Background: USP-46 deubiquitinates the C. elegans glutamate receptor GLR-1.
Results: WDR-20 and WDR-48 bind and activate USP-46 in vitro and increase the abundance of GLR-1 in neurons.
Conclusion: WD40-repeat proteins stimulate USP-46 activity, resulting in increased GLR-1 stability in neurons and alterations in glutamate-dependent behavior.
Significance: WD40-repeat protein regulation of DUB activity is important for glutamate receptor trafficking in vivo.
Keywords: Caenorhabditis elegans; Deubiquitination; Glutamate Receptors Ionotropic (AMPA, NMDA); Neurons; Synapses; Ubiquitin
Abstract
Ubiquitin-mediated endocytosis and degradation of glutamate receptors controls their synaptic abundance and is implicated in modulating synaptic strength. The deubiquitinating enzymes (DUBs) that function in the nervous system are beginning to be defined, but the mechanisms that control DUB activity in vivo are understood poorly. We found previously that the DUB USP-46 deubiquitinates the Caenorhabditis elegans glutamate receptor GLR-1 and prevents its degradation in the lysosome. The WD40-repeat (WDR) proteins WDR20 and WDR48/UAF1 have been shown to bind to USP46 and stimulate its catalytic activity in other systems. Here we identify the C. elegans homologs of these WDR proteins and show that C. elegans WDR-20 and WDR-48 can bind and stimulate USP-46 catalytic activity in vitro. Overexpression of these activator proteins in vivo increases the abundance of GLR-1 in the ventral nerve cord, and this effect is further enhanced by coexpression of USP-46. Biochemical characterization indicates that this increase in GLR-1 abundance correlates with decreased levels of ubiquitin-GLR-1 conjugates, suggesting that WDR-20, WDR-48, and USP-46 function together to deubiquitinate and stabilize GLR-1 in neurons. Overexpression of WDR-20 and WDR-48 results in alterations in locomotion behavior consistent with increased glutamatergic signaling, and this effect is blocked in usp-46 loss-of-function mutants. Conversely, wdr-20 and wdr-48 loss-of-function mutants exhibit changes in locomotion behavior that are consistent with decreased glutamatergic signaling. We propose that WDR-20 and WDR-48 form a complex with USP-46 and stimulate the DUB to deubiquitinate and stabilize GLR-1 in vivo.
Introduction
Regulation of the abundance of glutamate neurotransmitter receptors (GluRs)3 at synapses can modulate synaptic strength and is a key molecular mechanism involved in learning and memory (1). Posttranslational modification of GluRs by ubiquitin promotes receptor endocytosis and degradation, providing a mechanism to regulate synaptic GluR levels (2, 3). Ubiquitin is covalently attached to and removed from target proteins by a large family of ubiquitin ligases and deubiquitinating enzymes (DUBs), respectively (4). The balance of activity of specific ubiquitin ligases and DUBs controls the localization, function, and stability of target proteins, including neurotransmitter receptors (5–8).
In Caenorhabditis elegans, the abundance of the AMPA-type GluR, GLR-1, is regulated by ubiquitin (3). Ubiquitin is directly conjugated to GLR-1 and promotes receptor endocytosis and post-endocytic degradation in the lysosome (3, 9). Mammalian AMPA receptors are also regulated by ubiquitin (10, 11). Several ubiquitin ligases have been identified that regulate invertebrate GluRs (12–16) and mammalian AMPA receptors (17–20). DUBs counter the action of ubiquitin ligases by removing ubiquitin from substrates, but the specific DUBs that function at synapses are not well defined. We showed recently that the DUB USP-46 regulates the levels of GLR-1 at synapses in C. elegans. USP-46 promotes the abundance of GLR-1 by deubiquitinating the receptor and preventing its degradation in the lysosome (21).
USP-46 and its homologs have been implicated in regulating diverse cellular functions, including endocytosis, cell polarity, signal transduction, chromatin modification, and mitochondrial biogenesis (22–27). In some cases, the substrates of the DUB have been identified. For example, USP46 deubiquitinates the phosphatase PHLPP (PH domain and leucine-rich repeat protein phosphatase) to regulate Akt signaling in colon cancer cells (25). The USP46 homolog USP12 negatively regulates Notch levels in Drosophila and mammalian cells by deubiquitinating Notch and promoting its lysosomal degradation (27). USP12 and USP46 also deubiquitinate histones H2A and H2B to regulate development in Xenopus embryos (22). Although the role of USP46 in regulating GluRs has not been examined in the mammalian nervous system, usp46 mutant mice exhibit antidepressive-like behaviors and abnormalities in the GABA signaling system (28, 29). However, the mechanism by which USP46 affects the GABA system is not yet known. These studies illustrate that USP46 is involved in a variety of processes in multiple cell types, underscoring the importance of understanding the mechanisms involved in regulating USP46 activity in vivo.
Mammalian USP46 and its close homolog USP12 have very low intrinsic catalytic activity in vitro, indicating that they may be regulated by other factors in vivo (30–32). Research in systems from yeast to mammals has identified several proteins that interact with USP46 or USP12, including two WD40-repeat-containing proteins: WDR48 (also known as UAF-1) and WDR20 (22–24, 27, 30, 32, 33). Biochemical isolation of protein complexes shows that the catalytic activity of mammalian USP12 and USP46 can be stimulated by WDR48 (22, 30–32) and WDR20 (32) in vitro, which suggests that these proteins might be required for normal USP46 function in vivo. In mammalian cells, WDR48 can bind USP12 to promote deubiquitination of non-activated Notch (27). The WDR48 homolog Duf1 binds and stimulates the activity of Ubp9 (USP46) to regulate ATP synthase expression in Saccharomyces cerevisiae (23), and Bun107 (WDR20) and Bun 62 (WDR48) interact with Usp9 in Schizosaccharomyces pombe (24). The conservation of the interaction between the homologs of WDR-20, WDR-48, and USP-46 across phylogeny emphasizes the importance of this complex. However, the physiological relevance of this interaction in vivo remains to be fully defined.
In this study, we identified the C. elegans homologs of WDR20 and WDR48. As with their mammalian counterparts, C. elegans WDR-20 and WDR-48 can bind to and activate USP-46 in vitro. Overexpression of WDR-20 and WDR-48 in neurons increases the abundance and stability of GLR-1 in vivo. Importantly, the effect of the WDR proteins on GLR-1 results in a corresponding change in glutamate-mediated locomotion behavior that is dependent on usp-46. This study identifies a novel physiological role for WDR-20 and WDR-48 in neurons and shows that they can activate USP-46 to control GLR-1 levels in vivo, resulting in changes in glutamate-dependent behavior.
EXPERIMENTAL PROCEDURES
Strains
The following strains were used for experiments described in this manuscript: N2 (Bristol) wild type, nuIs24 (Pglr-1::glr-1::gfp), nuIs25 (Pglr-1::glr-1::gfp), pzIs25 (Pglr-1::wdr-20; Pglr-1::wdr-48), pzIs22 (Pglr-1::usp-46; Pglr-1::wdr-20; Pglr-1::wdr-48), pzEx230 (Pglr-1::wdr-20), pzEx231 (Pglr-1::wdr-48), usp-46 (ok2232), wdr-20 (gk547140), wdr-48 (gk173034), wdr-48 (tm4575), pzEx222 (Pglr-1::usp-46 (C>A)), pzEx224 (Pglr-1::usp-46), pzIs12 (Pglr-1::HA::glr-1::gfp), pzEx268 (Pwdr-20::gfp), pzEx246 (Pwdr-20::nls::gfp::lacZ), pzEx270 (Pwdr-48::gfp), nuEx993 (Pglr-1::lin-10::gfp), nuEx1004 (Pglr-1::magi-1::yfp), and nuIs125 (Pglr-1::snb-1::gfp). All strains were maintained at 20 °C as described previously (34).
Plasmids
Sequences encoding C. elegans USP-46, WDR-20, and WDR-48 were amplified from wild-type cDNA and subcloned into the mammalian expression vector pMT3 (a gift from Larry Feig) to generate the following epitope-tagged plasmids: pMT3-FLAG-USP-46 (FJ#66), pMT3-HA-WDR-20 (FJ#94), pMT3-His6X-WDR-20 (FJ#95), and pMT3-Myc-WDR-48 (FJ#96). The sequences for USP-46, WDR-20, and WDR-48 were subcloned into the C. elegans expression vector pV6 (35) for expression under the glr-1 promoter to create Pglr-1::usp-46 (FJ#1) (21), Pglr-1::wdr-20 (FJ#97), and Pglr-1::wdr-48 (FJ#98). Pglr-1::usp-46(C>A)(FJ#2) has been described previously (21). The WDR proteins were tagged at the N terminus with mCherry by inserting the sequence encoding mCherry in-frame immediately after the start codons for wdr-20 or wdr-48 downstream of the glr-1 promoter to generate Pglr-1::mCherry::wdr-20 (FJ#102) and Pglr-1::mCherry::wdr-48 (FJ#103), respectively. Pwdr-20::gfp (FJ#99) and Pwdr-48::gfp (FJ#100) were created by subcloning 2.7 kb of the 5′ untranslated region of C08B6.7 (WDR-20) or 3.5 kb of the 5′ untranslated region of F35G12.4 (WDR-48), respectively, upstream of the start codon of GFP in pPD95.75. Pwdr-20::nls::gfp::lacZ (FJ#101) was created by subcloning 2.7 kb of the 5′ untranslated region of C08B6.7 (WDR-20) upstream of the start codon of NLS::GFP::LAC-Z in pPD96.04.
Imaging
Fluorescence imaging of GLR-1::GFP was performed as described previously (21). Briefly, L4 larval-stage animals were immobilized using 30 mg/ml 2,3-butanedione monoxamine (Sigma-Aldrich), and the VNC was imaged in the anterior region of the animals just posterior to the RIG neuronal cell bodies. 1 μm (total depth) Z-series stacks were collected using a Carl Zeiss Axioscope M1 microscope with a ×100 Plan Apochromat (1.4 numerical aperture) objective equipped with GFP and Cy3.5 filters. Images were collected with an Orca-ER charge coupled device (CCD) camera (Hamamatsu) and MetaMorph (version 7.1) software (Molecular Devices). Maximum intensity projections of Z-series stacks were used for quantitative analyses of fluorescent puncta. Exposure settings and gain were adjusted to fill the 12-bit dynamic range without saturation and were identical for all images taken of each fluorescent marker. Line scans of ventral cord puncta were generated using MetaMorph (version 6.0) and were analyzed with custom-written software (Jeremy Dittman, Weill Cornell Medical College) in Igor Pro (version 5) (Wavemetrics) (3). Arc lamp output was monitored by measuring the intensities of 0.5-μm FluoSphere beads (Invitrogen). Puncta intensities for wild-type and mutant animals were normalized to the average bead intensity for the corresponding day before normalizing the wild-type intensities to 1. Puncta density was determined by quantifying the average number of puncta per 10 μm of the VNC. GFP reporter strains were imaged as above, except that young adult animals were imaged. Primary neuronal cultures were imaged as above but without 2,3-butanedione. The anti-HA-Alexa Fluor 594 punctum fluorescence intensity relative to the interpunctal neurite fluorescence was analyzed using Igor Pro (version 5) as described above.
