Background: Post-transcriptional regulation of p21 expression is still not fully understood.
Results: Rbm24 is induced by p53 and serves as a mediator of p53 to regulate p21 expression via binding to its 3′-UTR.
Conclusion: Rbm24, an RNA-binding protein and a target of p53, enhances p21 expression via mRNA stability.
Significance: A novel p53 target gene Rbm24 is identified to regulate p21 mRNA stability.
Keywords: Cancer Biology, Cell Cycle, mRNA Decay, p53, RNA-binding Protein
Abstract
p21, a cyclin-dependent kinase inhibitor, is necessary for proper control of the cell cycle and premature senescence. Thus, p21 expression needs to be tightly controlled. In this study, we found that Rbm24, an RNA-binding protein and a target gene of the p53 protein, can regulate p21 expression via mRNA stability. Specifically, we showed that Rbm24 is induced by DNA damage and Mdm2 inhibitor Nutlin-3. We also found that p53 protein binds to and activates the promoter of the Rbm24 gene. Moreover, we found that overexpression of Rbm24 increases, whereas knockdown of Rbm24 decreases, p21 mRNA and protein expression. In addition, we demonstrated that overexpression of Rbm24 enhances the half-life of p21 transcript. Consistent with this, we provided evidence that Rbm24 binds to the 3′-untranslated region (3′-UTR) of p21 transcript and an AU/U-rich element in the p21 3′-UTR is necessary for Rbm24 to increase p21 expression. Finally, we showed that the RNA recognition motif in Rbm24 is required for binding to p21 transcript and subsequently for inducing p21 expression. Altogether, we uncovered that Rbm24 is a novel player in the p53 pathway, which may be explored to restore proper cell cycle control in p53-deficient tumors via p21.
Introduction
The p53 tumor suppressor plays a central role in suppressing cancer development. The importance of p53 is evidenced as loss or inactivation of functional p53 in >50% of human cancers (1). In response to stress signals, such as DNA damage, hypoxia, or activated oncogenes, p53 is activated and functions as a transcriptional factor to induce a number of downstream targets, including p21waf1/cip1, PUMA, and DEC1. These p53 targets mediate diverse cellular functions of p53 tumor suppressor, including cell cycle arrest, senescence, and apoptosis (2–5).
RNA-binding proteins (RBPs)2 are known to play a key role in post-transcriptional regulation, including mRNA stabilization and translation (6–8). Many oncogenes and tumor suppressor genes are under the control of RBPs in mammalian cancer cell lines (9). Rbm38, also known as RNPC1, is an RNA-binding protein that has been found to be overexpressed in various types of cancer, including colon carcinoma (10), esophageal cancer (11), and lymphoma (10, 12). Rbm38 contains a highly conserved RNA recognition motif (RRM), which consists of two submotifs, RNP1 and RNP2 (13). Previous studies showed that Rbm38 is a target of the p53 family and regulates p21 mRNA stability by binding to AU-rich elements (AREs) in their 3′-untranslated regions (UTRs) (13, 14). Interestingly, we found that another RBP, Rbm24, shares high similarity with Rbm38 in the RRM region (Fig. 1A). Rbm24 has been found to be preferentially expressed in cardiac and skeletal muscle tissues (15, 16). Rbm24 is necessary for sarcomere assembly and heart contractility, suggesting that Rbm24 plays an important role in myogenic differentiation and heart development (15–17). Similarly, Rbm24 possesses one conserved RRM, including two submotifs, RNP1 and RNP2 (17). However, the role of Rbm24 in the p53 pathway is unclear.
FIGURE 1.
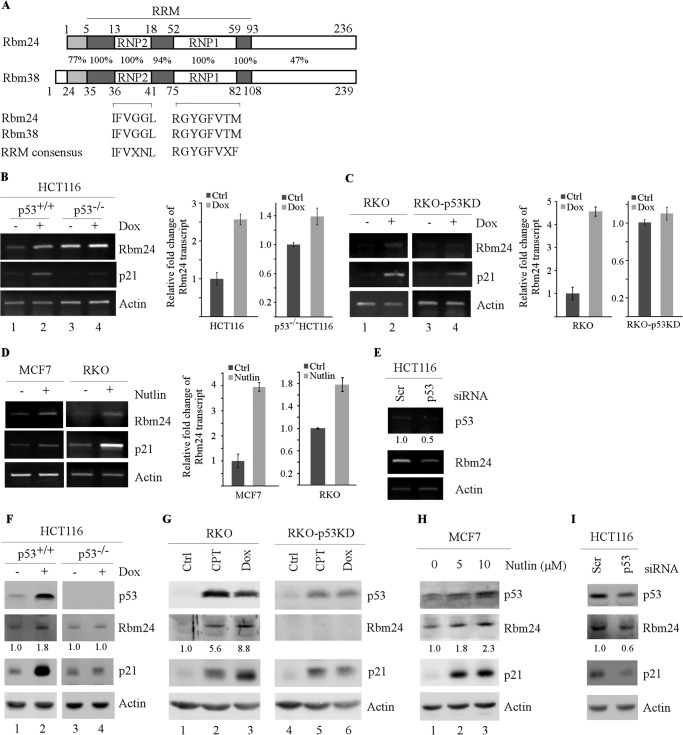
Rbm24 is induced by p53. A, sequence identity between human Rbm24 and Rbm38, and the consensus sequence for RNP1 and RNP2 submotifs within the RRM are shown. The sequence comparison was done using the online tool, BLAST. B–D, HCT116 (p53+/+), HCT116 (p53−/−), RKO, and RKO-p53-KD cells were untreated (−) or treated (+) with 125 μg/ml doxorubicin (Dox) for 24 h (B and C). Total RNAs were purified from cell extracts, and the levels of Rbm24 were quantified by RT-PCR. Left panels, semiquantitative RT-PCR analysis was performed to determine the level of Rbm24, p21, and actin. Right panels, quantitative RT-PCR analysis was performed in triplicate, and the level of Rbm24 transcript was normalized to that of actin control. Upon normalization to actin transcript levels, the relative -fold change of Rbm24 transcript was calculated by dividing the level of Rbm24 transcript in treated cells to that in untreated cells. D, MCF7 and RKO cells were untreated (−) or treated (+) with 10 μg/ml Nutlin-3 (Nutlin) for 24 h. The transcript levels of Rbm24, p21, and actin were determined as in B and C. E, HCT116 cells were transiently transfected with a control siRNA or p53 siRNA for 72 h. The mRNA levels of p53, Rbm24, and actin were measured through RT-PCR. F and G, HCT116, HCT116 (p53−/−), RKO, and RKO-p53-KD cells were untreated (−) or treated (+) with 125 μg/ml doxorubicin (Dox) or 250 nm camptothecin (CPT) for 24 h. Western blot analysis was used to determine the level of p53, Rbm24, p21, and actin proteins. H, MCF7 cells were untreated (−) or treated (+) with 5 μm and 10 μm Nutlin-3 for 24 h. Western blot analysis was used to determine the level of p53, Rbm24, p21, and actin proteins. I, the experiment was performed as in E except that the levels of p53, Rbm24, p21, and actin proteins were determined by Western blot analysis. Error bars indicate S.D.
The cyclin-dependent kinase inhibitor p21 is a major mediator of p53 to induce cell cycle arrest in G1 (5). In addition, p21 plays a vital role in cellular senescence and modulation of apoptosis and differentiation (5, 18–20). Due to these important functions of p21, tight control of p21 expression is essential to maintain proper cell cycle regulation in cells to combat tumorigenesis. Indeed, the level of p21 is highly regulated at both transcriptional and post-transcriptional levels (21). For example, the p53 family proteins, including p53, p63, and p73, regulate p21 transcriptionally (22–24), whereas several RBPs, including RRM-containing proteins Rbm38, HuD, and HuR, regulate p21 mRNA stability through binding of AREs in the p21 3′-UTR (13, 14, 25–27). Additionally, poly(C)-binding protein 1 (PCBP1), PCBP2, and PCBP4, which all contain several K homology domains, regulate p21 mRNA stability through binding of CU-rich elements in the p21 3′-UTR (28, 29). Previously, we found that Rbm38, a target of the p53 family, cooperates with HuR to regulate p21 mRNA stability by binding to AREs in the 3′-UTR (13, 14). Consistent with this finding, Rbm38 has been reported to regulate myogenic differentiation via stabilization of the p21 transcript (15). As Rbm24 and Rbm38 share a similarity in RRM, we hypothesize that Rbm24 may also be a target of p53 and serve as a mediator of p53 to regulate p21 post-transcriptionally.
