Background: It is unknown whether SUMOylation regulates myogenesis in skeletal muscle.
Results: SENP2 de-SUMOylates MEF2A, promoting myostatin expression and suppressing myogenesis in skeletal muscle.
Conclusion: SENP2 plays a critical role in the regulation of myostatin-induced inhibition of myogenesis.
Significance: SENP2 is a potential therapeutic target in skeletal muscle regeneration.
Keywords: Cell Signaling, Gene Transcription, Myogenesis, SUMOylation, Transforming Growth Factor β (TGFβ)
Abstract
Sentrin/small ubiquitin-like modifier (SUMO)-specific protease 2 (SENP2) has broad de-SUMOylation activities in vitro, which is essential for embryonic heart development. Here, we show that myostatin, a key factor in skeletal muscle development, is markedly reduced in Senp2−/− mouse embryonic fibroblast cells and embryos. SENP2 regulates the transcription of myostatin mainly through de-SUMOylation of MEF2A. Silencing SENP2 can reduce myostatin expression and, therefore, promote myogenesis of skeletal muscle. These results reveal the important role of SENP2 in the regulation of myostatin expression and myogenesis.
Introduction
The small ubiquitin-like modifier (SUMO)3 covalently modifies a large number of cellular proteins and regulates the localization, function, and protein-protein interaction of proteins (1–3). The transcription factors are the most popular targets for SUMO conjugation, which suggests an important role of SUMOylation in the regulation of gene expression (4, 5). SUMOylation is catalyzed by SUMO-specific E1s, E2s, and E3s, and is reversed by a family of Sentrin/SUMO-specific proteases (SENPs) (6, 7). Studies have shown that SENPs are important determinants of the SUMO modification status in cells (8–10). Although the biochemical properties of SENPs have been well documented, their targets with involved biological processes are largely unknown.
There are six mammal SENPs with different subcellular locations and substrate specificities (3, 9, 10). SENP2 was originally reported to be associated with the nuclear envelope through the binding to Nup153 (11, 12). We have shown previously that SENP2 contains both nuclear import and export signals that can shuttle between the nucleus and cytoplasm (13). SENP2 is the key regulator of Pc2/CBX4 function through the regulation of its SUMOylation status, which is critical for embryonic heart development (14).
Myostatin (growth and differentiation factor 8, GDF8) is a member of the TGF-β superfamily of secreted growth/differentiation factors (15–17). Myostatin is primarily expressed by skeletal muscle and acts in an autocrine manner to inhibit myoblast proliferation, differentiation, and protein synthesis (18, 19). Disruption of the myostatin gene in mice causes a large and widespread increase in skeletal muscle mass because of cell hyperplasia and hypertrophy (20–22). An inactivating mutation of the myostatin gene in a child has been reported to be hypermuscular (23). However, the overexpression or lack of active myostatin does not affect cardiac systolic function, although it reduces cardiac mass and cardiomyocyte proliferation (24).
Myostatin promoter activity is upregulated by differentiation. Both the myogenic regulatory factor and the myocyte enhancer factor 2 family of muscle transcription factors increase myostatin promoter activity (25, 26). Myostatin transcription is also upregulated by dexamethasone treatment (27). However, it is not currently known whether SUMOylation regulates myostatin expression and myogenesis. In this study, we found that the deletion of the Senp2 gene in mice diminished myostatin expression. We further confirmed that MEF2A is a target for SENP2 de-SUMOylation activity and that SENP2 promotes myostatin expression and inhibits myogenesis through MEF2A. These results reveal the critical role of SENP2 in muscle development through the MEF2A-myostatin pathway.
EXPERIMENTAL PROCEDURES
Cell Culture
The generation of MEF cells has been described previously (14). The C2C12 cell line was purchased from the ATCC. Satellite stem cells were isolated from the hind limb muscles of 6-week-old C57BL/6 mice. All cell lines were cultured in DMEM (Invitrogen) supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin.
Plasmids and Antibodies
Plasmids HA-SUMO-1 and FLAG-SENP2 and the FLAG-SENP2 mutation have been described previously (28, 29). The MEF2A, MEF2B, MEF2C, and MEF2D plasmids were gifts from Dr. Eric Olson (Southwestern Medical Center). siRNA against SENP2, myostatin, MEF2A, MEF2B, MEF2C, MEF2D, and nonspecific siRNA (NS-siRNA) were purchased from Sigma. Probes for SENP2 and myostatin were purchased from Applied Biosystems. Anti-MEF2A, anti-myosin heavy chain, and anti-PAX7 antibodies were purchased from Abcam. Anti-FLAG and anti-HA antibodies were purchased from Sigma. Anti-SUMO-1 antibody was from Zymed Laboratories Inc.