Detection of GLR-1::GFP by Immunoblot Analysis of Whole-animal Lysates
Total GLR-1::GFP protein levels were determined by immunoblot analysis of total worm lysates prepared from mixed-stage populations of animals expressing GLR-1::GFP (nuIs24). Lysates were made by resuspending worms in SDS sample buffer, followed by freezing at −80 °C and boiling for 2 min. After electrophoresis and transfer, the upper half of the nitrocellulose membrane was probed with monoclonal anti-GFP antibodies (JL-8, Covance), and the lower half was probed with polyclonal anti-tubulin antibodies (Abcam) for normalization of input levels.
Cell Culture
HEK293T cells (a gift from Grace Gill) were maintained in DMEM with 0.5% Fetal Clone II Serum (HyClone) and penicillin/streptomycin (Invitrogen). Cells were transfected with 0.5–3 μg of plasmid DNA using Lipofectamine 2000 according to the directions of the manufacturer. pCMV-GFP was cotransfected along with other plasmids as a control to monitor transfection efficiency.
Immunoprecipitation from Tissue Culture Cells
After 24 h, cells were washed once with PBS and lysed with IP buffer (50 mm Tris (pH7.5), 150 mm NaCl, 0.5% Nonidet P-40). Harvested cells were spun at 14,000 rpm for 10 min at 4 °C. Cleared lysate was incubated for 4–12 h with protein A- and G-Sepharose beads (GE Healthcare) and mouse anti-FLAG antibody (M2, Sigma) in the presence of protease inhibitors (10 μg/ml leupeptin, 5 μg/ml chymostatin, 3 μg/ml elastin, 1 μg/ml pepstatin A, and 1 mm PMSF) and 0.01 m DTT. Immunoprecipitated complexes were washed four times with lysis buffer and resuspended in 2× SDS-PAGE buffer. Samples were subjected to SDS-PAGE on 10% acrylamide gels and subsequently transferred to a nitrocellulose membrane. Immunoblots were blocked in Tris-buffered saline with Tween (TBS-T) and 5% milk and probed with the following antibodies: mouse anti-GFP (Covance), rabbit anti-FLAG (Rockland, a gift from Ira Herman), rat anti-HA (Roche, a gift from Ira Herman), mouse anti-Myc (Santa Cruz Biotechnology, a gift from Ira Herman), rabbit anti-FLAG (M2)-HRP-conjugated (Sigma, a gift from Grace Gill).
Ub-VME Reaction
HEK293T cells were transfected as above. FLAG·USP-46 complexes were immunoprecipitated with anti-FLAG antibody and resuspended in 30 μl Ub-VME reaction buffer (20 mm Tris (pH 8.0), 150 mm NaCl, and 1 mm DTT). Samples were divided into two separate tubes, and 1.5 μg of Ub-VME was added to one of them. 1.5 μl of reaction buffer was added to the other (control) sample. Samples were incubated at 37 °C for 30 min with intermittent, gentle shaking. Reactions were stopped by addition of 5 μl of 5× SDS-PAGE sample buffer.
Primary Culture of C. elegans Neurons
Cells were isolated and cultured as described in Ref. 36. Cells were grown for no more than 3 days at 20 °C prior to fixation and staining. Cells were washed two times with M9 buffer (phosphate-buffered saline with magnesium sulfate) and then fixed in 3% paraformaldehyde in PBS for 30 min at room temperature. Incubation for 2 min with 0.2% Triton X-100 in PBS was used to permeabilize the cells. After blocking with 0.2% BSA in PBS for 20 min, cells were incubated with mouse anti-HA, monoclonal 16B12, Alexa Fluor 594 conjugate (Invitrogen, a gift from Lars Dreier) at 1 μg/ml in blocking solution for 45 min at room temperature. Cells were washed four times with PBS prior to mounting and imaging.
Behavioral Assays
Spontaneous locomotion of young adult hermaphrodites was analyzed as described previously (21, 37). Briefly, reversals were defined as a backward movement that brought the tip of the nose farther back than the position of the large pharyngeal bulb at the start of the movement. Individual animals were monitored for 5 min each. For each experiment, all genotypes shown were tested on each assay day.
Immunoprecipitation and Detection of Ubiquitinated GLR-1::GFP
Immunoprecipitation of ubiquitinated GLR-1::GFP was performed on membrane preparations that were isolated from mixed-stage populations of wild-type (nuIs24) or mutant worms expressing USP-46, WDR-20, and WDR-48 (pzIs22) (3, 21). Lysates were prepared essentially as described in Ref. 38. Briefly, worms were washed in 100 mm NaCl, isolated by sucrose gradient centrifugation, and resuspended in freezing buffer (50 mm HEPES (pH 7.4), 1 mm EGTA, 1 mm MgCl2, 100 mm KCl, and 10% glycerol) plus 1 mm PMSF. After freezing in liquid nitrogen, the recovered frozen pellets were ground finely in a chilled mortar and pestle. Ground worm powder was resuspended in buffer A (50 mm HEPES (pH 7.7), 50 mm potassium acetate, 2 mm magnesium acetate, 250 mm sucrose, and 1 mm EDTA) plus protease inhibitors (10 μg/ml leupeptin, 5 μg/ml chymostatin, 3 μg/ml elastin, 1 μg/ml pepstatin A, and 1 mm PMSF). Membrane fractions were isolated by spinning the clarified lysates at 55,000 rpm in a Beckman Ti 60 rotor, followed by resuspension in buffer A plus protease inhibitors and 7 mm β-mercaptoethanol (3). Membranes were solubilized in 1 volume of SDS buffer (50 mm Tris-HCl (pH 8.5), 1% SDS, and 2 mm DTT) and were diluted with 5 volumes of 50 mm HEPES (pH 7.7), 250 mm NaCl, and 1% Nonidet P-40 plus protease inhibitors. GLR-1::GFP was immunoprecipitated with anti-GFP polyclonal antibodies (a gift from J. Kaplan) and protein A- and protein G-Sepharose beads (GE Healthcare) at 4 °C overnight (first IP). Immunoprecipitations were washed with 50 mm HEPES (pH 7.7), 150 mm NaCl, and 1% Nonidet P-40, and GLR-1::GFP was eluted by incubation in SDS buffer twice at 95 °C for 5 min/elution. Eluates were pooled and diluted with 5 volumes of 50 mm Tris (pH 7.6), 10 mm magnesium acetate, 1% Nonidet P-40, and 0.1 mg/ml IgG-free BSA (Sigma-Aldrich) plus protease inhibitors. Finally, ubiquitin-conjugated GLR-1::GFP was immunoprecipitated from the diluted eluate using anti-ubiquitin polyclonal antibodies (a gift from J. Kaplan) and protein A- and protein G-Sepharose beads for 3 h at 4 °C (second IP). Beads were washed in Tris-magnesium acetate-Nonidet P-40 buffer and eluted in SDS sample buffer (125 mm Tris (pH 6.8), 20% glycerol, 5% SDS, and 200 mm DTT). GLR-1::GFP bands on immunoblots were detected by incubating the blots with anti-GFP monoclonal antibodies (Clontech, catalog no. JL-8), anti-mouse-HRP secondary antibodies (GE Healthcare), and Pierce Femto Super Signal chemiluminescence solution and exposing them to film (Kodak). The films were scanned, integrated band intensities were calculated using ImageJ, and the relative ratios of ubiquitinated GLR-1::GFP/total GLR-1::GFP were determined. Three independent membrane preparations for each strain were made and tested in IP experiments. For measurement of ubiquitin-GLR-1 conjugates, boxes were drawn around the entire area of the ubiquitin-GLR-1 conjugates.
Quantitative PCR
cDNA was reverse-transcribed from RNA using established protocols. Amplification reactions were run on a Stratagene real-time cycler, MX4000, using SYBR Green (Invitrogen) reaction mixture with Rox as a reference dye. Amplification of usp-46, wdr-20, and wdr-48 was specific, as monitored by agarose gel and melting curves. The relative abundance of each gene product for each genotype was normalized to actin using the ΔΔCt method. All amplifications were performed in triplicate using cDNA from two to three independent RNA isolations.
RESULTS
Identification of C. elegans WDR-20 and WDR-48
We showed previously that the DUB USP-46 regulates the abundance of the glutamate receptor GLR-1 in the VNC of C. elegans (21). To study how USP-46 activity is regulated in the nervous system, we decided to characterize proteins that were known to interact with USP-46 or its close homolog, USP-12. Two WD40-repeat proteins, WDR20 and WDR48, were of particular interest because mammalian WDR20 and WDR48 can bind to and activate USP-46 and its close homolog, USP12, in vitro (30–33). We identified homologs of both WDR20 and WDR48 in the C. elegans genome (GenBankTM accession numbers NM_001269255.1 and NM_065530.5, respectively). The C. elegans ORF C08B6.7 is 43% similar and 33% identical to human WDR-20, and the ORF F35G12.4 is 55% similar and 43% identical to human WDR48. C. elegans WDR-20 and WDR-48 are predicted to contain the same number and organization of WD40 repeats as their mammalian counterparts (Fig. 1A). C08B6.7 is also similar to mammalian DMWD (Dystrophia Myotonica, WD repeat containing), another WD40-repeat protein that interacts with USP12/46 (33). However, C08B6.7 is closer in homology to WDR20. Hereafter, we refer to C. elegans C08B6.7 as WDR-20 and F35G12.4 as WDR-48.
FIGURE 1.