EXPERIMENTAL PROCEDURES
Reagents
Anti-Rbm24, raised in rabbit, was generated by Cocalico Biologicals (Reamstown, PA). Anti-p53, FL393, and anti-p21, SX118, were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-actin, proteinase inhibitor mixture, RNase A, and protein A/G beads were purchased from Sigma-Aldrich. Scrambled siRNA (GGC CGA UUG UCA AAU AAU U) and siRNA against Rbm24 (CAC UGG AGC UGC AUA CGC A) were purchased from Dharmacon RNA Technologies (Chicago, IL). siRNA against p53 (GGA AAU UUG CGU GUG GAG U) was purchased from Qiagen. Transfection reagent METAFECTENE® PRO was purchased from Biontex. Transfection agent ExpressFectTM was purchased from Denville (Metuchen, NJ). siLentFectTM lipid was purchased from Bio-Rad. TRIzol® agent was purchased from Invitrogen. MMLV reverse transcriptase was purchased from Promega (Madison, WI).
Plasmids
All lentiviral vectors (pLKO.1-puro) expressing shRNA of interest were purchased from Sigma-Aldrich. The targeting sequences are 5′-CGC TGA GTA CTT CGA AAT GTC-3′ for control luciferase shRNA, 5′-GCG AGC AAT ATG TAG CTT GAA-3′ for Rbm24 shRNA#1, and 5′-CCC ATC ATT GAT GGC AGA AAG-3′ for Rbm24#2.
To generate pcDNA3-HA-Rbm24, a PCR product was amplified using an expressed sequence tag clone as a template using forward primer, 5′-GGG GAA TTC ATG CAC ACG ACC CAG AAG-3′ and reverse primer, 5′-GGG CTC GAG CTA TTG CAT TCG GTC TGT CTG-3′, and then the EcoRI-XhoI fragment was inserted into pcDNA3-HA vector. To generate pcDNA3 vector expressing untagged Rbm24 (pcDNA3-Rbm24), a PCR product was amplified using pcDNA3-HA-Rbm24 as a template using forward primer, 5′-AAA AAG CTT CAC CAT GAT GCA CAC GAC CCA GAA GGA CAC GAC GTA CA-3′, and reverse primer used for pcDNA3-HA-Rbm24, and then inserted into pcDNA3 vector via HindIII and XhoI sites. To generate pcDNA4-Rbm24 vector, a similar strategy was used for pcDNA3-Rbm24, except that pcDNA4 vector was used.
To generate constructs expressing Rbm24 lacking RNP1 or RNP2, two-step PCRs were performed. The first step was performed to amplify two separate cDNA fragments. Fragment 1 was amplified with forward primer, 5′-AAA AAG CTT CAC CAT GAT GCA CAC GAC CCA GAA GGA CAC GAC GTA CA-3′ and reverse primer, 5′-GCG TCG GTG GTG TGG TAG GGC TTG GTG TAC GTC GTG TCC T-3′ for Rbm24 (ΔRNP2), or reverse primer, 5′-TCG GCA GCA GCC CGG TCA GCG GAC TTG CCC GTC TGC CGG TCG GTG-3′ for Rbm24 (ΔRNP1). Fragment 2 was amplified with forward primer, 5′-AGG ACA CGA CGT ACA CCA AGC CCT ACC ACA CCA CCG ACG CCA GCC-3′ for Rbm24 (ΔRNP2), or 5′-ACC GGC AGA CGG GCA AGT CCG CTG ACC GGG CTG CTG CCG AAA GGG-3′ for Rbm24 (ΔRNP1), and reverse primer, 5′-GGG CTC GAG CTA TTG CAT TCG GTC TGT CTG-3′. The second step PCR was performed using a mixture of fragments 1 and 2 as a template with forward primer for fragment 1 and the reverse primer for fragment 2, and resulting fragments were separately cloned and confirmed by sequencing. A HindIII-XhoI fragment containing the coding region for Rbm24 ΔRNP1 or RNP2 was cloned into pcDNA3.
To generate a luciferase reporter under the control of the Rbm24 promoter, two genomic DNA fragments were amplified by PCR. The fragment from nucleotides −3283 to +4 (the first nucleotide upstream of the putative transcription start site is designated as −1) was amplified with forward primer 5′-ACA GGT ACC GGA GAT GAC AAC CTT GTG GAA CC-3′ and reverse primer 5′-ATA CTC GAG CTT CGC ACC CGC CCC GCG GCT-3′. The fragment from nucleotides −1571 to +4 was amplified with forward primer 5′-GCG GGT ACC CAG GTC TGC TGG TAG AAA CCA CTG G-3′ and reverse primer 5′-ATA CTC GAG CTT CGC ACC CGC CCC GCG GCT-3′. The DNA fragments were cloned separately into pGL2-basic vectors via KpnI and XhoI. The fragments were confirmed by sequencing.
Similarly, to generate an OFluc luciferase reporter under the control of potential p53 response element (p53-RE) in the Rbm24 gene, a 242-bp DNA fragment (from nucleotide −3280 to −3038) was amplified by PCR with forward primer 5′-ATG GGA TCC AGA TGA CAA CCT TGT GGA ACC T-3′ and reverse primer 5′-GCG AAG CTT ATC AAA TCA TGT AGC CTT GGG G-3′. The DNA fragment was then cloned into the OFluc reporter vector upstream of a c-fos basic promoter and a luciferase reporter (30). The resulting vector was named OFluc-Rbm24-W. To generate the Rbm24 luciferase reporter carrying mutant p53-RE, the region flanking the p53-RE in the Rbm24 promoter was amplified by PCR, with forward primer containing mutated p53-RE (5′-GCA CAA GGC CTG TGA ATA TAA AAT TAT TCT CTG CC-3′) and the reverse primer used for amplifying wild-type p53-RE. The DNA fragment and OFluc-Rbm24-W were then digested with StuI and HindIII, and the smaller fragment in OFluc-Rbm24-W was replaced with the DNA fragment containing the mutated p53-RE through StuI and HindIII sites. The resulting vector was named OFluc-Rbm24-M. Wild-type and mutant p53-RE fragments were confirmed by DNA sequencing.
pcDNA3-p21 (ORF), a pcDNA3 vector that carries the human p21 coding region, was generated as described previously (31). To generate a pcDNA3 vector that carries the p21 coding region plus both 5′-UTR and 3′-UTR (pcDNA3-p21 (5′-UTR+ORF+3′-UTR)), the p21 cDNA fragment from pZL-WAF1 (32) was inserted into pcDNA3 through EcoRI. To generate pcDNA3 vector that carries the p21 coding region plus 5′-UTR or 3′-UTR, pcDNA3-p21 (ORF) and pcDNA3-p21 (5′-UTR+ORF+3′-UTR) were both digested with StuI. The smaller fragment from pcDNA3-p21 (ORF) was replaced with the fragment containing partial p21 ORF and 3′-UTR from pcDNA3-p21 (5′-UTR+ORF+3′-UTR) to generate pcDNA-p21 (ORF+3′-UTR) via StuI. Similarly, the smaller fragment from pcDNA-p21 (5′-UTR+ORF+3′-UTR) was replaced with the fragment containing partial p21 ORF from pcDNA3-p21 (ORF) to generate pcDNA-p21 (ORF+5′-UTR) via StuI. To generate a mutant p21 vector that lacks the entire ARE (pcDNA3-p21 (ΔARE)), two-step PCRs were performed. The first step was performed to amplify two separate fragments. Fragment 1 was amplified with forward primer, 5′-GCG GAA TTC GTT GTA TAT CAG GGC CGC GCT GAG-3′ and reverse primer, 5′-AGG ACT GCA GGC TTC CTG TGG GCG GAT TAG-3′. Fragment 2 was amplified with forward primer, 5′-CTG CAG TCC TGG AAG CAT CCC GTG TTC TCC TT-3′, and reverse primer, 5′-GCG GAA TTC AGG TCT GAG TGT CCA GGA AAG G-3′. The second step PCR was performed using a mixture of fragments 1 and 2 as a template with forward primer for fragment 1 and the reverse primer for fragment 2, and the resulting fragment lacking the ARE was cloned into pcDNA3 via EcoRI.