Real-time Quantitative PCR
Total RNA was isolated by an RNeasy kit (Qiagen) and treated with DNase (Promega). Complementary DNA was synthesized using the cDNA synthesis kit (Clontech) according to the instructions of the manufacturer. Fluorescence real-time PCR was performed with the TaqMan probe using the ABI Prism 7300 system (PerkinElmer Life Sciences). PCR was done in triplicate, and standard deviations representing experimental errors were calculated. All data were analyzed using ABI Prism SDS 2.0 software (PerkinElmer Life Sciences).
Mystatin Promoter-Luciferase Plasmids
A myostatin bacterial artificial chromosome (BAC) clone (BACPAC Resources Center) was used to amplify the myostatin promoter (Fig. 2A) with specific primers (the sequences of primers will be provided upon request). The primers included a cleavage recognition site for either KpnI (forward primer) or XhoI (reverse primer) to facilitate subcloning into the multiple cloning sites of the pGL3-basic vector.
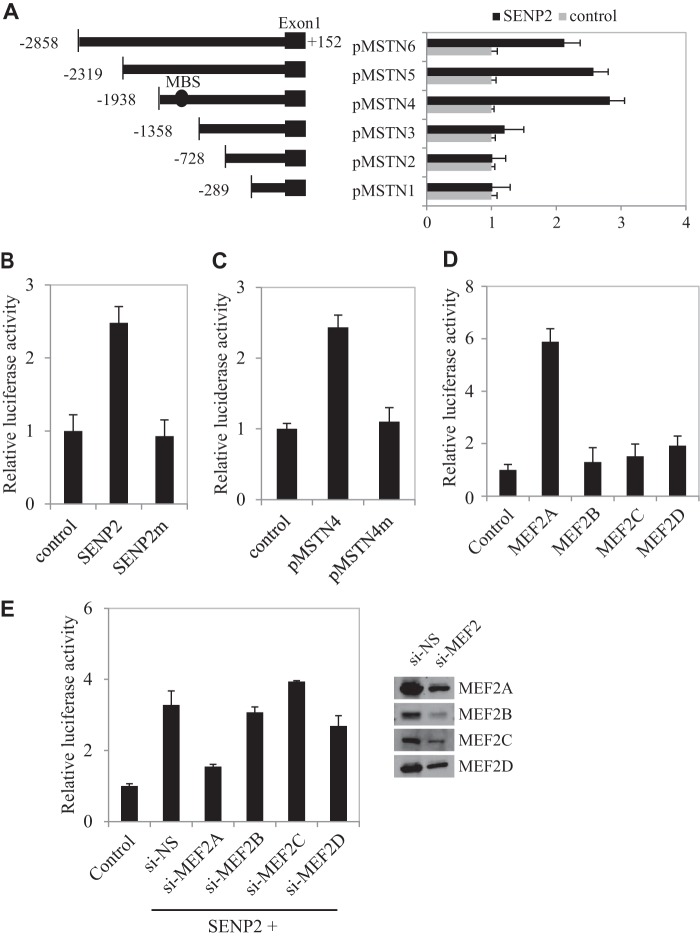
FIGURE 2.
MEF2A mediates SENP2 activity on the myostatin promoter. A, schematic of the various deletion fragments generated to identify the minimal promoter region required for myostatin activity. There is one MEF2 binding site (MBS) in pMSTN4 but not in pMSTN3. C2C12 cells were transfected with various deletion fragments and SENP2, and luciferase activity was measured 24 h after transfection. The data are presented as mean ± S.D. of three independent experiments. B, C2C12 cells were transfected with FLAG-tagged SENP2 or SENP2m plasmid, and relative luciferase activity was measured 24 h after transfection. The data are presented as mean ± S.D. of three independent experiments. Differences between SENP2 and SENP2m or control cells were significant (p < 0.05, Student's t test). C, C2C12 cells were transfected with pMSTN4 or pMSTN4m plasmid, and relative luciferase activity was measured 24 h after transfection. The data are presented as mean ± S.D. of three independent experiments. Differences between SENP2 and SENP2m or control cells were significant (p < 0.05, Student's t test). D, C2C12 cells were transfected with MEF2A, MEF2B, MEF2C, or MEF2D plasmid, and luciferase activity was measured 24 h after transfection. The data are presented as mean ± S.D. of three independent experiments. Differences between MEF2A and MEF2B, MEF2C, MEF2D, or the control were significant (p < 0.05, Student's t test). E, C2C12 cells were transfected with NS-siRNA (si-NS), MEF2A siRNA (si-MEF2A), MEF2B siRNA (si-MEF2B), MEF2C siRNA (si-MEF2C), or MEF2D siRNA (si-MEF2D), and the expression of MEF2 proteins was analyzed by Western blot analysis (right panel). Luciferase activity was measured 24 h after transfection. The data are presented as mean ± S.D. of three independent experiments.