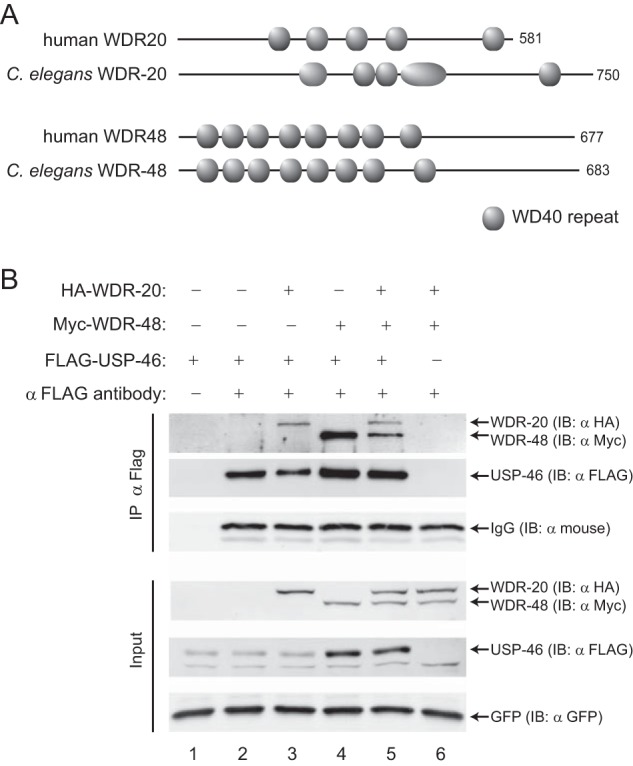
C. elegans homologs of WDR-20, WDR-48, and USP-46 interact in vitro. A, domain conservation between the human and C. elegans WD-40-repeat proteins WDR-20 and WDR-48. WD-40 domains were assigned using ProSite and SMART (String Matching Algorithms Research Tool). B, representative immunoblot (IB) analysis of the immunoprecipitation of HA·WDR-20 and Myc·WDR-48 by FLAG·USP-46 from HEK293T cells. FLAG·USP-46 was expressed alone (lanes 1 and 2) or in the presence of HA·WDR-20 and/or Myc·WDR-48 (lanes 3–5), and the resulting isolated complexes were assayed by immunoblot analysis. GFP is included as an expression and loading control for each condition. Controls are shown for antibody specificity (lane 1) and background binding (lane 6). Similar results were obtained in more than five independent experiments.
WDR-20 and WDR-48 Bind to and Activate USP-46 in Vitro
To test whether C. elegans WDR-20 and WDR-48 can interact with USP-46, we expressed and immunoprecipitated an epitope-tagged version of USP-46 (FLAG-USP-46) either alone or together with WDR-20 (HA·WDR-20) and/or WDR-48 (Myc·WDR-48) in HEK293T cells (Fig. 1B). We found that FLAG·USP-46 could coimmunoprecipitate with either HA·WDR-20 (Fig. 1B, lane 3) or Myc·WDR-48 (lane 4) or both together (lane 5). Neither HA·WDR-20 nor Myc·WDR-48 was immunoprecipitated by anti-FLAG antibody in the absence of FLAG·USP-46 (Fig. 1B, lane 6). Interestingly, expression of Myc·WDR-48 with or without HA·WDR-20 increased the amount of FLAG·USP-46 present in whole cell lysates (Fig. 1B, Input, compare lanes 1 and 2 with lanes 4 and 5), suggesting that WDR-48 may regulate the expression of USP-46. These results indicate that, similar to their mammalian counterparts, C. elegans WDR-20 and WDR-48 can interact with USP-46 in vitro.
We next tested whether C. elegans WDR-20 and WDR-48 could activate the catalytic activity of USP-46 using an in vitro DUB assay. The catalytic activity of a population of USP enzymes can be estimated by incubating the enzyme with HA epitope-tagged ubiquitin-vinyl methyl ester (HA-Ub-VME), a non-catalyzable ubiquitin suicide substrate (39). The active site cysteine of USP family DUBs forms an irreversible covalent bond with HA-Ub-VME, which results in an ∼9-kDa increase in the apparent molecular mass of the enzyme. We measured the effect of WDR-48 and WDR-20 on USP-46 activity by immunoprecipitating FLAG·USP-46 from HEK293T cells expressing FLAG·USP-46 alone (Fig. 2A, lanes 1 and 2), together with Myc·WDR-48 (lanes 3 and 4), His6·WDR-20 (lanes 7 and 8), or Myc·WDR-48 and His6·WDR-20 (lanes 5 and 6) and incubating the enzyme with HA-Ub-VME. Subsequent analysis by SDS-PAGE and immunoblotting for FLAG-USP-46 indicate that FLAG·USP-46 has a low level of activity (Fig. 2A, lane 2) and that this was unaffected by coexpression of Myc·WDR-48 or His6·WDR-20 alone (lanes 4 and 8). In contrast, coexpression of Myc·WDR-48 together with His6·WDR-20 increased the proportion of catalytically active USP-46 (Fig. 2A, lane 6). As expected, no increase in FLAG·USP-46 molecular weight was observed in the absence of HA-Ub-VME substrate (Fig. 2A, lanes 1, 3, 5, and 7). The low level of USP-46 activity observed in the absence of the WDR proteins (Fig. 2A, lane 2) could represent low intrinsic enzyme activity, as reported previously (30, 32), or could be attributed to the presence of endogenous human WDR20 and WDR48 in HEK293T cells. To ensure that the change in apparent molecular weight was due to the catalytic activity of USP-46, we repeated the experiment with a catalytically inactive version of USP-46 in which the catalytic cysteine was mutated to alanine (USP-46 (C>A)). Incubation of FLAG·USP-46(C>A) with HA-Ub-VME did not cause a change in molecular weight either in the absence or presence of the WDR proteins (Fig. 2B). These data show that C. elegans WDR-20 and WDR-48 can form a complex with USP-46 and enhance its catalytic activity, demonstrating that the regulation of USP-46 by these WD40-repeat proteins in vitro is conserved.
FIGURE 2.
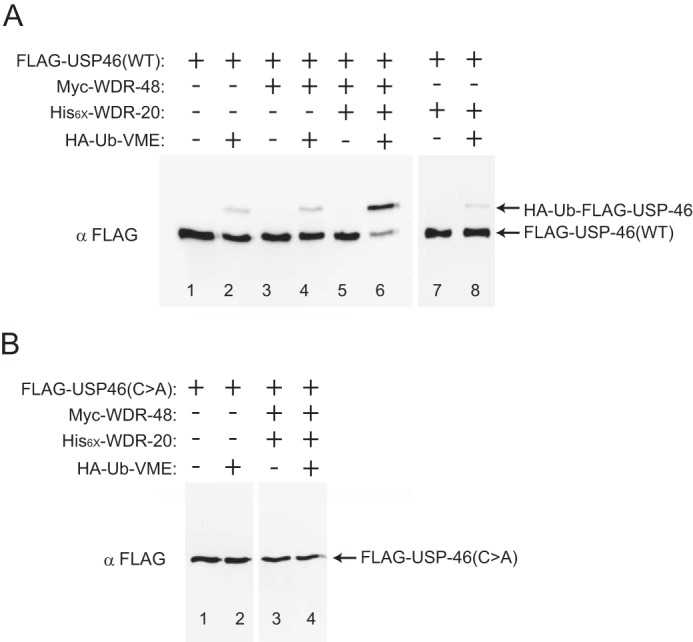
Stimulation of USP-46 catalytic activity by WDR-20 and WDR-48 in vitro. Representative immunoblot analysis showing the effect of Ub-VME on FLAG·USP-46 complexes isolated from HEK293T cells. A, the wild type (FLAG·USP-46(WT)) was transfected either alone or together with WDR-48 alone, WDR-20 and WDR-48 together, or WDR-20 alone. B, FLAG·USP-46(C>A) (catalytically inactive) was transfected either alone or together with WDR-20 and WDR-48. Wild-type (FLAG·USP-46(WT)) (A) or catalytically inactive (FLAG·USP-46(C>A)) (B) complexes were immunoprecipitated and incubated with or without HA-Ub-VME. After halting the reaction, adduct formation was assayed by immunoblotting against FLAG·USP-46. Similar results were obtained in more than three independent experiments.
WDR-20 and WDR-48 Promote GLR-1 Abundance in the VNC
We showed previously that USP-46 regulates the abundance of GLR-1 in the VNC of C. elegans (21). GLR-1 is most similar to mammalian AMPA-type glutamate receptors and is expressed in a subset of ventral nerve cord interneurons (40–43), where it localizes to sensory-interneuron and interneuron-interneuron synapses (3, 35). GLR-1 is required for several glutamate-dependent behaviors, including mechanosensory avoidance and spontaneous reversals during locomotion (41, 42, 44). We analyzed the abundance of GLR-1 in the VNC by imaging a GFP-tagged version of GLR-1 (GLR-1::GFP) expressed under the control of the glr-1 promoter (35). GLR-1::GFP localizes to puncta in the VNC that are closely apposed by presynaptic markers such as synaptobrevin (3, 35). GLR-1::GFP can rescue behavioral defects observed in glr-1 null mutants, indicating that GFP-tagged GLR-1 is functional (35).
usp-46 null mutants exhibit a decreased abundance of GLR-1::GFP in the VNC (21). This effect is dependent on USP-46 catalytic activity because the reduction in GLR-1::GFP can be rescued by expression of wild-type USP-46 but not catalytically inactive USP-46 (USP-46 (C>A)). We found that overexpression of USP-46 under control of the glr-1 promoter in either an usp-46 mutant or wild-type background does not increase the abundance of GLR-1::GFP in the VNC over wild-type levels (21) (Fig. 3, F and K). Overexpression of the usp-46 transgene was confirmed using quantitative PCR (∼9-fold increase versus the wild type). These results suggest that expression of USP-46 is not limiting and that USP-46 activity may be tightly controlled in vivo.
FIGURE 3.