Cell Culture
MCF7, RKO, HCT116, and H1299 cells were cultured in DMEM supplemented with 10% fetal bovine serum as described previously (13). Primary mouse embryonic fibroblasts (MEFs) were cultured in DMEM supplemented with 10% fetal bovine serum, 1× nonessential amino acids, and 55 mm β-mercaptoethanol. HCT116 (p53−/−) cells are derivatives of HCT116 in which the p53 gene was somatically knocked out (33, 34). RKO-p53-KD is a derivative of RKO in which p53 was stably knocked down by RNA interference (RNAi) (13, 35). MCF7, HCT116, and HCT116 (p53−/−) cell lines, which inducibly express Rbm24, were generated as described previously (36).
Lentiviral Knockdown
293T cells were cultured in DMEM supplemented with 10% FBS. All recombinant lentiviruses were produced by transient transfection of 293T cells according to standard protocols (ExpressFectTM). Briefly, 293T packing cells were cotransfected with a lentiviral vector, pLKO.1-puro expressing shRNA of interest, along with packing plasmids, pCMV-VSVG (5 μg), pRSV-REV (5 μg), and pMDL-g/pRRE (5 μg). After 16 h the medium was changed, and recombinant lentiviral vectors were harvested twice 24 h and 48 h after transfection. The supernatant containing shRNA-expressing lentivirus was then filtered and concentrated by ultracentrifugation (25,000 rpm, 4 °C, 1.5 h). Cells were then transduced with the concentrated lentiviral particles in the presence of Polybrene. The medium was removed and replaced with fresh medium the following day, followed by puromycin selection (1 μg/ml) for 3 days.
Western Blot Analysis
Whole cell lysates were prepared using 1× SDS sample buffer, incubated at 95 °C for 6 min, and run on 10% acrylamide gel. The blots were transferred to a nitrocellulose membrane and probed with the appropriate antibodies, followed by ECL detection.
Reverse Transcription-PCR (RT-PCR)
A reverse transcription assay was performed as described previously (37). Briefly, RNA was purified using TRIzol reagent following the manufacturer's protocol (Invitrogen). Reverse transcription was performed using MMLV reverse transcriptase, and PCR was run using GoTaq DNA Polymerase (Promega). Primers used to amplify p53, p21, Rbm24, actin, and GAPDH transcripts are listed in Table 1.
TABLE 1.
Primers used for RT-PCR and ChIP assay
| Primer name | Sequence |
|---|---|
| Actin-RT-F | 5′-CTGAAGTACCCCATCGAGCACGGCA-3′ |
| Actin-RT-R | 5′-GGATAGCACAGCCTGGATAGCAACG-3′ |
| p53-RT-F | 5′-GACCGGCGCACAGAGGAAGAGAATC-3′ |
| p53-RT-R | 5′-GAGTTTTTTATGGCGGGAGGTAGAC-3′ |
| p21-RT-F | 5′-CCATGTGGACCTGTCACTGT-3′ |
| p21-RT-R | 5′-AAGATGTAGAGCGGGCCTTT-3′ |
| Rbm24-RT-F | 5′-AGCCTGCGCAAGTACTTCG-3′ |
| Rbm24-RT-R | 5′-CAGGCCCTTTCGGCAGCAG-3′ |
| pre-p21-F | 5′-GACACTCCATAATACCCCTC-3′ |
| pre-p21-R | 5′-CTGAGACTAAGGCAGAAGATG-3′ |
| pre-GAPDH-F | 5′-GGACTGGCTTTCCCATAATTT-3′ |
| pre-GAPDH-R | 5′-AAGGTCATCCCTGAGCTGAAC-3′ |
| Mdm2-ChIP-F | 5′-GGGAGTTCAGGGTAAAGGTCA-3′ |
| Mdm2-ChIP-R | 5′-CCTTTTACTGCAGTTTCG-3′ |
| Rbm24-RE-ChIP-F | 5′-TGGAGATGACAACCTTGTGG-3′ |
| Rbm24-RE-ChIP-R | 5′-ATGTAGCCTTGGGGCCATA-3′ |
| GAPDH-ChIP-F | 5′-AAAAGCGGGGAGAAAGTAGG-3′ |
| GAPDH-ChIP-R | 5′ AAGAAGATGCGGCTGACTGT 3′ |
RNA-immunoprecipitation
RNA-immunoprecipitation and RT-PCR were performed as described previously (13). Briefly, cells were uninduced or induced to express Rbm24 for 36 h. Cell extracts were prepared with lysis buffer (100 mm KCl, 5 mm MgCl2, 10 mm Hepes, 1 mm DTT, and 0.5% Nonidet P-40), and then incubated with 2 μg of anti-Rbm24 or isotype control IgG at 4 °C overnight. The RNA-protein immunocomplexes were brought down by protein A/G beads followed by RT-PCR analysis.
RNA Electrophoretic Mobility Shift Assay (RNA EMSA)
Recombinant Rbm24-GST and GST proteins were expressed in BL21 and purified by glutathione-Sepharose beads. The p21 3′-UTR was PCR-amplified using primers containing T7 promoter. p21 probes were made by in vitro transcription using T7 RNA polymerase and radiolabeled with [α-32P]UTP. RNA EMSA was performed as previous described (14). Briefly, 32P-labeled probes were incubated with recombinant protein in a binding buffer (10 mm Hepes-KOH, at pH 7.5, 90 mm potassium acetate, 1.5 mm magnesium acetate, 2.5 mm DTT, 40 units of RNase inhibitor (Ambion, Austin, TX)) at 30 °C for 30 min. The RNA-protein complexes were resolved on a 5% native PAGE gel, and radioactive signals were detected by autoradiography.
Luciferase Assay
A dual luciferase reporter assay was performed according to the manufacturer's instructions (Promega). Briefly, H1299 cells were plated at ∼1 × 105 cells/well in a 24-well plate and allowed to recover overnight. Cells were then cotransfected with 250 ng of OFluc-Rbm24 reporter, together with 250 ng of pcDNA3 or pcDNA3 vector expressing wild-type p53 or mutant p53 (R249S). As an internal control, 3 ng of pRL-CMV, a Renilla luciferase vector (Promega), was also cotransfected per well. Thirty-six hours after transfection, luciferase activity was measured with the dual luciferase kit and Turner Designs luminometer. The -fold increase in relative luciferase activity is a product of the luciferase activity induced by a wild-type or mutant p53 divided by that induced by empty pcDNA3 vector.
RESULTS
Identification of Rbm24 as a Novel Target Gene of p53
DNA damage stabilizes and activates tumor suppressor p53, which then induces p53 target genes. Our previous results showed that Rbm38, a target of the p53 family, serves as a mediator of p53 to regulate p21 post-transcriptionally (13). Upon searching Rbm38-related RNA-binding proteins, we found that Rbm24 contains a conserved RRM region (amino acids 5–93). Surprisingly, sequence alignment of RRMs between Rbm24 and Rbm38 showed that Rbm24 and Rbm38 share a high similarity in the RNA-binding domain, including two highly conserved submotifs, RNP1 and RNP2 (Fig. 1A). Thus, we speculate that Rbm24 and Rbm38 constitute a family of RNA-binding proteins, designated as RNPC. As a member of the RNPC family, we hypothesize that Rbm24 may be a novel target of p53 and regulates p21 expression.
To test this, semiquantitative and quantitative RT-PCR analysis was performed using HCT116 and RKO cell lines in which endogenous p53 is wild type, and HCT116 (p53−/−) and RKO-p53-KD cell lines in which endogenous p53 was knocked out and knocked down, respectively (13). We found that the levels of Rbm24 transcript were increased by 2.6-fold in HCT116 cells and 4.6-fold in RKO cells upon treatment with DNA-damaging agent doxorubicin (Fig. 1, B and C, Rbm24 panel). p21 expression was also found to be increased (Fig. 1, B and C, compare lanes 1 with 2). However, little or no induction of Rbm24 was detected in p53-KD RKO cells (Fig. 1C, compare lane 3 with 4). Interestingly, Rbm24 was slightly induced by 1.39-fold upon DNA damage in HCT116 (p53−/−) cells, suggesting that other mechanisms are potentially responsible for induction of Rbm24 independent of p53 (Fig. 1B, compare lane 3 with 4).