Skeletal Myogenesis
C2C12 cells were cultured in growth medium (DMEM supplemented with 10% FBS). When cells reached about 80–90% confluence, the growth medium was aspirated, and cells were switched to differentiation medium (DMEM supplemented with 2% horse serum). Refeeding with fresh differentiation medium was done every 48 h. After 6 days of differentiation, the cells were stained with myosin heavy chain antibody to show myotube formation. DAPI was used to stain the nuclei.
Satellite Stem Cell Differentiation
Satellite stem cells were isolated from C57BL/6 mouse skeletal muscle. Cells with round morphological characteristics were selected and defined as primitive muscle satellite stem cells. The genetic marker PAX7 was used to identify satellite stem cells. To induce satellite stem cell differentiation, the cells were grown to 80–90% confluence, growth medium was aspirated, and cells were switched to differentiation medium (DMEM supplied with 2% horse serum). Refeeding with fresh differentiation medium was done every 48 h. After 6 days of differentiation, the cells were stained with myosin heavy chain antibody to show myotube formation. DAPI was used to stain the nuclei.
Cachexia Mouse Model
106 LLC cells in 0.1 ml PBS and PBS control were injected into the tail veins of 8-week-old C57BL/6 mice. Six weeks after LLC cell injection, when tumors appeared in the lungs, body weight was measured, and the skeletal muscle was harvested. The SENP2 and myostatin mRNA expression levels were measured via real-time PCR.
Statistical Analysis
Results were presented as mean ± S.D. Student's t test was used to compare the differences between two groups.
RESULTS
SENP2 Modulates Myostatin Expression
Because SENP2 is highly expressed in the dermomyotome in mouse embryos (data not shown), we reasoned that SENP2 might be an important regulator in myogenesis. Microarray profiles from Senp2+/+ and Senp2−/− embryos at embryonic day 10.5 showed that myostatin mRNA in Senp2−/− embryos was reduced significantly compared with that in Senp2+/+ embryos. We evaluated this observation in Senp2−/− MEF cells and embryos by using real-time PCR, and the evaluation showed that in both MEFs and embryos, myostatin mRNA was decreased in comparison with that in the Senp2+/+ controls (Fig. 1A). To further determine the role of SENP2 in myostatin expression, we overexpressed SENP2 in murine myoblast C2C12 cells, which showed that the overexpression of SENP2 increased myostatin expression (Fig. 1B). Interestingly, the SENP2 catalytic mutant did not have such an effect (Fig. 1B), suggesting that the de-SUMOylation activity is essential for SENP2 to regulate myostatin expression. We also confirmed the role of SENP2 in the regulation of myostatin expression by using the SENP2 siRNA approach. As shown in Fig. 1C, silencing SENP2 significantly reduced myostatin expression. Collectively, these results show a crucial role of SENP2 in myostatin expression.
FIGURE 1.
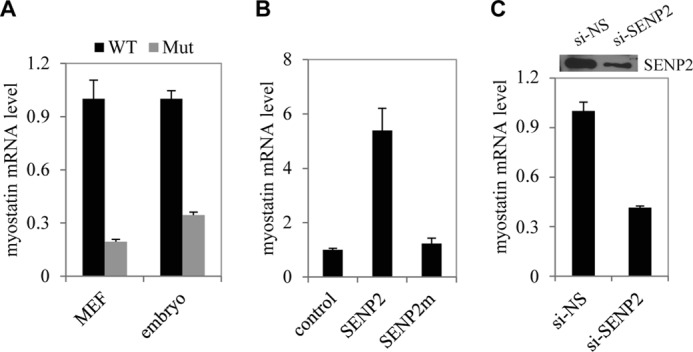
SENP2 specifically regulates myostatin expression. A, mRNA levels of myostatin in MEF cells and embryos of Senp2+/+ (WT) and Senp2−/− (Mut) mice were measured by real-time PCR. The data are presented as mean ± S.D. of three independent experiments. Differences between Senp2+/+ and Senp2−/− mice were significant (p < 0.05, Student's t test). B, mRNA levels of myostatin in C2C12 cells transfected with FLAG-tagged SENP2 or SENP2m plasmid were measured by real-time PCR. The data are presented as mean ± S.D. of three independent experiments. Differences between SENP2 and SENP2m or control cells were significant (p < 0.05, Student's t test). C, mRNA levels of myostatin in C2C12 cells transfected with NS-siRNA (si-NS) or SENP2 siRNA (si-SENP2) were measured by real-time PCR. The data are presented as mean ± S.D. of three independent experiments. Differences between SENP2 and SENP2m or control cells were significant (p < 0.05, Student's t test).