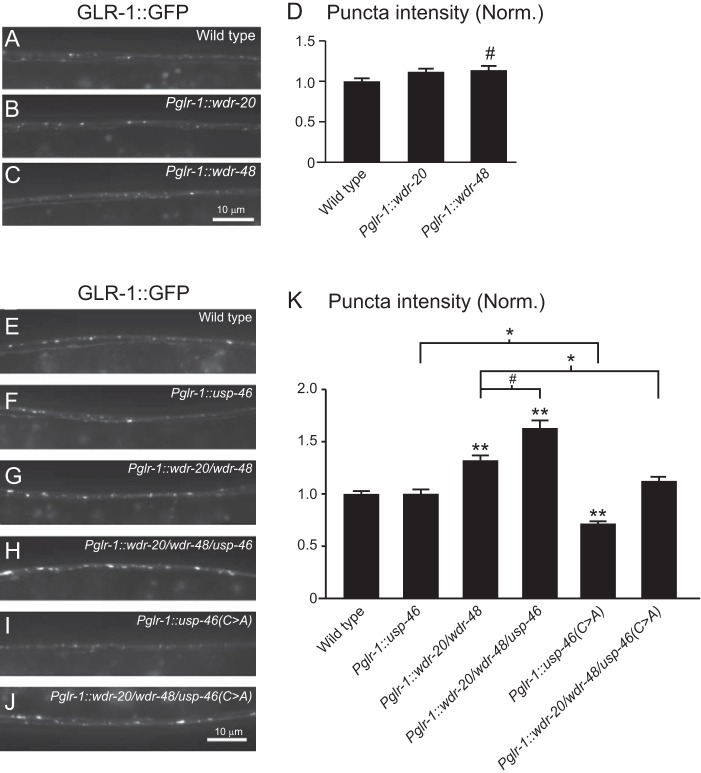
Effect of WDR-20, WDR-48, and USP-46 on GLR-1::GFP levels in vivo. A–C, representative images of the VNCs of L4 larval animals harboring a GLR-1::GFP transgene expressed under the control of the glr-1 promoter (nuIs24). The following genotypes are shown: wild-type (A), WDR-20 under control of the glr-1 promoter (pzEx230) (B), and WDR-48 under control of the glr-1 promoter (pzEx231) (C). D, quantification of GLR-1::GFP punctum intensities (normalized, norm.) for the strains pictured in A–C. Shown are the means ± S.E. for n = 38 (wild-type), n = 30 (Pglr-1::wdr-20), and n = 34 (Pglr-1::wdr-48). E–J, representative images of the VNCs of L4 larval animals harboring a GLR-1::GFP transgene expressed under the control of the glr-1 promoter (nuIs24). The following genotypes are shown: wild-type (E), usp-46 under the control of the glr-1 promoter (pzEx224) (F), wdr-20 and wdr-48 under the control of the glr-1 promoter (pzIs25) (G), wdr-20, wdr-48 and usp-46 under the control of the glr-1 promoter (pzIs25; pzEx224) (H), usp-46(C>A) under the control of the glr-1 promoter (pzEx222) (I), and wdr-20, wdr-48 and usp-46(C>A) under the control of the glr-1 promoter (pzIs25; pzEx222) (J). K, quantification of GLR-1::GFP punctum intensities (normalized) for the strains pictured in E–J. Shown are the means ± S.E. for n = 118 (wild-type), n = 56 (Pglr-1::usp-46), n = 76 (Pglr-1::wdr-20/wdr-48), n = 46 (Pglr-1::wdr-20/wdr-48/usp-46), n = 56 (Pglr-1::usp-46(C>A)), and n = 37 (Pglr-1::wdr-20/wdr-48/usp-46(C>A)). Values that differ significantly from the wild type (Tukey-Kramer) are indicated as follows: #, p < 0.05; *, p < 0.01; **, p < 0.001. Other comparisons are marked by brackets.
Because our biochemical data show that WDR-20 and WDR-48 can bind to and activate USP-46 in vitro (Figs. 1 and 2), we tested whether expression of the WDR proteins could affect the ability of USP-46 to regulate GLR-1 abundance in vivo. We found that expression of WDR-20 in interneurons under control of the glr-1 promoter (Pglr-1::wdr-20) had no effect on the amount of GLR-1::GFP in the VNC, as measured by the fluorescence intensity of GLR-1::GFP puncta (Fig. 3, B and D) (see “Experimental Procedures”). Expression of WDR-48 in interneurons (Pglr-1::wdr-48) resulted in a slight increase in GLR-1::GFP puncta fluorescence intensity (Fig. 3, C and D). However, coexpression of both WDR-20 and WDR-48 in interneurons (Pglr-1::wdr-20/wdr-48) resulted in a 32% increase in GLR-1::GFP puncta fluorescence intensity in the VNC (Fig. 3, G and K). This increase in GLR-1::GFP fluorescence was enhanced further by coexpression of USP-46 (Pglr-1::wdr-20/wdr-48/usp-46) (Fig. 3, H and K). Overexpression of usp-46 (∼2.2-fold), wdr-20 (∼3-fold), and wdr-48 (∼5-fold) was confirmed by quantitative PCR. The catalytic activity of USP-46 is required for the increase in GLR-1::GFP because this further increase was blocked by expression of the catalytically inactive form of USP-46 (USP-46 (C>A)) (Fig. 3, J and K). We also found that expression of USP-46 (C>A) alone in wild-type animals results in decreased GLR-1::GFP punctum intensity in the VNC (Fig. 3, I and K), suggesting that USP-46 (C>A) might act as a dominant negative, perhaps by disrupting endogenous WDR-20·WDR-48·USP-46 complexes. The effects of the WDR proteins on GLR-1 are relatively specific and unlikely to be due to changes in synapse development because overexpression of WDR-20, WDR-48, and USP-46 (pzIs22) had no effect on the density of the synaptic vesicle-associated protein synaptobrevin-GFP (average number of SNB-1::GFP puncta/10 μm ± S.E.: wild type, 2.03 ± 0.12; pzIs22, 2.12 ± 0.14; p > 0.05) or the synaptic PDZ (Postsynaptic density-95, Discs large, Zonula occludens-1) proteins LIN-10::GFP (35) (average number of LIN-10::GFP puncta/10 μm ± S.E.: wild type, 2.95 ± 0.15; pzIs22, 3.14 ± 0.13; p > 0.05) and MAGI-1::YFP (45) (average number of MAGI-1::YFP puncta/10 μm ± S.E.: wild-type, 3.00 ± 0.13; pzIs22, 3.17 ± 0.12; p > 0.05). From these data, we conclude that coexpression of WDR-20, WDR-48, and USP-46 increases GLR-1 levels in the VNC and that this effect is dependent on the catalytic activity of USP-46.
Expression of WDR-20, WDR-48, and USP-46 Promotes GLR-1 Deubiquitination
We next tested whether WDR-20, WDR-48, and USP-46 could regulate the overall expression levels of GLR-1::GFP. Immunoblot analysis of whole worm lysates indicate that total protein levels of GLR-1::GFP compared with tubulin were increased in animals overexpressing WDR-20 and WDR-48 (Pglr-1::wdr-20/wdr-48) (Fig. 4A, left panel) and in animals overexpressing WDR-20, WDR-48, and USP-46 (Pglr-1::wdr-20/wdr-48/usp-46) (right panel) under control of the glr-1 promoter.
FIGURE 4.
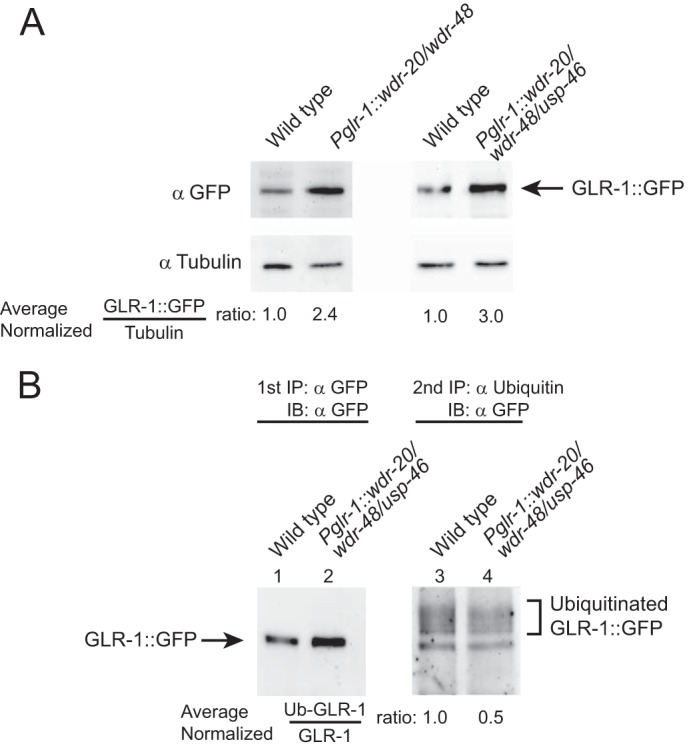
Overexpression of the WDR proteins in neurons results in increased levels of total GLR-1 protein and reduced levels of ubiquitin-GLR-1 conjugates. A, representative immunoblot analyses against GLR-1::GFP and tubulin (as a loading control) in lysates from mixed-stage animals harboring a GLR-1::GFP transgene expressed under the control of the glr-1 promoter (nuIs24). Lysates are from wild-type animals, animals expressing wdr-20 and wdr-48 under the control of the glr-1 promoter (pzIs25) (left panel), or wdr-20, wdr-48 and usp-46 under the control of the glr-1 promoter (pzIs22) (right panel). The numbers listed below the images indicate the average normalized ratio of GLR-1::GFP to tubulin control. Experiments were repeated at least twice with independent biological replicates. B, representative immunoprecipitation experiments detecting the relative amounts of ubiquitinated GLR-1::GFP in membranes prepared from mixed-stage populations of animals expressing the GLR-1::GFP transgene (nuIs24). Membranes were prepared from wild-type animals and animals expressing wdr-20, wdr-48, and usp-46 under the control of the glr-1 promoter (pzIs24). Total GLR-1::GFP was first immunoprecipitated using polyclonal anti-GFP antibodies (first IP, lanes 1 and 2) and either directly immunoblotted (IB) with anti-GFP antibodies (lanes 1 and 2) or subjected to a second, sequential immunoprecipitation of ubiquitin-GLR-1 conjugates using polyclonal anti-ubiquitin antibodies (second IP, lanes 3 and 4). Immunoblot analyses were used to detect immunoprecipitated proteins from each IP. Immunoprecipitated GLR-1::GFP from the first IP and ubiquitin-GLR-1::GFP conjugates immunoprecipitated in the second IP were detected with monoclonal anti-GFP antibodies. 120 times more material was loaded for the anti-GFP blot of the second IP than for the anti-GFP blot of the first IP. The numbers listed below the right panel indicate the average normalized ratio of the amount of ubiquitinated GLR-1 (right panel) to the amount of GLR-1::GFP isolated in the first IP (left panel) (Ub-GLR-1-GLR-1) from three independent experiments (p < 0.05).