To further determine whether Rbm24 is a target of p53, we investigated whether Rbm24 can be induced by activated endogenous p53. To test this, tumor cells with endogenous wild-type p53 were treated with Nutlin-3, an inhibitor of Mdm2, which activates p53 protein without causing DNA damage (38). Indeed, we showed that upon treatment with Nutlin-3, the level of Rbm24 transcript was increased by 3.9-fold in MCF7 cells and 1.7-fold in RKO cells (Fig. 1D, Rbm24 panel). Similarly, the levels of p21 were also induced (Fig. 1D, p21 panel). Furthermore, upon knockdown of p53 in HCT116 cells transfected with siRNA against p53, the level of Rbm24 transcript was decreased (Fig. 1E).
Next, to determine whether up-regulation of Rbm24 transcript correlates with an increase in the level of Rbm24 protein, Western blot analysis was performed. We found that the level of Rbm24 protein was significantly increased in HCT116 cells upon treatment with doxorubicin (Fig. 1F, compare lane 1 with 2). Similarly, the level of Rbm24 protein was increased in RKO cells upon treatment with camptothecin and doxorubicin (Fig. 1G, compare lane 1 with 2 and 3). However, Rbm24 expression was not significantly increased in HCT116 (p53−/−) cells (Fig. 1F, compare lane 3 with 4) and RKO-p53-KD cells (Fig. 1G, compare lane 4 with 5 and 6). Moreover, we found that the level of Rbm24 protein was markedly increased in MCF7 cells upon treatment with various doses of Nutlin-3 (Fig. 1H, compare lane 1 with 2 and 3). We also found that upon knockdown of p53 in HCT116 cells, the level of Rbm24 protein was decreased (Fig. 1I). The expression of p21 protein was measured as a positive control and the level of actin measured as a loading control (Fig. 1, F–I).
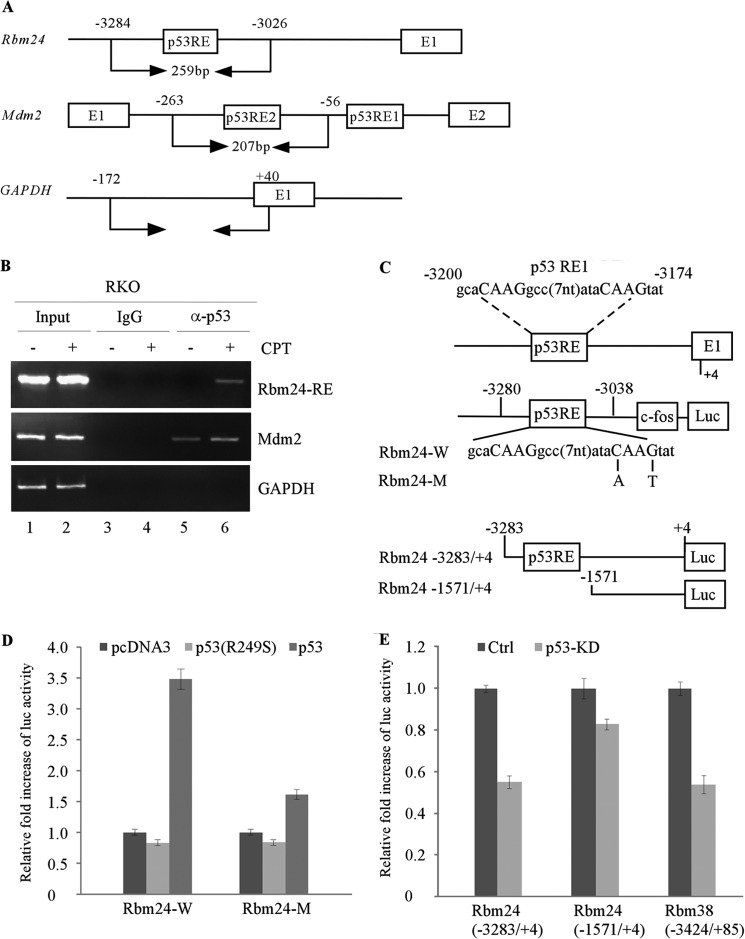
p53 activates its target genes expression via binding to specific DNA sequences in the promoter or intron. Upon searching for a p53-RE in the genomic locus of the Rbm24 gene, we found one potential binding site from nucleotide −3200 to −3174 in the Rbm24 promoter (Fig. 2C). To examine whether the p53-RE in the Rbm24 gene is recognized by p53, chromatin immunoprecipitation (ChIP) assay was performed with primers shown in Fig. 2A. RKO cells were untreated or treated with camptothecin to induce p53 expression, followed by cross-linking with formaldehyde and immunoprecipitation with anti-p53 antibody along with IgG as a control. We found that endogenous p53 bound to the p53-RE in the Rbm24 promoter, but not to the control GAPDH promoter (Fig. 2B). The binding of p53 to the Mdm2 promoter was measured as a positive control (Fig. 2B). Moreover, to examine whether the p53-RE in the Rbm24 promoter is responsive to p53, we generated two luciferase reporters carrying either wild-type or mutant p53-RE upstream of a c-fos minimal promoter and a luciferase reporter (Fig. 2C, upper panel). We found that the luciferase activity under the control of wild-type p53-RE (Rbm24-W), but not mutant p53-RE (Rbm24-M), was markedly increased by wild-type p53 (Fig. 2D). Mutant p53 (p53 R249S) had no effect on luciferase activity (Fig. 2D). To further investigate whether the endogenous promoter is responsive to p53, two DNA fragments from the Rbm24 promoter, in which the p53-RE is retained (−3283/+4) or deleted (−1571/+4), were cloned into pGL2-basic luciferase reporter, respectively (Fig. 2C, lower panel). We found that the luciferase activity for the full-length Rbm24 promoter (−3283/+4) and the full-length Rbm38 promoter (−3424/+85) promoter (13), but not the Rbm24 promoter without the p53-RE (−1571/+4), was markedly decreased upon knockdown of p53 by siRNA (Fig. 2E). Overall, these data suggest that Rbm24 is likely to be a direct target of p53.
FIGURE 2.
Identification of potential p53-REs in the Rbm24 gene. A, schematic presents the Rbm24, Mdm2, and GAPDH promoters along with the locations of the potential p53-RE and the primers used for ChIP assay. B, ChIP assay was performed with RKO cells untreated (−) or treated (+) with 200 nm camptothecin (CPT) for 12 h. The p53-DNA complexes were captured with anti-p53 or a control IgG, and the binding of p53 to the p53-RE in the Rbm24 or Mdm2 promoter was quantified by PCR. C, upper panel, schematic presents the Rbm24 locus and the OFluc luciferase constructs carrying wild-type (Rbm24-W) or mutant p53-RE (Rbm24-M) from the Rbm24 promoter. Lower panel, schematic presents luciferase constructs carrying the p53-RE (−3283/+4) or without the p53-RE (−1571/+4). D and E, the p53-RE in the Rbm24 is responsive to p53 but not mutant p53 (R249S). D, the luciferase activity was measured from the luciferase construct Rbm24-W or Rbm24-M, which was cotransfected in H1299 cells with an empty vector or pcDNA3 expressing wild-type p53 or mutant p53 (R249S) for 24 h. E, the luciferase activity was measured from the luciferase construct Rbm24 (−3283/+4) or Rbm24 (−1571/+4), which was transfected in HCT116 cells after p53 was knocked down by siRNA for 48 h. The luciferase activity of Rbm38 (−3424/+85) was measured as a positive control. The experiment was performed in triplicate. Error bars indicate S.D.