MEF2A Mediates the Effect of SENP2 on Myostatin Expression
To explore how SENP2 regulates myostatin expression, we generated a myostatin promoter (−2858 to +152 bp of myostatin genomic DNA)-luciferase report gene (Fig. 2A, pMSTN6). Cotransfection of SENP2 significantly induced the transcription activity of the myostatin promoter (Fig. 2B). To further map the SENP2 response element of the myostatin promoter, we tested a series of truncated forms of myostatin promoter-luciferase plasmids and found that the SENP2-responsive element was located between −1938 to −1358 bp of the myostatin promoter (Fig. 2A). Consistent with the observation in Fig. 1B, we found that SENP2 catalytic activity was also required to activate myostatin promoter transcription (Fig. 2B). The TESS program predicted a MEF2 binding site located in this area (−1336 to −1326 bp of the myostatin promoter) (Fig. 2A). We tested whether this MEF2 binding site might mediate SENP2 action on myostatin promoter activity by a mutation approach. As expected, we found that the SENP2 activation on the myostatin promoter was almost abolished when the MEF2 binding site was mutated (Fig. 2C).
The MEF2 family includes four members, denoted as MEF2A, MEF2B, MEF2C, and MEF2D. Coexpression of MEF2A, but not of other MEF2 members, could promote the transactivation of myostatin in a reporter gene assay (Fig. 2D). Thus, we speculated that MEF2A was essential for SENP2 in the regulation of myostatin expression. Indeed, silencing only MEF2A, no other MEF2 genes blocked SENP2 activation of the myostatin promoter. These data suggest a critical role of MEF2A in SENP2 activation of myostatin transcription.
SENP2 De-SUMOylates MEF2A
Given that MEF2A is a SUMOylated protein, the induction of myostatin might be through the de-SUMOylation of MEF2A by SENP2. Therefore, we first assessed whether SUMOylation of MEF2A would have an effect on myostatin transcription. Indeed, the SUMOylation site mutant of MEF2A induced more activity of the myostatin promoter than the MEF2A wild type (Fig. 3A). We further showed that the overexpression of SUMO-1 suppressed the activity of the MEF2A wild type, not the MEF2A mutant, on the myostatin promoter (Fig. 3A). These results suggest that SUMOylation can negatively regulate MEF2A transactivation on the myostatin promoter.
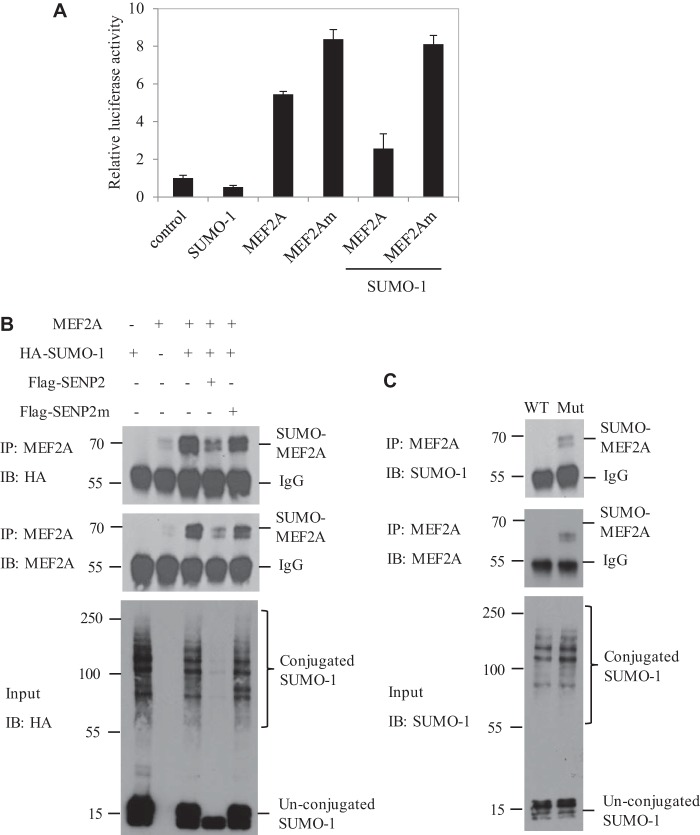
FIGURE 3.