Because USP-46 can deubiquitinate and stabilize GLR-1 (21), we tested whether the increase in GLR-1::GFP that we observed in animals overexpressing WDR-20, WDR-48, and USP-46 was due to a decrease in the amount of ubiquitinated receptor. Levels of ubiquitinated GLR-1 are very low in vivo (3), so we used a double IP protocol to enhance our isolation of ubiquitin-GLR-1 conjugates, as described previously (3, 12, 13, 21). We compared the levels of ubiquitinated GLR-1::GFP in wild-type controls and in animals overexpressing WDR-20, WDR-48, and USP-46 (Pglr-1::wdr-20/wdr-48/usp-46). Ubiquitinated GLR-1::GFP and the amount of GLR-1::GFP isolated in the first IP were detected by immunoblotting with anti-GFP antibodies (Fig. 4B). Compared with wild-type controls, we found that animals overexpressing WDR-20, WDR-48, and USP-46 have a decreased proportion of ubiquitin-GLR-1::GFP conjugates (Fig. 4B, right panel) relative to the amount of GLR-1::GFP isolated in the first IP (left panel). Saturation of the anti-ubiquitin antibodies with purified ubiquitin during the second IP eliminates detection of the ubiquitin-GLR-1 conjugates (data not shown) (3, 13, 21). These data suggest that overexpression of WDR-20, WDR-48, and USP-46 in the VNC results in increased deubiquitination of GLR-1::GFP in vivo.
WDR-20- and WDR-48-mediated Regulation of GLR-1::GFP in the VNC Is Partially Dependent on USP-46
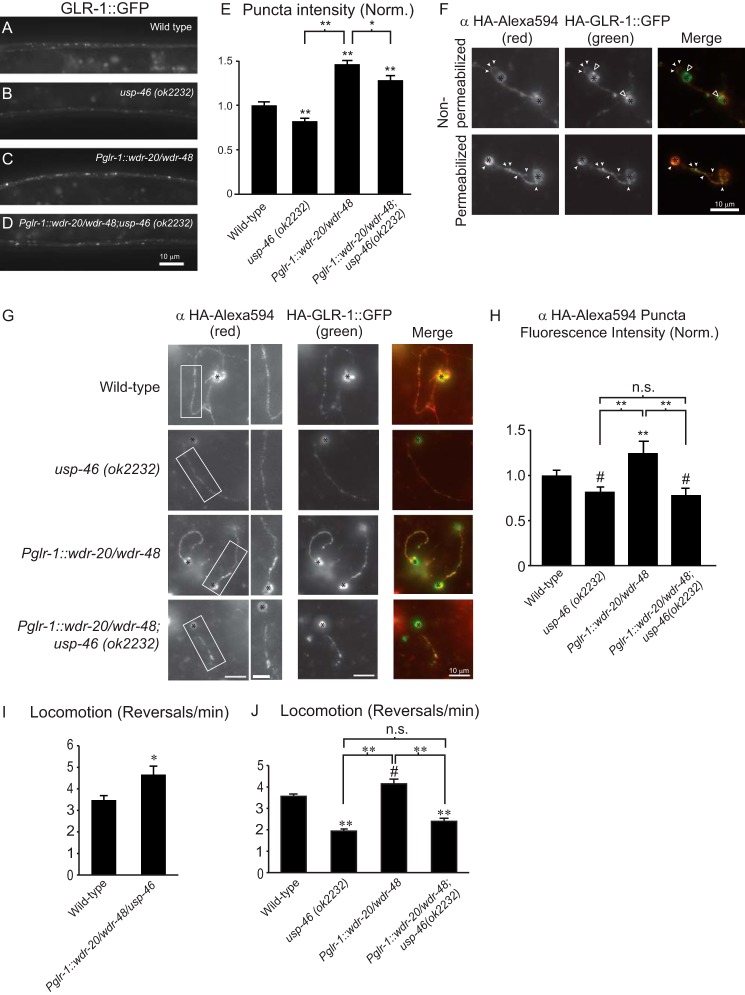
We next tested whether the increased accumulation of GLR-1::GFP in the VNC of animals overexpressing WDR-20 and WDR-48 was dependent on usp-46 by repeating our overexpression experiments in a usp-46 null mutant background (Fig. 5, A–E). As reported previously (21), we found that GLR-1::GFP punctum intensity decreased in the VNC of usp-46 null mutants compared with wild-type animals (Fig. 5, B and E). The increase in GLR-1::GFP punctum intensity observed in the VNC of animals overexpressing WDR-20 and WDR-48 (Pglr-1::wdr-20/wdr-48) (Fig. 5, C–E) was partially reduced in the usp-46 null mutant background (D and E). These data reveal that the WDR-20/WDR-48-induced increase in GLR-1::GFP in the VNC is partially dependent on usp-46.
FIGURE 5.
Analysis of GLR-1::GFP abundance and glr-1-dependent behaviors in animals expressing WDR-20 and WDR-48 in the presence or absence of endogenous usp-46. A–D, representative images of the VNCs of L4 larval animals harboring a GLR-1::GFP transgene expressed under the control of the glr-1 promoter (nuIs24). The following genotypes are shown: wild-type (A), usp-46 (ok2232) (B), wdr-20 and wdr-48 under the control of the glr-1 promoter (pzIs25) (C), and wdr-20 and wdr-48 under the control of the glr-1 promoter (pzIs25) in the usp-46 (ok2232) mutant background (D). E, quantification of GLR-1::GFP punctum intensities (normalized, norm.) for the strains pictured in A–D. Shown are the means ±S.E. for n = 31 (wild-type), n = 36 (usp-46 (ok2232)), n = 34 (Pglr-1::wdr-20/wdr-48), and n = 35 (Pglr-1::wdr-20/wdr-48; usp-46 (ok2232)). F, representative images of primary, glr-1-expressing neurons isolated from wild-type C. elegans expressing HA-GLR-1::GFP (pzIs12). Cells were fixed under non-permeabilizing conditions (top row) or permeabilized (bottom row) and then fixed prior to immunostaining with anti-HA-Alexa Fluor 594 antibodies. Images of anti-HA-Alexa Fluor 594 staining (left column) and GFP (center column) are shown separately or merged (right column). White arrowheads indicate examples of anti-HA-Alexa Fluor 594-stained surface GLR-1 puncta that colocalized with the GFP signal of HA·GLR-1::GFP. Open arrowheads indicate internal GLR-1::GFP in the non-permeabilized cell bodies. The black asterisks mark cell bodies. G, representative images of primary neurons from animals of the indicated genotypes. Cells were fixed and stained with anti-HA-Alexa Fluor 594 antibodies under non-permeabilizing conditions. Images of anti-HA-Alexa Fluor 594 immunostaining with higher magnification images of the boxed regions (left column) or GFP (center column) are shown separately or merged (right column). Black asterisks mark cell bodies. H, quantification of anti-HA-Alexa Fluor 594 punctum intensities relative to the interpunctal intensity of the neurite (normalized). Means ± S.E. are shown for individual neurites analyzed. n = 48 (wild type), n = 22 (usp-46 (ok2232)), n = 35 (Pglr-1::wdr-20/wdr-48), and n = 35 (Pglr-1::wdr-20/wdr-48; usp-46 (ok2232)). I, average number of reversals per minute for spontaneous locomotion assays performed on animals of the following genotypes: n = 14 (wild type) and n = 15 (Pglr-1::usp-46/wdr-20/wdr-48). J, average number of reversals per minute for spontaneous locomotion assays performed on worms of the following genotypes: n = 37 (wild type), n = 37 (usp-46 (ok2232)), n = 38 (Pglr-1::wdr-20/wdr-48), and n = 37 (Pglr-1::wdr-20/wdr-48;usp-46 (ok2232)). Values that differ significantly from the wild type (Tukey-Kramer test) are denoted above each bar. Other comparisons are marked by brackets. #, p < 0.05; *, p < 0.01; **, p < 0.001; n.s., no significant difference between the indicated strains (p ≥ 0.05). Error bars show S.E.
WDR-20 and WDR-48 Increase the Levels of GLR-1 at the Plasma Membrane in a usp-46-dependent Manner
GLR-1::GFP fluorescence in the VNC likely represents pools of receptors at the plasma membrane and in internal compartments. To measure the amount of GLR-1 at the cell surface, we cultured primary neurons from wild-type C. elegans expressing an N-terminal (extracellular) HA-tagged GLR-1::GFP (HA-GLR-1::GFP) and measured the levels of HA-GLR::GFP under permeabilized and non-permeabilized conditions, as described previously (36, 46). Immunostaining with anti-HA-Alexa Fluor 594 antibodies of non-permeabilized neurons revealed GLR-1 puncta (surface receptors) in the cell body and neurites that overlapped well (Fig. 5F, top row, solid white arrowheads), but not completely, with the GFP signal (surface and internal receptors) (Fig. 5F, top row; internal receptors are indicated by open arrowheads). As expected, permeabilization of the cultured neurons resulted in almost complete overlap of the anti-HA-Alexa Fluor 594 antibody and the GFP signal (Fig. 5F, bottom row, solid white arrowheads), consistent with previous reports (46).
We next tested whether expression of WDR-20 and WDR-48 could increase GLR-1 levels at the cell surface and, if so, whether this effect was dependent on usp-46. We generated primary neuronal cultures from transgenic animals expressing HA·GLR-1::GFP (pzIs12) in four different genetic backgrounds: wild type, usp-46 (ok2232) mutants, animals overexpressing WDR-20 and WDR-48 (Pglr-1::wdr-20/wdr-48), and animals overexpressing WDR-20 and WDR-48 in usp-46 mutants (Pglr-1::wdr-20/wdr-48; usp-46(ok2232)) (Fig. 5G). We measured the amount of anti-HA-Alexa Fluor 594 immunostaining and GFP signal under non-permeabilized conditions as described above. We found that the anti-HA-Alexa Fluor 594 signal was reduced in usp-46 mutants and increased in neurons expressing WDR-20 and WDR-48 compared with wild-type controls (Fig. 5, G and H). Interestingly, the WDR-20- and WDR-48-induced increase in anti-HA-Alexa Fluor 594 signal was completely blocked in usp-46 mutant neurons (Pglr-1::wdr-20/wdr-48; usp-46 (ok2232)) (Fig. 5, G and H). These data suggest that expression of WDR-20 and WDR-48 in neurons increases the surface levels of GLR-1 in an usp-46-dependent manner.