p21 Expression Is Regulated by Rbm24
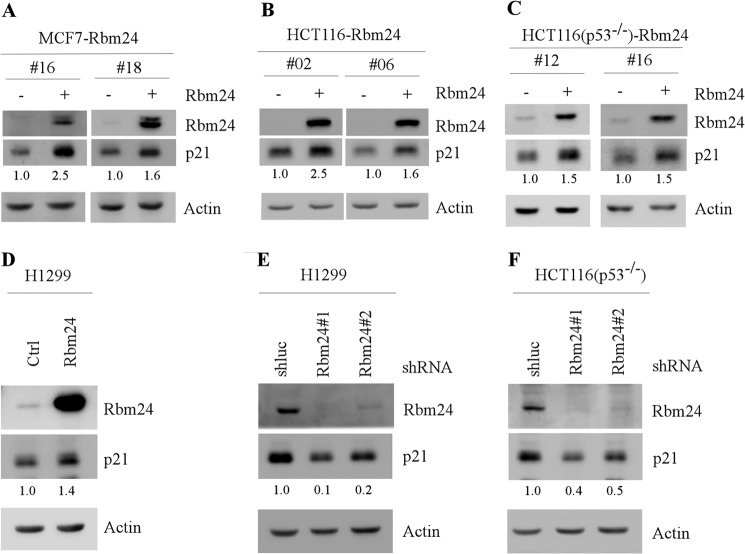
p21 plays a major role in the cell cycle control. Thus, the expression of p21 needs to be tightly controlled. Rbm38 is known to regulate p21 mRNA stability via binding to ARE in the p21 3′-UTR (13, 14). Because Rbm24 and Rbm38 share a high sequence similarity in the RNA-binding domain, we examined whether Rbm24 can regulate p21 expression post-transcriptionally. To test this, we generated MCF7, HCT116, and HCT116 (p53−/−) cell lines that can inducibly express Rbm24 under the control of the tetracycline-regulated promoter. We found that upon induction of Rbm24, p21 protein levels were markedly increased in MCF7 and HCT116 cell lines (Fig. 3, A and B). To determine whether Rbm24 regulates p21 expression in the absence of p53, HCT116 (p53−/−) and p53-null H1299 cells were used. We showed that the level of p21 was up-regulated upon induction of Rbm24 in HCT116 (p53−/−) cell (Fig. 3C). Consistent with this, p21 expression was also increased upon transient expression of Rbm24 for 48 h in H1299 cells (Fig. 3D).
FIGURE 3.
Ectopic expression of Rbm24 increases whereas knockdown of Rbm24 decreases the level of p21 protein. A–C, MCF7, HCT116, and HCT116 (p53−/−) cells were uninduced (−) or induced (+) with 0.5 μg/ml tetracycline to express Rbm24 for 48 h. The levels of Rbm24, p21, and actin proteins were then measured by Western blot analysis. D, H1299 cells were transiently transfected with a control vector or a vector expressing Rbm24 for 48 h. The protein levels of Rbm24, p21, and actin were determined by Western blot analysis. E and F, H1299 (E) and HCT116 (p53−/−) (F) cells were transduced with a lentivirus expressing a control luciferase shRNA (shluc) or Rbm24 shRNA (shRbm24) and selected by puromycin (1 μg/ml) for 3 days prior to Western blot analysis.
Next, to determine whether endogenous Rbm24 regulates p21 expression, the levels of p21 protein were measured in HCT116 (p53−/−) and H1299 cells in which Rbm24 expression was knocked down with shRNA targeting Rbm24 via lentiviral transduction. shRNA targeting luciferase was used as a negative control. We found that upon knockdown of Rbm24, the levels of p21 protein were reduced in HCT116 (p53−/−) and H1299 cells (Fig. 3, E and F).
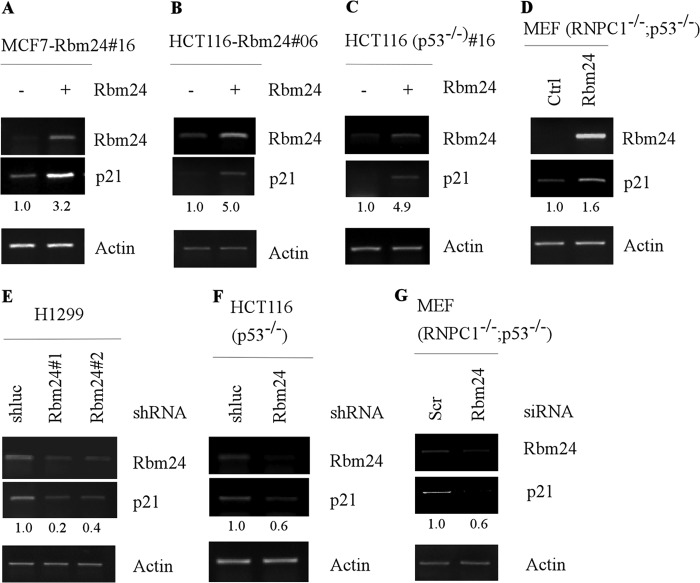
As an RNA-binding protein, Rbm24 may directly bind to p21 mRNA and consequently increase the level of p21 transcript. Thus, RT-PCR was performed and showed that the levels of p21 transcript were increased by Rbm24 in MCF7, HCT116, and HCT116 (p53−/−) cells (Fig. 4, A–C). We also found that knockdown of Rbm24 decreased the level of p21 transcript in HCT116 (p53−/−) and H1299 cells (Fig. 4, E and F). To further verify this, the level of p21 transcript was determined in MEFs isolated from Rbm38−/−; p53−/−embryos. We showed that the level of p21 transcript was increased by ectopic expression of Rbm24, but decreased by knockdown of Rbm24 in MEF (Rbm38−/−; p53−/−) cells (Fig. 4, D and G).
FIGURE 4.
Ectopic expression of Rbm24 increases whereas knockdown of Rbm24 decreases the level of p21 mRNA. A–C, MCF7 (A), HCT116 (B), and HCT116 (p53−/−) (C) cells were uninduced (−) or induced (+) with 0.5 μg/ml tetracycline to express Rbm24 for 48 h. The levels of Rbm24, p21, and actin transcripts were measured by RT-PCR analysis. D, MEFs (p53−/−;RNPC1−/−) were transiently transfected with a control vector or a vector expressing Rbm24 for 48 h. RT-PCR analysis was used to determine the mRNA levels of Rbm24, p21, and actin. E and F, H1299 (E) and HCT116 (p53−/−) (F) cells were transduced with a lentivirus expressing a control luciferase shRNA (shluc) or Rbm24 shRNA (shRbm24) and selected by puromycin (1 μg/ml) for 3 days. The levels of Rbm24, p21, and actin transcripts were measured by RT-PCR analysis. G, MEFs (p53−/−;RNPC1−/−) were transiently transfected with a control or Rbm24 siRNA for 72 h. The levels of Rbm24, p21, and actin transcripts were determined by RT-PCR analysis.
p21 mRNA Stability Is Increased by Rbm24
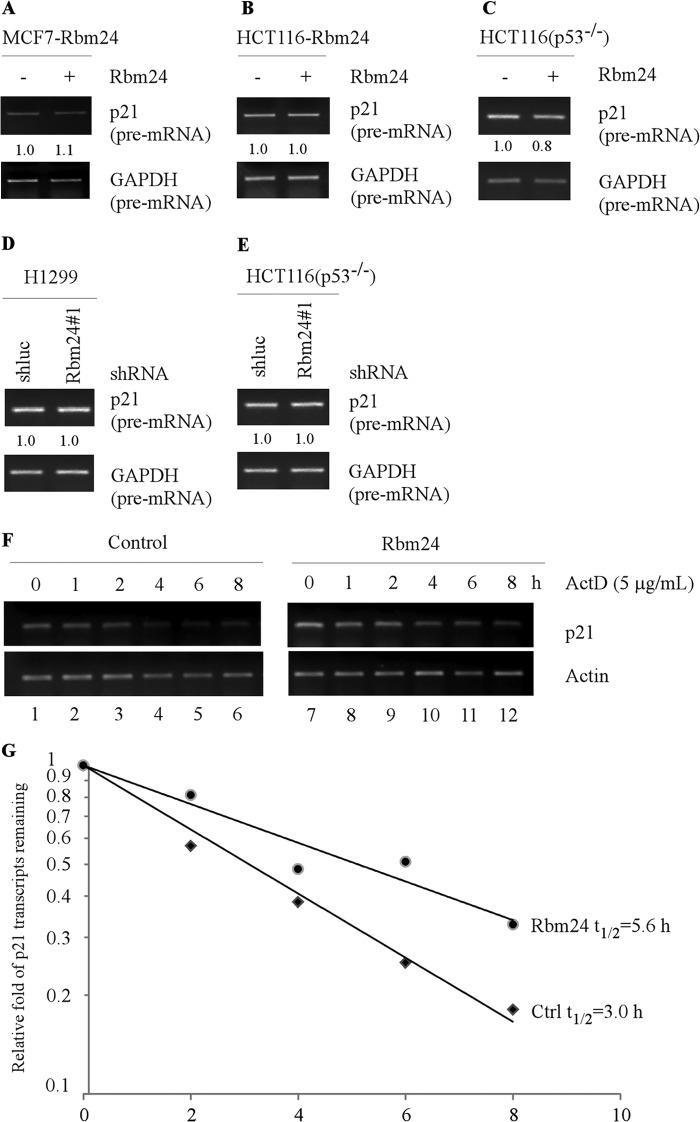
RNA-binding proteins are known to regulate gene expression at post-transcriptional levels including mRNA stability (39). To explore how Rbm24 regulates p21 expression, the level of precursor p21 mRNA (pre-mRNA) was measured and found not to be significantly altered by ectopic expression of Rbm24 in MCF7, HCT116, and HCT116 (p53−/−) cells (Fig. 5, A–C). Similarly, knockdown of Rbm24 in HCT116 (p53−/−) and H1299 cells had no effect on the level of p21 pre-mRNA (Fig. 5, D and E). These data suggest that Rbm24 regulates p21 expression mainly through a post-transcriptional mechanism. To test this, we examined whether the mRNA stability of p21 transcript is altered by Rbm24 by measuring the half-life of p21 transcript. Specifically, HCT116 (p53−/−) cells were uninduced or induced to express Rbm24 for 48 h, followed by treatment with a transcription inhibitor actinomycin-D (5 μg/ml) to inhibit de novo RNA synthesis. p21 mRNA levels were examined by RT-PCR (Fig. 5F), followed by quantification by density analysis. We found that overexpression of Rbm24 in HCT116 (p53−/−) cells increased the half-life of p21 from ∼3.0 h in control cells to ∼5.6 h in Rbm24-producing cells (Fig. 5, F and G). Together, these data suggest that Rbm24 is able to stabilize the p21 transcript.