MEF2A is a target of SENP2. A, C2C12 cells were transfected with the SUMO-1, MEF2A, and MEF2Am plasmids, as indicated, and luciferase activity was measured 24 h after transfection. The data are presented as mean ± S.D. of three independent experiments. B, C2C12 cells were transfected with MEF2A, HA-SUMO-1, FLAG-SENP2, or SENP2m, as indicated. MEF2A proteins were pulled down by MEF2A antibody from these cell lysates. Bound proteins were blotted with anti-HA (top panel) or anti-MEF2A antibody (center panel). Cell lysate (Input) was immunoblotted (IB) with anti-HA antibody (bottom panel). IP, immunoprecipitation. C, the proteins extracted from Senp2+/+ (WT) and Senp2−/− (Mut) MEF cells were pulled down with anti-MEF2A antibody and immunoblotted with anti-SUMO-1 (top panel) or anti-MEF2A antibody (center panel). Cell lysate was immunoblotted with anti-SUMO-1 antibody (bottom panel).
We next determined whether SENP2 de-SUMOylated MEF2A in vivo and in vitro. In the in vitro assay, MEF2A SUMOylation was detected in C2C12 cells transfected with MEF2A and HA-SUMO-1. The overexpression of SENP2, not the SENP2 mutant, deconjugated SUMO-MEF2A (Fig. 3B). The role of SENP2 in de-SUMOylation of MEF2A was also confirmed in Senp2+/+ and Senp2−/− MEF cells. SUMO-MEF2A was detected easily detected and accumulated in Senp2−/− MEF cells (Fig. 3C). These data indicate that SENP2 can de-SUMOylate MEF2A and enhance MEF2A transactivation on the myostatin promoter.
SENP2 Inhibits Skeletal Myogenesis
Because SENP2 promotes myostatin expression, we speculated that SENP2 would repress myogenesis as myostatin does. We tested this hypothesis in two cell models. We first used C2C12 cells transfected with SENP2 siRNA, MEF2A, or myostatin siRNA (Fig. 4A). Myostatin expression was reduced significantly in these cells compared with si-NS control cells (Fig. 4, A and B). These cells were induced into myogenesis by 2% horse serum in culture. Myotube formation, as a marker for myogenesis, was checked in these cells by using immunofluorescence staining with anti-myosin heavy chain antibody. As shown in Fig. 4C, silencing of MEF2A or SENP2 enhanced myotube formation in C2C12 cells as myostatin siRNA did. Another cell model used was satellite stem cells isolated from C57BL/6 mouse skeletal muscle. The isolated satellite stem cells were confirmed by PAX7 immunostaining (data not shown). Satellite stem cells transfected with MEF2A, SENP2, or myostatin siRNA were induced into differentiation by addition of 2% horse serum. As observed in the C2C12 cell model, myostatin expression was decreased significantly after MEF2A or SENP2 knockdown in satellite stem cells (Fig. 4B). Differentiation was shown by immunostaining with anti-myosin heavy chain antibody in myostatin siRNA- or SENP2 siRNA-transfected satellite stem cells (Fig. 4C). Like myostatin, SENP2 is a negative regulator in myogenesis.