Overexpression of WDR-20 and WDR-48 Affects GLR-1-dependent Locomotion Behavior
Changes in the levels of GLR-1 at the cell surface, and at synapses in particular, might be expected to alter GLR-1 signaling and, consequently, affect GLR-1-dependent locomotion. Because WDR-20 and WDR-48 increase surface levels of GLR-1 in neuronal cultures, we tested whether overexpression of WDR-20 and WDR-48 affected the frequency of spontaneous reversals during locomotion. In C. elegans, spontaneous locomotion is characterized by periods of forward movement interspersed with brief periods of backward movement (44). The amount of glutamatergic transmission in VNC interneurons controls the frequency of these reversals. For example, animals with reduced glutamatergic signaling, such as those with mutations in the vesicular glutamate transporter eat-4/VGLUT or glr-1, exhibit decreased reversal frequencies compared with wild-type animals (3, 44, 47). In contrast, mutants with increased glutamatergic signaling have increased reversal frequencies (3, 13, 16, 37, 44). We found that overexpression of WDR-20, WDR-48, and USP-46 (Pglr-1::wdr-20/wdr-48/usp-46) increased spontaneous reversal frequencies compared with wild-type animals, which is consistent with increased glutamatergic signaling (Fig. 5I). Conversely, usp-46 (ok2232) mutants have decreased reversal frequencies (Fig. 5J), consistent with previous results (21). Interestingly, overexpression of WDR-20 and WDR-48 (Pglr-1::wdr-20/wdr-48) significantly increased reversal frequencies compared with controls, and this effect was completely blocked in usp-46 null mutants (Pglr-1::wdr-20/wdr-48; usp-46 (ok2232)) (Fig. 5J). These data indicate that overexpression of WDR-20 and WDR-48 proteins affect GLR-1-dependent locomotion behavior in an usp-46-dependent manner.
Effects of wdr-20 and wdr-48 Loss-of-function Mutants on GLR-1::GFP and GLR-1-dependent Locomotion Behavior
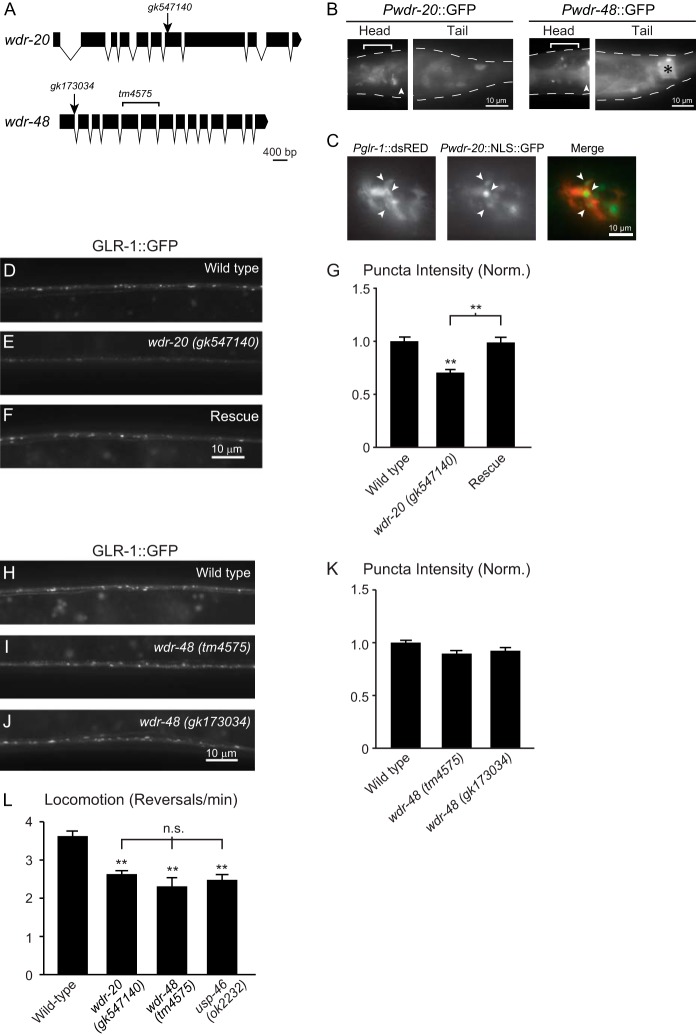
We next tested whether the WDR proteins are expressed in the nervous system by analyzing the expression pattern of wdr-20 and wdr-48 transcriptional reporters (i.e. GFP under control of the wdr-20 or wdr-48 promoters, respectively). We found that wdr-20 is expressed in several neurons in the head and tail (Fig. 6B, left panel), including several glr-1-expressing neurons on the basis of overlap with a glr-1 transcriptional reporter (Pglr-1::dsRED) (Fig. 6C), and in VNC processes (data not shown). Although expression of the wdr-48 transcriptional reporter was weak, we were able to detect GFP expression in several head neurons and cells in the tail, including the anal depressor cell (Fig. 6B, right panel). These transcriptional reporters may not encompass the full endogenous expression pattern of the wdr genes; however, these data suggest that both wdr-20 and wdr-48 are expressed in the nervous system.
FIGURE 6.
Effects of mutations in wdr-20 and wdr-48 on GLR-1::GFP accumulation and glr-1-dependent behavior. A, schematic of the structure of the coding and intronic sequences of wdr-20 (top panel) and wdr-48 (bottom panel). Locations of the mutations in gk547140, gk173034 and tm4575 are shown. Single nucleotide changes are indicated by arrows, and the deletion in tm4575 is denoted by a bracket. Schematics were generated using the Exon-Intron Graphic Maker (Wormweb). B, transgenic animals expressing the wdr-20 or wdr-48 transcriptional reporters Pwdr-20::GFP (left panel) or Pwdr-48::GFP (right panel), respectively, were imaged. The dotted white line outlines the worm body. Head neurons are marked by a white bracket. The posterior pharyngeal bulb is marked by a white arrowhead for orientation, and the anal depressor cell is marked by a black asterisk. C, transgenic animals expressing Pglr-1::dsRED and Pwdr-20::NLS::GFP were imaged. White arrowheads mark head neurons that express both reporters. D–F, representative images of the VNCs of L4 larval animals harboring a GLR-1::GFP transgene expressed under the control of the glr-1 promoter (nuIs25). The following genotypes are shown: wild type (D), wdr-20 (gk547140) (E), and wdr-20 (gk547140) expressing WDR-20 under the control of the glr-1 promoter (F, Rescue). The wdr-20 (gk547140) allele was backcrossed at least eight times prior to analysis. G, quantification of GLR-1::GFP punctum intensities (normalized, norm.) for the strains pictured in D--F. Shown are the means and S.E. for n = 30 (wild type), n = 27 (wdr-20 (gk547140)), and n = 27 (Rescue). H–J, representative images of the VNCs of L4 larval animals harboring a GLR-1::GFP transgene expressed under the control of the glr-1 promoter (nuIs25). The following genotypes are shown: wild type (H), wdr-48 (tm4575) (I), and wdr-48 (gk173034) (J). The wdr-48 (gk173034) allele was backcrossed at least six times prior to analysis. K, quantification of GLR-1::GFP punctum intensities (normalized) for the strains pictured in H–J. Shown are the means ± S.E. for n = 127 (wild type), n = 60 (wdr-48 (tm4575)), and n = 51 (wdr-48 (gk173034)). L, average number of reversals per minute for spontaneous locomotion assays performed on worms of the following genotypes: n = 28 (wild type), n = 16 (wdr-20 (gk547140)), n = 15 (wdr-48 (tm4575)), n = 13 (usp-46 (ok2232)). Values that differ significantly from the wild type (Tukey-Kramer test) are denoted above each bar. Other comparisons are marked by brackets. **, p < 0.001; n.s., no significant difference between the indicated strains (p ≥ 0.05). Error bars show S.E.
To determine whether WDR-20 and WDR-48 are required to maintain normal levels of GLR-1::GFP, we analyzed the distribution of GLR-1::GFP in the VNC of wdr-20 (gk547140) and wdr-48 (tm4575) loss-of-function mutants. The wdr-20 (gk547140) allele contains a non-sense mutation (G>A) that results in a premature stop codon at residue 288 of the WDR-20 protein. The wdr-48 (tm4575) allele deletes 132 amino acids from the WDR-48 coding region, disrupting WD40 repeat domains 5–7, and is predicted to represent a null allele because of the introduction of a premature stop codon downstream of the deletion (Fig. 6A). We found that wdr-20 mutant animals exhibited a 25% decrease in GLR-1::GFP punctum fluorescence intensity in the VNC (Fig. 6, E and G). This effect could be rescued by expression of wild-type wdr-20 cDNA in interneurons under control of the glr-1 promoter (Fig. 6, F and G, Rescue). These data indicate that wdr-20 functions in interneurons to maintain normal GLR-1::GFP levels in the VNC. In contrast, we observed no change in GLR-1::GFP punctum fluorescence intensity in the VNC of wdr-48 (tm4575) mutants compared with wild-type controls (Fig. 6, H, I, and K). Similarly, we found no alteration in GLR-1::GFP punctum intensity in another independent wdr-48 mutant, wdr-48 (gk173034) (Fig. 6, J and K). The wdr-48 (gk173034) allele contains a missense mutation (G>A) at a putative splice donor site of the first intron (Fig. 6A), creating alternative splice variants that ultimately result in premature stop codons (data not shown). The genetic lesions described above were verified through amplification and sequencing of cDNA and/or genomic DNA. These results show that wdr-20, but not wdr-48, is required for maintaining total levels of GLR-1::GFP in the VNC, suggesting that the WDR proteins regulate GLR-1 via distinct mechanisms.
Because GLR-1::GFP fluorescence intensity in the VNC likely represents both surface and internal pools of the receptor, and because our data indicate that overexpression of WDR-20 and WDR-48 increases surface levels of GLR-1 (Fig. 5G) and the rate of spontaneous reversals (I and J), we tested the effects of wdr-20 and wdr-48 loss-of-function mutation on locomotion behavior. Interestingly, we found that both wdr-20 and wdr-48 mutant animals exhibited decreased reversal frequencies, consistent with decreased glutamatergic signaling. In addition, the magnitude of this decrease was similar to that observed in usp-46 mutant animals (Fig. 6L). These data demonstrate that both wdr-20 and wdr-48 are required for normal GLR-1-dependent locomotion behavior. These data also suggests that, although wdr-48 mutants do not exhibit decreased total GLR-1::GFP levels in the VNC, these mutants likely have reduced surface levels or function of GLR-1 at the synapse. Our data suggest a model whereby WDR-20 and WDR-48 act together to regulate USP-46 activity, which, in turn, controls GLR-1 levels at the synapse.