FIGURE 5.
Rbm24 stabilizes p21 transcript. A–C, ectopic expression of Rbm24 has no obvious effect on the level of p21 pre-mRNA. MCF7 (A), HCT116 (B), and HCT116 (p53−/−) (C) cells were uninduced (−) or induced (+) with 0.5 μg/ml tetracycline to express Rbm24 for 48 h. Total RNAs were isolated and then subjected to RT-PCR analysis to examine the level of p21 pre-mRNA. GAPDH pre-mRNA was measured as a control. D and E, the level of p21 pre-mRNA remains relatively even upon knockdown of Rbm24. H1299 (D) and HCT116 (p53−/−) (E) cells were transfected with a lentivirus containing either control luciferase shRNA (shluc) or Rbm24 shRNA (shRbm24) and then selected by puromycin (1 μg/ml) for 3 days prior to RT-PCR analysis to determine the level of p21 pre-mRNA. GAPDH pre-mRNA was measured as a control. F, HCT116 (p53−/−) cells were treated with actinomycin-D (5 μg/ml) over an 8-h period at 2-h intervals. Total RNAs were purified, and the levels of p21 and actin transcripts were analyzed using RT-PCR. G, the relative levels of p21 transcript were normalized with the levels of actin transcript and plotted along with time to calculate the relative half-life of p21 mRNA in the presence or absence of Rbm24. The x axis represents the time after addition of actinomycin-D (5 μg/ml), and the y axis represents the relative levels of remaining p21 transcript in the cells.
An AU-rich Element in the p21 3′-UTR Is Recognized and Responsive to Rbm24
To further investigate the underlying mechanism by which Rbm24 stabilizes p21 transcript, we examined whether Rbm24 can physically interacts with p21 transcript. To do this, an RNA immunoprecipitation assay followed by RT-PCR was performed using extracts from HCT116 cells that were induced to express Rbm24. We found that p21 mRNA was detectable in Rbm24, but not control IgG immunoprecipitates (Fig. 6A). As a control, actin transcripts were not found to interact with Rbm24 or IgG (Fig. 6A).
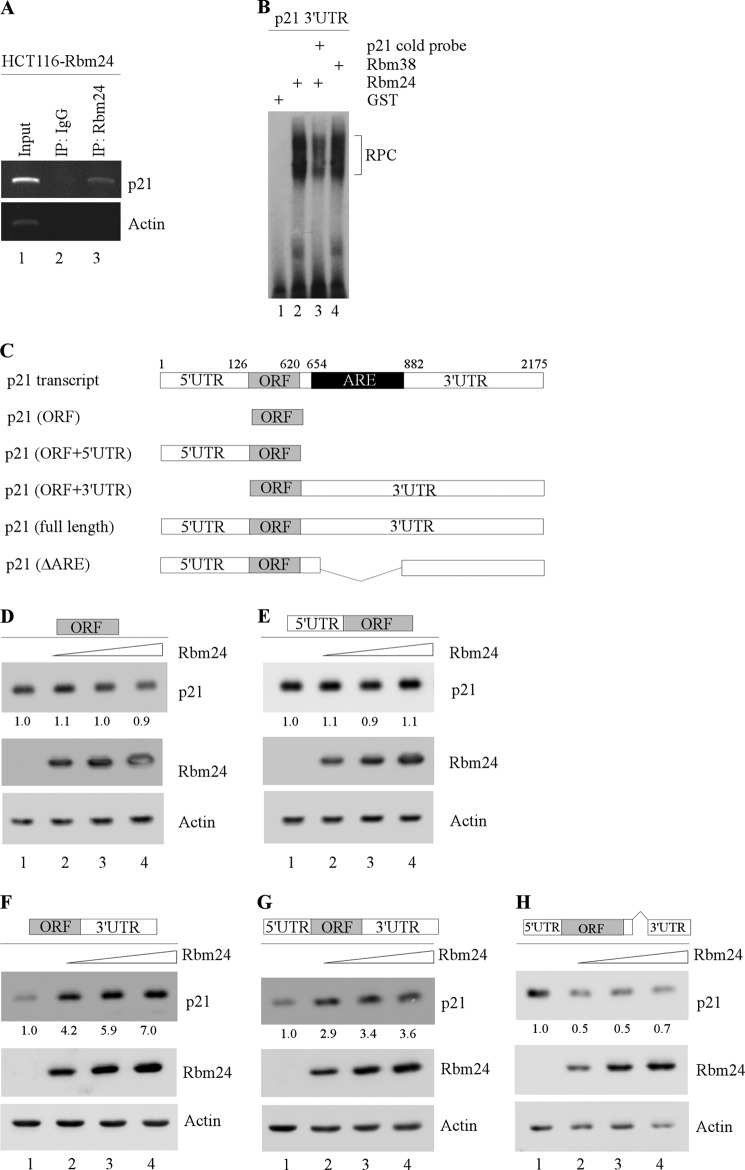
FIGURE 6.
The p21 3′-UTR is bound by and responsive to Rbm24. A, Rbm24-expressing HCT116 cell extracts were immunoprecipitated (IP) with a control IgG or Rbm24 antibody followed by RT-PCR to determine the transcript levels of p21 and actin within Rbm24 or IgG immunocomplexes. B, RNA EMSA was performed by incubating 32P-labeled p21 3′-UTR with GST alone, GST-fused Rbm24, or GST-fused Rbm38 (as a positive control). Unlabeled p21 was used as cold probe for competition. The bracket labeled as RPC represents an RNA-protein complex. C, schematic diagrams of the p21 transcript and p21-expresing vectors, including p21 coding region (ORF) alone or in combination with the 5′-UTR, the 3′-UTR, or both. p21 (ΔARE) represents mutant p21-expressing vector that lacks the entire AU-/U-rich elements. D–H, the ARE in the 3′-UTR is required for Rbm24 to induce p21 expression. Various amounts of Rbm24 expression vector were transfected into H1299 cells along with a fixed amount of p21 expression vector that contains the ORF alone (D), ORF plus 5′-UTR (E), ORF plus 3′-UTR (F), ORF plus both 5′ and 3′-UTRs (G), or p21 (ΔARE) (H). The levels of p21, Rbm24, and actin proteins were analyzed by Western blotting.
Next, to verify that the p21 3′-UTR is bound by Rbm24, an RNA EMSA was performed by using a radiolabeled probes derived from the p21 3′-UTR. We found that recombinant GST-fused Rbm24 but not GST alone was able to form a complex with the probe (Fig. 6B, compare lane 1 with 2). The specificity of the binding of Rbm24 to the p21 3′-UTR was further confirmed by a competition assay in which unlabeled p21 3′-UTR cold probe was added to the reaction mixture. Indeed, the RNA-protein complex was significantly decreased in the presence of p21 cold probe (Fig. 6B, compare lane 1 with 3). The complex between Rbm38 and the p21 3′-UTR was also measured and used as a positive control (Fig. 6B, compare lane 1 with 4).