FIGURE 4.
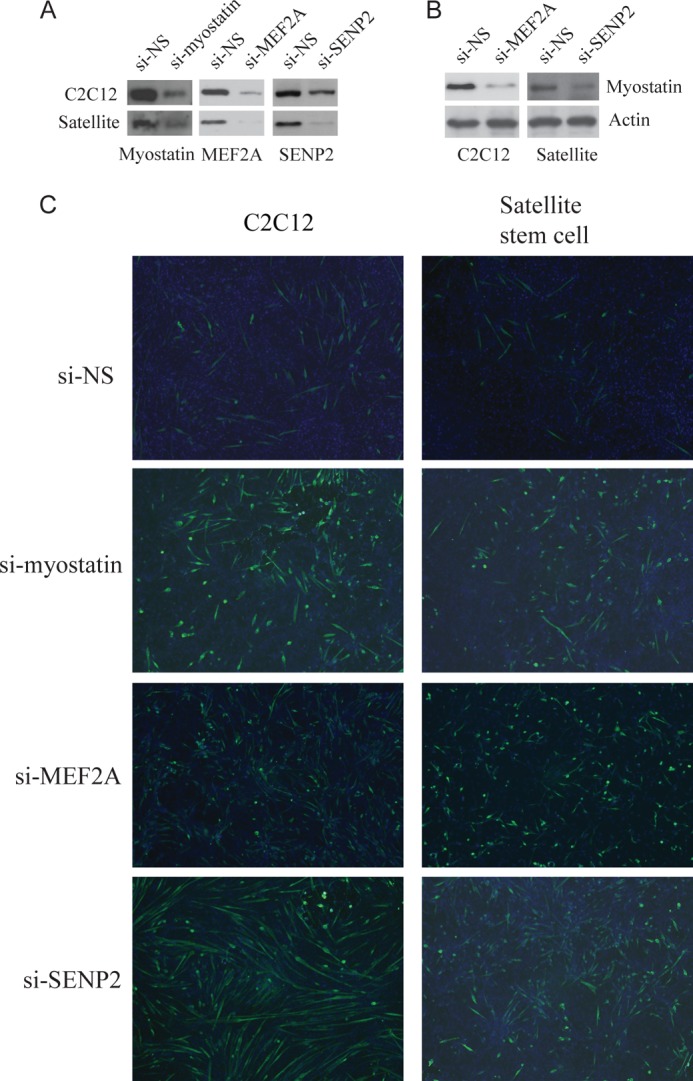
SENP2 and myostatin repress skeletal myogenesis in C2C12 and satellite stem cells. Differentiation of C2C12 cells or satellite stem cells was induced with 2% horse serum, and then NS-siRNA (si-NS), MEF2A siRNA (si-MEF2A), SENP2 siRNA (si-SENP2), or myostatin siRNA (si-myostatin) were used for transfection. The expression of myostatin, MEF2A, or SENP2 in these cells was analyzed by Western blot analysis (A and B). Skeletal myogenesis was analyzed by staining with anti-myosin heavy chain (green) (C). DAPI (blue) was used to stain the cell nucleus.
The Correlation of Senp2 and Myostatin Expression in Cachexia
Because myostatin expression is increased and related to weight loss and muscle atrophy during cachexia, we reasoned that SENP2 might be involved in cachexia through the regulation of myostatin expression. To test this possibility, we created a cancer cachexia mouse model by injecting LLC cells via the tail vein. Six weeks after LLC cell injection, the mice not only grew lung tumors (data not shown) but also showed a body weight loss of 45%, which is a typical cachexia condition (Fig. 5A). Interestingly, the expression of both Senp2 and myostatin mRNA showed a remarkable increase in the skeletal muscle of the cachexia mice (Fig. 5, B and C), suggesting that SENP2 might promote cachexia through the regulation of myostatin expression in cancer-bearing mice.
FIGURE 5.
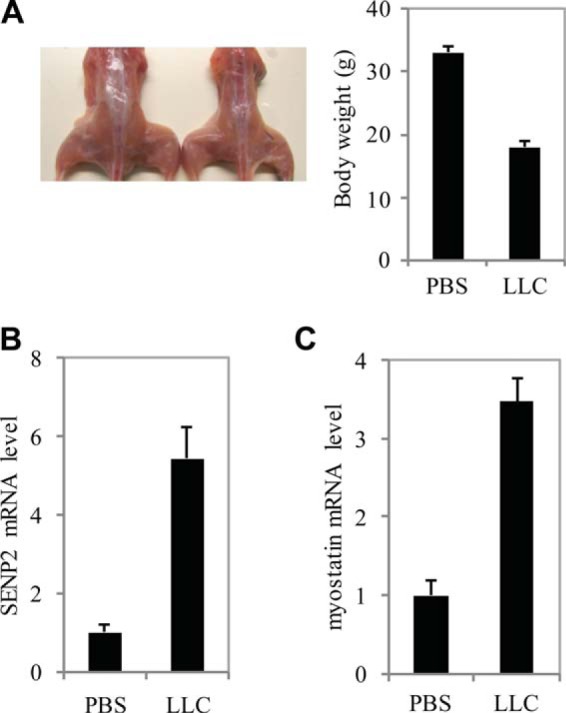
The role of SENP2 and myostatin in a cachexia mouse model. A, 8-week-old C57BL/6 mice were injected with PBS or Lewis lung carcinoma cells. The skeletal muscle morphology and body weight of PBS- or LLC-injected mice were measured 6 weeks after injection. mRNA levels of Senp2 (B) and myostatin (C) in skeletal muscle were measured by real-time PCR. The data are presented as mean ± S.D. of three independent experiments. Differences between LLC- and PBS-injected mice were significant (p < 0.05, Student's t test).
DISCUSSION
In this study, we found that SENP2 contributes to the regulation of myostatin expression in muscle cells. We also observed SENP2 acting as a negative regulator in myogenesis, like myostatin. We further observed that MEF2A, a specific transcription factor of myostatin, mediates SENP2 promotion for myostatin expression through de-SUMOylation. We detected the increase of both the expression of SENP2 and the expression of myostatin in a tumor-bearing cachexia model, suggesting that SENP2 plays a critical role in cancer-induced cachexia.