DISCUSSION
Ubiquitin has emerged as a critical regulator of synapse development and function (48–50). Although the specific DUBs that function in the nervous system are beginning to be described (8, 51), the mechanisms that regulate DUB activity in vivo are understood poorly. A large scale proteomics study revealed that 36% of mammalian DUBs interact with WD40-repeat proteins (33). In particular, the WD40-repeat proteins WDR-20 and WDR-48/UAF-1 can interact with and stimulate the catalytic activity of USP12 and USP46 in vitro (30, 32, 33). In this study, we identified C. elegans homologs of WDR20 and WDR48. We showed that WDR-20 and WDR-48 can form a complex with USP-46 in heterologous cells and activate its catalytic activity in vitro (Figs. 1 and 2). Overexpression of WDR-20 and WDR-48 in neurons results in increased abundance of GLR-1 in the VNC, and this effect was further enhanced by coexpression of USP-46 but not by catalytically inactive USP-46 (C>A) (Fig. 3). Coexpression of WDR-20, WDR-48, and USP-46 results in an increase in total GLR-1 protein and a corresponding decrease in the levels of ubiquitin-GLR-1 conjugates (Fig. 4), suggesting that the WDR proteins stimulate USP-46 to deubiquitinate and stabilize GLR-1. In addition, overexpression of WDR-20 and WDR-48 increases the levels of GLR-1 on the cell surface in an usp-46-dependent manner (Fig. 5G). Importantly, overexpression of WDR-20 and WDR-48 affects GLR-1-dependent locomotion behavior in a manner that is consistent with increased glutamatergic signaling, and this effect is blocked by usp-46 mutation (Fig. 5J). Conversely, wdr-20 and wdr-48 loss-of-function mutants have decreased spontaneous locomotion reversals, which is similar to usp-46 null mutants and consistent with decreased glutamatergic signaling (Fig. 6L) (21). Together, our results show that the WD40-repeat proteins WDR-20 and WDR-48 function together with USP-46 to promote the stability and synaptic abundance of GLR-1 in vivo.
Although we found that the effect of overexpressing WDR-20 and WDR-48 on surface expression of GLR-1::GFP and on locomotion behavior was completely dependent on usp-46 (Fig. 5, G, H, and J), their effect on GLR-1::GFP levels in the VNC was only partially dependent on usp-46 (A–E). These data suggest that the WDR proteins likely act through USP-46 to regulate functional GLR-1 levels at the synaptic surface. In support of this idea, we found that, similar to usp-46 mutants, wdr-20 and wdr-48 loss-of-function mutants exhibit defects in locomotion behavior consistent with decreased glutamatergic signaling (Fig. 6L). In addition, although wdr-20 and usp-46 loss-of-function mutants have decreased levels of GLR-1::GFP in the VNC, wdr-48 loss-of-function mutants did not affect the abundance of GLR-1::GFP (Fig. 6, H–K). These data suggest that, although WDR-20 and WDR-48 may have differential effects on GLR-1 trafficking, both WDR-20 and WDR-48 regulate GLR-1 at the synaptic surface to impact locomotion behavior.
Because GLR-1::GFP fluorescence in the VNC represents both surface and internal pools of receptors, our data suggest that WDR-20 and WDR-48 may regulate internal pools of GLR-1 via an usp-46-independent mechanism. The WDR proteins may interact directly with GLR-1 to regulate receptor trafficking. Alternatively, WDR-20 and WDR-48 may activate another DUB to control GLR-1. Indeed, several other studies indicate that WDR-20 and WDR-48 or their homologs can function together with other DUBs. For example, mammalian WDR48/UAF-1 can bind and stimulate USP1 activity (30, 33, 52), and yeast Bun107/WDR20 and Bun62/WDR48 both interact with Ubp15, the S. pombe homolog of mammalian USP7 (24). Thus, USP-46 may function together with other DUBs downstream of WDR-20 and WDR-48 to control GLR-1 levels. Consistent with this idea, USP-46 and its homologs have been shown to function together with other DUBs in several systems. USP-46 functions together with USP-47 and MATH-33/USP7 to regulate cell polarity in the C. elegans embryo (26), and the USP46 homolog Ubp9 functions redundantly with several other DUBs to regulate endocytosis and cell polarity in S. pombe (24) and mitochondrial function in S. cerevisiae (23). Further genetic and biochemical analysis will be required to identify the usp-46-independent mechanisms by which the WDR proteins regulate GLR-1.
Our data are consistent with other in vitro studies showing that both WDR20 and WDR-48/UAF1 are required for maximal activation of mammalian USP12 and USP46 (32). A recent study on ubiquitin chain specificity of DUBs showed that WDR48-USP46 complexes have a preference for cleaving lysine 6- and lysine 11-linked ubiquitin chains (31). It will be interesting in the future to test whether different combinations of activator proteins may also alter the ubiquitin chain specificity of USP46. It is possible that WDR20 may not only fully activate the catalytic activity of the DUB but may also determine its ubiquitin chain specificity. Regarding subcellular localization, we showed previously that USP-46 partially colocalizes with the endosomal marker RAB5 in the cell body and VNC (21). In yeast, the WDR proteins Bun107 and Bun62 can regulate the subcellular localization of Ubp9p (24), and it is possible that WDR-20 and WDR-48 regulate the subcellular localization of USP-46 in neurons. However, we found that mCherry-tagged WDR-20 and WDR-48 (under control of the glr-1 promoter) were predominantly diffuse throughout the cell body and VNC (data not shown). Because the WDR proteins can bind and activate USP-46 to regulate GLR-1 levels in the VNC, our localization data suggest that the WDR proteins may either transiently localize and activate USP-46, or, perhaps, even low levels of WDR proteins at the synapse are sufficient to activate the DUB. Alternatively, the mCherry-tagged WDR proteins may not accurately reflect the subcellular distribution of the endogenous proteins. Although other physiological targets for WDR20 have not yet been identified, activation of USP46 or USP12 by WDR48 toward specific substrates has been described in Xenopus laevis, S. cerevisiae, and mammalian cell culture (22, 23, 27). In light of the conservation of WDR-20 between species, it is likely that future studies in diverse systems and cell types will uncover additional targets for WDR20 in conjunction with WDR48.
Concluding Remarks
Regulation of AMPA receptor levels at the synapse is an important mechanism for controlling synaptic strength, and much research has focused on understanding the proteins involved in regulating endo- and exocytosis of GluRs at the synapse. After endocytosis, AMPA receptors can be targeted to the lysosome for degradation or recycled back to the synaptic cell surface in an activity-dependent manner (53, 54). However, the mechanisms that control this critical sorting decision are understood poorly. We showed previously that USP-46 deubiquitinates GLR-1 and prevents its degradation in the lysosome (21). Here we identify WDR-20 and WDR-48 as regulators of USP-46 activity and show that expression of both WDR proteins in neurons increases the abundance of GLR-1 in vivo. Our data suggest that controlling the expression levels of WDR-20 and WDR-48 may provide a mechanism to regulate USP-46 DUB activity and, consequently GLR-1 stability and function at synapses. Future studies will be focused on identifying upstream signals, such as changes in synaptic activity, that might regulate WDR-20 or WDR-48 expression and/or localization. Furthermore, WDR protein regulation of USP46 and GluRs may be conserved in the mammalian nervous system because the mammalian homologs of WDR-20, WDR-48, and USP-46 have highly overlapping patterns of expression in the mouse brain, including regions such as the hippocampus that are involved in learning and memory (Allen Brain Atlas). Because 36% of mammalian DUBs can interact with WD40-repeat family proteins (33), understanding how WDR-20 and WDR-48 regulate USP-46 may be informative for understanding the regulatory mechanisms that control a large number of other DUBs.
Acknowledgments
We thank Shohei Mitani (National Bioresource), the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN), and the Million Mutation Project (Simon Fraser University) for strains. We thank Jennifer Kowalski and members of the Juo laboratory for discussions and critical reading of this manuscript. We thank Villu Maricq (University of Utah) and Larry Feig (Tufts University) for plasmids, and Josh Kaplan (Massachusetts General Hospital) and Lars Dreier (University of California Los Angeles) for antibodies. We also thank Steven Garafalo for help with anti-HA antibody labeling experiments, members of the Gill laboratory (Tufts University) for reagents and help with tissue culture experiments, and the Herman laboratory (Tufts University) for reagents.
This work was supported, in whole or in part, by National Institutes of Health Grants R01NS059953 (to P. J.) and 5K12GM074869 (Training in Education and Critical Research Skills Postdoctoral Program) (to C. L. D.). This work was also supported by a Natalie V. Zucker Research Grant (to C. L. D.) and the Tufts Center for Neuroscience Research Grant P30NS047243.
- GluR
- glutamate receptor
- DUB
- deubiquitinating enzyme
- WDR
- WD40-repeat
- VNC
- ventral nerve cord
- IP
- immunoprecipitation
- Ub
- ubiquitin
- VME
- vinyl methyl ester
- USP
- ubiquitin-specific protein.