To determine whether the 5′- and 3′-UTRs are necessary for Rbm24 to induce p21 expression, we generated five expression vectors that contain the p21 coding region (ORF) alone or in combination with the 5′-UTR, the 3′-UTR, or both. We showed that the level of p21 was drastically increased by Rbm24 in a dose-dependent manner as long as the p21 expression vector carried the 3′-UTR (Fig. 6, F and G). In contrast, Rbm24 had no effect on expression of p21 transcripts that carried 5′-UTR or only contained the coding region (Fig. 6, D and E). Finally, to verify that the AU-/U-rich elements in the p21 3′-UTR are required for Rbm24 to induce p21 expression, a p21 expression vector that lacks the entire ARE (nucleotides 654 to 882) (14) was generated (Fig. 6C). We found that p21 expression was not increased, but instead decreased by Rbm24 (Fig. 6H). These data suggest that the AU-/U-rich element in the p21 3′-UTR is necessary for Rbm24 induction of p21.
The RNA-binding Domain in Rbm24 Is Required for Inducing p21 Expression
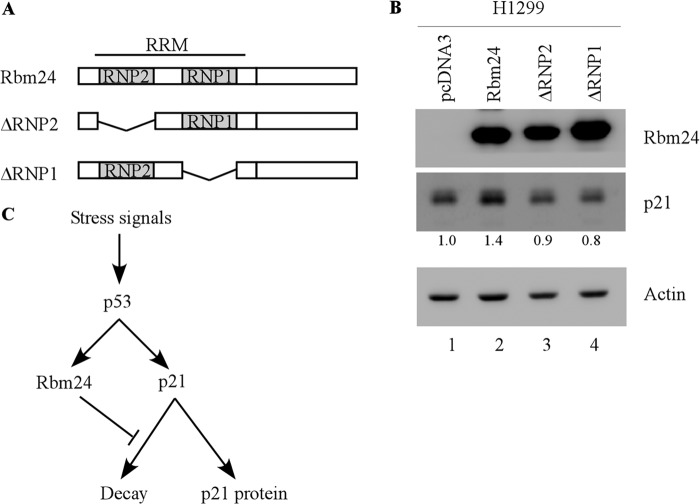
The RNA-binding domain in Rbm24 is composed of two putative subdomains, RNP1 and RNP2 (Fig. 7A). To determine which subdomain of RNPs is required for regulating p21 expression, two mutant forms of Rbm24, which lack either RNP1 or RNP2 (ΔRNP1 or ΔRNP2), were generated (Fig. 7A). We found that both ΔRNP1 and ΔRNP2 were unable to increase p21 expression compared with wild-type Rbm24 in H1299 cells (Fig. 7B).
FIGURE 7.
The RNA-binding domain is required for Rbm24 to induce p21 expression. A, schematic represents wild-type Rbm24, ΔRNP2, and ΔRNP1. B, the RNA-binding domain is required for Rbm24 to increase p21 expression. H1299 cells were transiently transfected with a control vector or a vector expressing Rbm24, ΔRNP1, or ΔRNP2 for 48 h. The levels of Rbm24, p21, and actin proteins were determined by Western blot analysis. C, model shows the role of Rbm24 in the p53 pathway.
DISCUSSION
In this study, we showed that Rbm24, an RNA-binding protein that shares a highly similar RRM with Rbm38, is a novel target of p53 and regulates p21 expression via mRNA stability. Specifically, we showed that Rbm24 can be induced upon DNA damage in a p53-dependent manner. We also showed that p53 binds to the promoter of the Rbm24 gene and transcriptionally regulates Rbm24 through a p53-responsive element found in the Rbm24 promoter. Moreover, we found that overexpression of Rbm24 increases, whereas knockdown of Rbm24 decreases, p21 expression independent of p53. Importantly, we provided evidence that Rbm24 is required for the maintenance of the stability of p21 transcript through binding to the ARE in the p21 3′-UTR. Finally, we found that the RRM of Rbm24 is required for stabilizing p21 transcript. Together, we hypothesize that in response to stress signals, p53 is activated and induces its target gene Rbm24, which in turn regulates p21 mRNA stability, leading to enhanced cell cycle arrest (Fig. 7C).
The 3′-UTR of p21 transcript can be directly recognized by several RNA-binding proteins, including Rbm38, HuR, and HuD (14, 25, 26). Previously, we showed that Rbm38 and HuR can bind the ARE in the p21 3′-UTR, and Rbm38 can modulate and cooperate with HuR to regulate p21 mRNA stability (14). However, Rbm38 may compete with HuD in binding to the ARE in the p21 3′-UTR (14). The similarity in the regulation of p21 by Rbm24 versus Rbm38 is probably due to their similarity in the RNA-binding domain and the surrounding sequence (Fig. 1A). Moreover, RRM is also known to be involved in protein-protein interaction (40). Therefore, it is possible that Rbm24 may cooperate or compete with other AU-/CU-rich binding and RRM-containing RBPs, including Rbm38 and HuD, to bind to their corresponding sequences and consequently regulate p21 mRNA stability. In addition, p21 is known to be regulated by PCBP1, PCBP2, and PCBP4 (28, 29). Thus, further studies are warranted to examine whether Rbm24 cooperates or antagonizes with the PCBP proteins to regulate p21. Interestingly, our study implies that other Rbm38-regulated targets, such as p53, Mdm2, p63, and p73, may also be regulated by Rbm24. However, several regions, especially the C terminus in Rbm24, are quite different from that in Rbm38 (Fig. 1A), suggesting that their biological functions and the mechanism by which these two RBPs regulate the p53 pathway may be different.
Rbm38 is known to be overexpressed in various human cancers, such as colon carcinoma (30), lymphoma (29), and esophageal cancer (31), but down-regulated due to hypermethylation in the promoter region in some breast tumors (26). Both Rbm38 and Rbm24 have been shown to play a role in muscle development (15), and Rbm24 is also found to play a role in myogenic differentiation, sarcomere assembly, and heart contractility (15–17). Here, we have shown that Rbm24 can directly bind to p21 mRNA and regulate its stability, which is consistent with its role in myogenesis (15). However, the Rbm24 gene has not been found to be altered in cancer. Due to loss of p53 or other factors required for p21 expression, tumor cells may undergo uncontrolled proliferation and eventually form malignant tumors (41). Because proper cell cycle control in tumors is an active approach to suppress cell transformation and tumorigenesis (42), the regulation of p21 expression by Rbm24 may be explored to restore normal cell cycle control in p53-deficient tumor cells.
Acknowledgments
We thank Tiffany Yin, Seong-Jun Cho, and Wensheng Yan for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants CA076069 and CA121137.
- RBP
- RNA-binding protein
- ARE
- AU-rich element
- MEF
- mouse embryonic fibroblast
- PCBP
- poly(C)-binding protein
- p53-RE
- p53 response element
- RRM
- RNA recognition motif.