Senp2-null mice died from embryonic heart failure at around embryonic day 10 (14, 30). Further evidence showed that the knockout of Senp2 decreased cardiomyocyte proliferation and heart formation in mouse embryonic hearts (14). Interestingly, here we found that silencing SENP2 would promote the differentiation and myogenesis in skeletal muscle progenitor cells. The two phenotypes in Senp2 deficiency cells look contradictory. However, SENP2 acts with different mechanisms in these two different processes. In cardiomyocytes, SENP2 regulates Gata4/6 expression through the PcG complex, which is essential for heart development (14). However, the function of SENP2 in skeleton muscle is through the induction of myostatin, which suppresses myogenesis. These observations suggest that SENP2 has multiple functions in regulating biological processes involving different targets. In a future study, it will be interesting to identify by which mechanism SENP2 targets specific proteins in different signaling pathways.
Satellite stem cells are quiescent muscle stem cells (31–33). How to activate the differentiation process of satellite stem cells into new myofibers is still a critical issue for skeletal muscle regeneration. It seems that SENP2 functions in maintaining the quiescent status of satellite stem cells. When SENP2 expression is silenced or SENP2 activity is inactivated, satellite stem cells can differentiate into myocytes. Thus, SENP2 is a potential therapeutic target in skeletal muscle regeneration.
This work was supported by National Basic Research Program of China (973 Program) Grants 2010CB912104 (to J. C.) and 2012CB910102 (to Y. Z.), by National Natural Science Foundation of China Grant 91019021 (to J. C.), and by Shanghai Committee of Science and Technology Grants 11XD1403200 (to J. C.) and 1DZ2260200.
- SUMO
- small ubiquitin-like modifier
- SENP
- sentrin/small ubiquitin-like modifier-specific protease
- NS
- nonspecific
- LLC
- Lewis lung carcinoma.
REFERENCES
- 1. Geiss-Friedlander R., Melchior F. (2007) Concepts in sumoylation. A decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 2. Hay R. T. (2005) SUMO. A history of modification. Mol. Cell 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 3. Yeh E. T. (2009) SUMOylation and de-SUMOylation. Wrestling with life's processes. J. Biol. Chem. 284, 8223–8227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gill G. (2003) Post-translational modification by the small ubiquitin-related modifier SUMO has big effects on transcription factor activity. Curr. Opin. Genet. Dev. 13, 108–113 [DOI] [PubMed] [Google Scholar]
- 5. Ouyang J., Valin A., Gill G. (2009) Regulation of transcription factor activity by SUMO modification. Methods Mol. Biol. 497, 141–152 [DOI] [PubMed] [Google Scholar]
- 6. Yeh E. T., Gong L., Kamitani T. (2000) Ubiquitin-like proteins. New wines in new bottles. Gene 248, 1–14 [DOI] [PubMed] [Google Scholar]
- 7. Schwienhorst I., Johnson E. S., Dohmen R. J. (2000) SUMO conjugation and deconjugation. Mol. Gen. Genet. 263, 771–786 [DOI] [PubMed] [Google Scholar]
- 8. Cheng J., Kang X., Zhang S., Yeh E. T. (2007) SUMO-specific protease 1 is essential for stabilization of HIF1α during hypoxia. Cell 131, 584–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hay R. T. (2007) SUMO-specific proteases. A twist in the tail. Trends Cell Biol. 17, 370–376 [DOI] [PubMed] [Google Scholar]
- 10. Mukhopadhyay D., Dasso M. (2007) Modification in reverse. The SUMO proteases. Trends Biochem. Sci. 32, 286–295 [DOI] [PubMed] [Google Scholar]
- 11. Chow K. H., Elgort S., Dasso M., Ullman K. S. (2012) Two distinct sites in Nup153 mediate interaction with the SUMO proteases SENP1 and SENP2. Nucleus 3, 349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hang J., Dasso M. (2002) Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 277, 19961–19966 [DOI] [PubMed] [Google Scholar]
