living matter. Science. 1978 Jun 2;200(4345):1012–1018. doi: 10.1126/science.653353. [DOI] [PubMed] [Google Scholar]
- Tomita M., Marchesi V. T. Amino-acid sequence and oligosaccharide attachment sites of human erythrocyte glycophorin. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2964–2968. doi: 10.1073/pnas.72.8.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]