REFERENCES
- 1. Vogelstein B., Lane D., Levine A. J. (2000) Surfing the p53 network. Nature 408, 307–310 [DOI] [PubMed] [Google Scholar]
- 2. el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. (1993) WAF1, a potential mediator of p53 tumor suppression. Cell 75, 817–825 [DOI] [PubMed] [Google Scholar]
- 3. Jeffers J. R., Parganas E., Lee Y., Yang C., Wang J., Brennan J., MacLean K. H., Han J., Chittenden T., Ihle J. N., McKinnon P. J., Cleveland J. L., Zambetti G. P. (2003) Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 4, 321–328 [DOI] [PubMed] [Google Scholar]
- 4. Qian Y., Zhang J., Yan B., Chen X. (2008) DEC1, a basic helix-loop-helix transcription factor and a novel target gene of the p53 family, mediates p53-dependent premature senescence. J. Biol. Chem. 283, 2896–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. el-Deiry W. S., Harper J. W., O'Connor P. M., Velculescu V. E., Canman C. E., Jackman J., Pietenpol J. A., Burrell M., Hill D. E., Wang Y. (1994) WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 54, 1169–1174 [PubMed] [Google Scholar]
- 6. Kiledjian M., Wang X., Liebhaber S. A. (1995) Identification of two KH domain proteins in the α-globin mRNP stability complex. EMBO J. 14, 4357–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Collier B., Goobar-Larsson L., Sokolowski M., Schwartz S. (1998) J. Biol. Chem. 273, 22648–22656 [DOI] [PubMed] [Google Scholar]
- 8. Krecic A. M., Swanson M. S. (1999) hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 11, 363–371 [DOI] [PubMed] [Google Scholar]
- 9. Wurth L. (2012) Versatility of RNA-binding proteins in cancer. Comp. Funct. Genomics 2012, 178525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carvalho B., Postma C., Mongera S., Hopmans E., Diskin S., van de Wiel M. A., van Criekinge W., Thas O., Matthäi A., Cuesta M. A., Terhaar Sive Droste J. S., Craanen M., Schröck E., Ylstra B., Meijer G. A. (2009) Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression. Gut 58, 79–89 [DOI] [PubMed] [Google Scholar]
- 11. Hötte G. J., Linam-Lennon N., Reynolds J. V., Maher S. G. (2012) Radiation sensitivity of esophageal adenocarcinoma: the contribution of the RNA-binding protein RNPC1 and p21-mediated cell cycle arrest to radioresistance. Radiat. Res. 177, 272–279 [DOI] [PubMed] [Google Scholar]
- 12. Zhang J., Cho S. J., Shu L., Yan W., Guerrero T., Kent M., Skorupski K., Chen H., Chen X. (2011) Translational repression of p53 by RNPC1, a p53 target overexpressed in lymphomas. Genes Dev. 25, 1528–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shu L., Yan W., Chen X. (2006) RNPC1, an RNA-binding protein and a target of the p53 family, is required for maintaining the stability of the basal and stress-induced p21 transcript. Genes Dev. 20, 2961–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cho S. J., Zhang J., Chen X. (2010) RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability. Nucleic Acids Res. 38, 2256–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miyamoto S., Hidaka K., Jin D., Morisaki T. (2009) RNA-binding proteins Rbm38 and Rbm24 regulate myogenic differentiation via p21-dependent and -independent regulatory pathways. Genes Cells 14, 1241–1252 [DOI] [PubMed] [Google Scholar]
- 16. Maragh S., Miller R. A., Bessling S. L., McGaughey D. M., Wessels M. W., de Graaf B., Stone E. A., Bertoli-Avella A. M., Gearhart J. D., Fisher S., McCallion A. S. (2011) Identification of RNA binding motif proteins essential for cardiovascular development. BMC Dev. Biol. 11, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Poon K. L., Tan K. T., Wei Y. Y., Ng C. P., Colman A., Korzh V., Xu X. Q. (2012) RNA-binding protein RBM24 is required for sarcomere assembly and heart contractility. Cardiovasc. Res. 94, 418–427 [DOI] [PubMed] [Google Scholar]
- 18. Erhardt J. A., Pittman R. N. (1998) Ectopic p21WAF1 expression induces differentiation-specific cell cycle changes in PC12 cells characteristic of nerve growth factor treatment. J. Biol. Chem. 273, 23517–23523 [DOI] [PubMed] [Google Scholar]
- 19. Dulić V., Drullinger L. F., Lees E., Reed S. I., Stein G. H. (1993) Altered regulation of G1 cyclins in senescent human diploid fibroblasts: accumulation of inactive cyclin E-Cdk2 and cyclin D1-Cdk2 complexes. Proc. Natl. Acad. Sci. U.S.A. 90, 11034–11038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Y., Blandino G., Givol D. (1999) Induced p21waf expression in H1299 cell line promotes cell senescence and protects against cytotoxic effect of radiation and doxorubicin. Oncogene 18, 2643–2649 [DOI] [PubMed] [Google Scholar]
- 21. Jung Y. S., Qian Y., Chen X. (2010) Examination of the expanding pathways for the regulation of p21 expression and activity. Cell. Signal. 22, 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaghad M., Bonnet H., Yang A., Creancier L., Biscan J. C., Valent A., Minty A., Chalon P., Lelias J. M., Dumont X., Ferrara P., McKeon F., Caput D. (1997) Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 90, 809–819 [DOI] [PubMed] [Google Scholar]
- 23. Yang A., Kaghad M., Wang Y., Gillett E., Fleming M. D., Dötsch V., Andrews N. C., Caput D., McKeon F. (1998) p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2, 305–316 [DOI] [PubMed] [Google Scholar]
- 24. Zhu J., Jiang J., Zhou W., Chen X. (1998) The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer Res. 58, 5061–5065 [PubMed] [Google Scholar]
- 25. Joseph B., Orlian M., Furneaux H. (1998) p21waf1 mRNA contains a conserved element in its 3′-untranslated region that is bound by the Elav-like mRNA-stabilizing proteins. J. Biol. Chem. 273, 20511–20516 [DOI] [PubMed] [Google Scholar]
- 26. Wang W., Furneaux H., Cheng H., Caldwell M. C., Hutter D., Liu Y., Holbrook N., Gorospe M. (2000) HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 20, 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang X., Wang W., Fan J., Lal A., Yang D., Cheng H., Gorospe M. (2004) Prostaglandin A2-mediated stabilization of p21 mRNA through an ERK-dependent pathway requiring the RNA-binding protein HuR. J. Biol. Chem. 279, 49298–49306 [DOI] [PubMed] [Google Scholar]
- 28. Scoumanne A., Cho S. J., Zhang J., Chen X. (2011) The cyclin-dependent kinase inhibitor p21 is regulated by RNA-binding protein PCBP4 via mRNA stability. Nucleic Acids Res. 39, 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Waggoner S. A., Johannes G. J., Liebhaber S. A. (2009) Depletion of the poly(C)-binding proteins αCP1 and αCP2 from K562 cells leads to p53-independent induction of cyclin-dependent kinase inhibitor (CDKN1A) and G1 arrest. J. Biol. Chem. 284, 9039–9049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johansen F. E., Prywes R. (1994) Two pathways for serum regulation of the c-fos serum response element require specific sequence elements and a minimal domain of serum response factor. Mol. Cell. Biol. 14, 5920–5928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen X., Zhang J., Zhang M., Liu S., Yan W., Jung J. (2012) Serine 123 phosphorylation modulates p21 protein stability and activity by suppressing ubiquitin-independent proteasomal degradation. J. Biol. Chem. 287, 34410–34418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen X., Bargonetti J., Prives C. (1995) p53, through p21WAF1/CIP1, induces cyclin D1 synthesis. Cancer Res. 55, 4257–4263 [PubMed] [Google Scholar]
- 33. Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J. P., Sedivy J. M., Kinzler K. W., Vogelstein B. (1998) Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282, 1497–1501 [DOI] [PubMed] [Google Scholar]
- 34. Waldman T., Lengauer C., Kinzler K. W., Vogelstein B. (1996) Uncoupling of S phase and mitosis induced by anticancer agents in cells lacking p21. Nature 381, 713–716 [DOI] [PubMed] [Google Scholar]
- 35. Liu G., Chen X. (2006) DNA polymerase η, the product of the xeroderma pigmentosum variant gene and a target of p53, modulates the DNA damage checkpoint and p53 activation. Mol. Cell. Biol. 26, 1398–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dohn M., Nozell S., Willis A., Chen X. (2003) Tumor suppressor gene-inducible cell lines. Methods Mol. Biol. 223, 221–235 [DOI] [PubMed] [Google Scholar]
- 37. Xu E., Zhang J., Chen X. (2013) MDM2 expression is repressed by the RNA-binding protein RNPC1 via mRNA stability. Oncogene 32, 2169–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., Fotouhi N., Liu E. A. (2004) In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 [DOI] [PubMed] [Google Scholar]
- 39. Zhang J., Chen X. (2008) Posttranscriptional regulation of p53 and its targets by RNA-binding proteins. Curr. Mol. Med. 8, 845–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cléry A., Blatter M., Allain F. H. (2008) RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 18, 290–298 [DOI] [PubMed] [Google Scholar]
- 41. Collins K., Jacks T., Pavletich N. P. (1997) The cell cycle and cancer. Proc. Natl. Acad. Sci. U.S.A. 94, 2776–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vogelstein B., Kinzler K. W. (2004) Cancer genes and the pathways they control. Nat. Med. 10, 789–799 [DOI] [PubMed] [Google Scholar]