- 13. Itahana Y., Yeh E. T., Zhang Y. (2006) Nucleocytoplasmic shuttling modulates activity and ubiquitination-dependent turnover of SUMO-specific protease 2. Mol. Cell. Biol. 26, 4675–4689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kang X., Qi Y., Zuo Y., Wang Q., Zou Y., Schwartz R. J., Cheng J., Yeh E. T. (2010) SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol. Cell 38, 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McPherron A. C., Lee S. J. (1997) Double muscling in cattle due to mutations in the myostatin gene. Proc. Natl. Acad. Sci. U.S.A. 94, 12457–12461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee S. J., McPherron A. C. (1999) Myostatin and the control of skeletal muscle mass. Curr. Opin. Genet. Dev. 9, 604–607 [DOI] [PubMed] [Google Scholar]
- 17. Lee S. J., McPherron A. C. (2001) Regulation of myostatin activity and muscle growth. Proc. Natl. Acad. Sci. U.S.A. 98, 9306–9311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Artaza J. N., Bhasin S., Magee T. R., Reisz-Porszasz S., Shen R., Groome N. P., Meerasahib M. F., Fareez M. M., Gonzalez-Cadavid N. F. (2005) Myostatin inhibits myogenesis and promotes adipogenesis in C3H 10T(1/2) mesenchymal multipotent cells. Endocrinology 146, 3547–3557 [DOI] [PubMed] [Google Scholar]
- 19. McCroskery S., Thomas M., Maxwell L., Sharma M., Kambadur R. (2003) Myostatin negatively regulates satellite cell activation and self-renewal. J. Cell Biol. 162, 1135–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Walsh F. S., Celeste A. J. (2005) Myostatin. A modulator of skeletal-muscle stem cells. Biochem. Soc. Trans. 33, 1513–1517 [DOI] [PubMed] [Google Scholar]
- 21. Bogdanovich S., Krag T. O., Barton E. R., Morris L. D., Whittemore L. A., Ahima R. S., Khurana T. S. (2002) Functional improvement of dystrophic muscle by myostatin blockade. Nature 420, 418–421 [DOI] [PubMed] [Google Scholar]
- 22. Zimmers T. A., Davies M. V., Koniaris L. G., Haynes P., Esquela A. F., Tomkinson K. N., McPherron A. C., Wolfman N. M., Lee S. J. (2002) Induction of cachexia in mice by systemically administered myostatin. Science 296, 1486–1488 [DOI] [PubMed] [Google Scholar]
- 23. Schuelke M., Wagner K. R., Stolz L. E., Hübner C., Riebel T., Kömen W., Braun T., Tobin J. F., Lee S. J. (2004) Myostatin mutation associated with gross muscle hypertrophy in a child. N. Engl. J. Med. 350, 2682–2688 [DOI] [PubMed] [Google Scholar]
- 24. Artaza J. N., Reisz-Porszasz S., Dow J. S., Kloner R. A., Tsao J., Bhasin S., Gonzalez-Cadavid N. F. (2007) Alterations in myostatin expression are associated with changes in cardiac left ventricular mass but not ejection fraction in the mouse. J. Endocrinol. 194, 63–76 [DOI] [PubMed] [Google Scholar]
- 25. Deng B., Wen J., Ding Y., Gao Q., Huang H., Ran Z., Qian Y., Peng J., Jiang S. (2012) Functional analysis of pig myostatin gene promoter with some adipogenesis- and myogenesis-related factors. Mol. Cell. Biochem. 363, 291–299 [DOI] [PubMed] [Google Scholar]
- 26. Du R., An X. R., Chen Y. F., Qin J. (2007) Some motifs were important for myostatin transcriptional regulation in sheep (Ovis aries). J. Biochem. Mol. Biol. 40, 547–553 [DOI] [PubMed] [Google Scholar]
- 27. Qin J., Du R., Yang Y. Q., Zhang H. Q., Li Q., Liu L., Guan H., Hou J., An X. R. (2013) Dexamethasone-induced skeletal muscle atrophy was associated with upregulation of myostatin promoter activity. Res. Vet. Sci. 94, 84–89 [DOI] [PubMed] [Google Scholar]
- 28. Gong L., Millas S., Maul G. G., Yeh E. T. (2000) Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J. Biol. Chem. 275, 3355–3359 [DOI] [PubMed] [Google Scholar]
- 29. Kamitani T., Nguyen H. P., Kito K., Fukuda-Kamitani T., Yeh E. T. (1998) Covalent modification of PML by the sentrin family of ubiquitin-like proteins. J. Biol. Chem. 273, 3117–3120 [DOI] [PubMed] [Google Scholar]
- 30. Chiu S. Y., Asai N., Costantini F., Hsu W. (2008) SUMO-specific protease 2 is essential for modulating p53-Mdm2 in development of trophoblast stem cell niches and lineages. PLoS Biol. 6, e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yin H., Price F., Rudnicki M. A. (2013) Satellite cells and the muscle stem cell niche. Physiol. Rev. 93, 23–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brack A. S., Rando T. A. (2012) Tissue-specific stem cells. Lessons from the skeletal muscle satellite cell. Cell Stem Cell 10, 504–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zammit P. S., Partridge T. A., Yablonka-Reuveni Z. (2006) The skeletal muscle satellite cell. The stem cell that came in from the cold. J. Histochem. Cytochem. 54, 1177–1191 [DOI] [PubMed] [Google Scholar]