REFERENCES
- 1. Shepherd J. D., Huganir R. L. (2007) The cell biology of synaptic plasticity. AMPA receptor trafficking. Annu. Rev. Cell Dev. Biol. 23, 613–643 [DOI] [PubMed] [Google Scholar]
- 2. Schwarz L. A., Patrick G. N. (2012) Ubiquitin-dependent endocytosis, trafficking and turnover of neuronal membrane proteins. Mol. Cell Neurosci. 49, 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burbea M., Dreier L., Dittman J. S., Grunwald M. E., Kaplan J. M. (2002) Ubiquitin and AP180 regulate the abundance of GLR-1 glutamate receptors at postsynaptic elements in C. elegans. Neuron 35, 107–120 [DOI] [PubMed] [Google Scholar]
- 4. Nijman S. M., Luna-Vargas M. P., Velds A., Brummelkamp T. R., Dirac A. M., Sixma T. K., Bernards R. (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786 [DOI] [PubMed] [Google Scholar]
- 5. Clague M. J., Coulson J. M., Urbé S. (2012) Cellular functions of the DUBs. J. Cell Sci. 125, 277–286 [DOI] [PubMed] [Google Scholar]
- 6. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 7. Komander D., Clague M. J., Urbé S. (2009) Breaking the chains. Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 [DOI] [PubMed] [Google Scholar]
- 8. Kowalski J. R., Juo P. (2012) The role of deubiquitinating enzymes in synaptic function and nervous system diseases. Neural Plast. 2012, 892749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chun D. K., McEwen J. M., Burbea M., Kaplan J. M. (2008) UNC-108/Rab2 regulates postendocytic trafficking in Caenorhabditis elegans. Mol. Biol. Cell 19, 2682–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Colledge M., Snyder E. M., Crozier R. A., Soderling J. A., Jin Y., Langeberg L. K., Lu H., Bear M. F., Scott J. D. (2003) Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron 40, 595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Patrick G. N., Bingol B., Weld H. A., Schuman E. M. (2003) Ubiquitin-mediated proteasome activity is required for agonist-induced endocytosis of GluRs. Curr. Biol. 13, 2073–2081 [DOI] [PubMed] [Google Scholar]
- 12. Dreier L., Burbea M., Kaplan J. M. (2005) LIN-23-mediated degradation of beta-catenin regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Neuron 46, 51–64 [DOI] [PubMed] [Google Scholar]
- 13. Juo P., Kaplan J. M. (2004) The anaphase-promoting complex regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Curr. Biol. 14, 2057–2062 [DOI] [PubMed] [Google Scholar]
- 14. Park E. C., Glodowski D. R., Rongo C. (2009) The ubiquitin ligase RPM-1 and the p38 MAPK PMK-3 regulate AMPA receptor trafficking. PloS ONE 4, e4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Roessel P., Elliott D. A., Robinson I. M., Prokop A., Brand A. H. (2004) Independent regulation of synaptic size and activity by the anaphase-promoting complex. Cell 119, 707–718 [DOI] [PubMed] [Google Scholar]
- 16. Schaefer H., Rongo C. (2006) KEL-8 is a substrate receptor for CUL3-dependent ubiquitin ligase that regulates synaptic glutamate receptor turnover. Mol. Biol. Cell 17, 1250–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fu A. K., Hung K. W., Fu W. Y., Shen C., Chen Y., Xia J., Lai K. O., Ip N. Y. (2011) APC(Cdh1) mediates EphA4-dependent downregulation of AMPA receptors in homeostatic plasticity. Nat. Neurosci. 14, 181–189 [DOI] [PubMed] [Google Scholar]
- 18. Lin A., Hou Q., Jarzylo L., Amato S., Gilbert J., Shang F., Man H. Y. (2011) Nedd4-mediated AMPA receptor ubiquitination regulates receptor turnover and trafficking. J. Neurochem. 119, 27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lussier M. P., Herring B. E., Nasu-Nishimura Y., Neutzner A., Karbowski M., Youle R. J., Nicoll R. A., Roche K. W. (2012) Ubiquitin ligase RNF167 regulates AMPA receptor-mediated synaptic transmission. Proc. Natl. Acad. Sci. U.S.A. 109, 19426–19431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schwarz L. A., Hall B. J., Patrick G. N. (2010) Activity-dependent ubiquitination of GluA1 mediates a distinct AMPA receptor endocytosis and sorting pathway. J. Neurosci. 30, 16718–16729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kowalski J. R., Dahlberg C. L., Juo P. (2011) The deubiquitinating enzyme USP-46 negatively regulates the degradation of glutamate receptors to control their abundance in the ventral nerve cord of Caenorhabditis elegans. J. Neurosci. 31, 1341–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Joo H. Y., Jones A., Yang C., Zhai L., Smith A. D., 4th, Zhang Z., Chandrasekharan M. B., Sun Z. W., Renfrow M. B., Wang Y., Chang C., Wang H. (2011) Regulation of histone H2A and H2B deubiquitination and Xenopus development by USP12 and USP46. J. Biol. Chem. 286, 7190–7201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kanga S., Bernard D., Mager-Heckel A. M., Erpapazoglou Z., Mattiroli F., Sixma T. K., Léon S., Urban-Grimal D., Tarassov I., Haguenauer-Tsapis R. (2012) A deubiquitylating complex required for neosynthesis of a yeast mitochondrial ATP synthase subunit. PloS ONE 7, e38071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kouranti I., McLean J. R., Feoktistova A., Liang P., Johnson A. E., Roberts-Galbraith R. H., Gould K. L. (2010) A global census of fission yeast deubiquitinating enzyme localization and interaction networks reveals distinct compartmentalization profiles and overlapping functions in endocytosis and polarity. PLoS Biol. 8, e1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li X., Stevens P. D., Yang H., Gulhati P., Wang W., Evers B. M., Gao T. (2013) The deubiquitination enzyme USP46 functions as a tumor suppressor by controlling PHLPP-dependent attenuation of Akt signaling in colon cancer. Oncogene 32, 471–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McCloskey R. J., Kemphues K. J. (2012) Deubiquitylation machinery is required for embryonic polarity in Caenorhabditis elegans. PLoS Genet. 8, e1003092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moretti J., Chastagner P., Liang C. C., Cohn M. A., Israël A., Brou C. (2012) The ubiquitin-specific protease 12 (USP12) is a negative regulator of Notch signaling acting on Notch receptor trafficking toward degradation. J. Biol. Chem. 287, 29429–29441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Imai S., Mamiya T., Tsukada A., Sakai Y., Mouri A., Nabeshima T., Ebihara S. (2012) Ubiquitin-specific peptidase 46 (Usp46) regulates mouse immobile behavior in the tail suspension test through the GABAergic system. PloS ONE 7, e39084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tomida S., Mamiya T., Sakamaki H., Miura M., Aosaki T., Masuda M., Niwa M., Kameyama T., Kobayashi J., Iwaki Y., Imai S., Ishikawa A., Abe K., Yoshimura T., Nabeshima T., Ebihara S. (2009) Usp46 is a quantitative trait gene regulating mouse immobile behavior in the tail suspension and forced swimming tests. Nat. Genet. 41, 688–695 [DOI] [PubMed] [Google Scholar]
- 30. Cohn M. A., Kee Y., Haas W., Gygi S. P., D'Andrea A. D. (2009) UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J. Biol. Chem. 284, 5343–5351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Faesen A. C., Luna-Vargas M. P., Geurink P. P., Clerici M., Merkx R., van Dijk W. J., Hameed D. S., El Oualid F., Ovaa H., Sixma T. K. (2011) The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem. Biol. 18, 1550–1561 [DOI] [PubMed] [Google Scholar]
- 32. Kee Y., Yang K., Cohn M. A., Haas W., Gygi S. P., D'Andrea A. D. (2010) WDR20 regulates activity of the USP12 x UAF1 deubiquitinating enzyme complex. J. Biol. Chem. 285, 11252–11257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rongo C., Whitfield C. W., Rodal A., Kim S. K., Kaplan J. M. (1998) LIN-10 is a shared component of the polarized protein localization pathways in neurons and epithelia. Cell 94, 751–759 [DOI] [PubMed] [Google Scholar]
- 36. Strange K., Christensen M., Morrison R. (2007) Primary culture of Caenorhabditis elegans developing embryo cells for electrophysiological, cell biological and molecular studies. Nat. Prot. 2, 1003–1012 [DOI] [PubMed] [Google Scholar]
- 37. Juo P., Harbaugh T., Garriga G., Kaplan J. M. (2007) CDK-5 regulates the abundance of GLR-1 glutamate receptors in the ventral cord of Caenorhabditis elegans. Mol. Biol. Cell 18, 3883–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cheeseman I. M., Desai A. (2005) A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE 2005, pl1. [DOI] [PubMed] [Google Scholar]
- 39. Ovaa H., Kessler B. M., Rolén U., Galardy P. J., Ploegh H. L., Masucci M. G. (2004) Activity-based ubiquitin-specific protease (USP) profiling of virus-infected and malignant human cells. Proc. Natl. Acad. Sci. U.S.A. 101, 2253–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brockie P. J., Madsen D. M., Zheng Y., Mellem J., Maricq A. V. (2001) Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC-42. J. Neurosci. 21, 1510–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hart A. C., Sims S., Kaplan J. M. (1995) Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature 378, 82–85 [DOI] [PubMed] [Google Scholar]
- 42. Maricq A. V., Peckol E., Driscoll M., Bargmann C. I. (1995) Mechanosensory signalling in C. elegans mediated by the GLR-1 glutamate receptor. Nature 378, 78–81 [DOI] [PubMed] [Google Scholar]
- 43. Wang R., Mellem J. E., Jensen M., Brockie P. J., Walker C. S., Hoerndli F. J., Hauth L., Madsen D. M., Maricq A. V. (2012) The SOL-2/Neto auxiliary protein modulates the function of AMPA-subtype ionotropic glutamate receptors. Neuron 75, 838–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zheng Y., Brockie P. J., Mellem J. E., Madsen D. M., Maricq A. V. (1999) Neuronal control of locomotion in C. elegans is modified by a dominant mutation in the GLR-1 ionotropic glutamate receptor. Neuron 24, 347–361 [DOI] [PubMed] [Google Scholar]
- 45. Emtage L., Chang H., Tiver R., Rongo C. (2009) MAGI-1 modulates AMPA receptor synaptic localization and behavioral plasticity in response to prior experience. PloS ONE 4, e4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zheng Y., Mellem J. E., Brockie P. J., Madsen D. M., Maricq A. V. (2004) SOL-1 is a CUB-domain protein required for GLR-1 glutamate receptor function in C. elegans. Nature 427, 451–457 [DOI] [PubMed] [Google Scholar]
- 47. Brockie P. J., Mellem J. E., Hills T., Madsen D. M., Maricq A. V. (2001) The C. elegans glutamate receptor subunit NMR-1 is required for slow NMDA-activated currents that regulate reversal frequency during locomotion. Neuron 31, 617–630 [DOI] [PubMed] [Google Scholar]
- 48. Bingol B., Sheng M. (2011) Deconstruction for reconstruction. The role of proteolysis in neural plasticity and disease. Neuron 69, 22–32 [DOI] [PubMed] [Google Scholar]
- 49. Tai H. C., Schuman E. M. (2008) Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 9, 826–838 [DOI] [PubMed] [Google Scholar]
- 50. Yi J. J., Ehlers M. D. (2007) Emerging roles for ubiquitin and protein degradation in neuronal function. Pharmacol. Rev. 59, 14–39 [DOI] [PubMed] [Google Scholar]
- 51. Todi S. V., Paulson H. L. (2011) Balancing act. Deubiquitinating enzymes in the nervous system. Trends Neurosci. 34, 370–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cohn M. A., Kowal P., Yang K., Haas W., Huang T. T., Gygi S. P., D'Andrea A. D. (2007) A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell 28, 786–797 [DOI] [PubMed] [Google Scholar]
- 53. Ehlers M. D. (2000) Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron 28, 511–525 [DOI] [PubMed] [Google Scholar]
- 54. Lin J. W., Ju W., Foster K., Lee S. H., Ahmadian G., Wyszynski M., Wang Y. T., Sheng M. (2000) Distinct molecular mechanisms and divergent endocytotic pathways of AMPA receptor internalization. Nat. Neurosci. 3, 1282–1290 [DOI] [PubMed] [Google Scholar]