Background: The regulation and function of USP19 is unknown.
Results: We identify Hsp90 as a USP19 interactor and regulator. USP19 is predominantly localized in the cytosol despite having a transmembrane domain.
Conclusion: USP19 is a Hsp90-regulated deubiquitinase dispensable for ERAD.
Significance: The study reveals a novel means of DUB regulation involving chaperone association and membrane integration.
Keywords: Deubiquitination, ER-associated Degradation, Hsp90, Membrane Proteins, Ubiquitin, USP19, Tail-anchored Protein Biogenesis
Abstract
Deubiquitinating enzymes (DUBs) regulate various cellular processes ranging from protein degradation to cellular signaling. USP19, the only DUB containing a carboxyl-terminal transmembrane domain, was proposed to function in endoplasmic reticulum-associated degradation (ERAD). Here we characterize the function and regulation of USP19. We identify Hsp90 as a specific partner that binds the catalytic domain of USP19 to promote substrate association. Intriguingly, although overexpressed USP19 interacts with Derlin-1 and other ERAD machinery factors in the membrane, endogenous USP19 is mostly in the cytosol where it binds Hsp90. Accordingly, we detect neither interaction of endogenous USP19 with Derlin-1 nor significant effect on ERAD by USP19 depletion. The USP19 transmembrane domain appears to be partially stabilized in the cytosol by an interaction with its own catalytic domain, resulting in auto-inhibition of its deubiquitinating activity. These results clarify the role of USP19 in ERAD and suggest a novel DUB regulation that involves chaperone association and membrane integration. Moreover, our study indicates that the localization of tail-anchored membrane proteins can be subject to regulation in cells.
Introduction
In eukaryotic cells, ubiquitination, a powerful post-translational modification, regulates a plethora of cellular processes. Ubiquitination occurs when the small polypeptide ubiquitin is conjugated to a lysine or serine/threonine residues in a substrate protein, which often alters the fate of the modified protein (1, 2). This reaction requires three enzymes, a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3) (3). Ubiquitination is reversible due to the presence of a large family of deubiquitinases (DUBs)2 in cells, which remove ubiquitin conjugates from substrates and disassemble them. In humans, there are ∼100 such DUBs grouped into 5 subfamilies (4). The USP family is the largest DUB subfamily with more than 50 members. Given that DUBs are essentially proteases that cleave isopeptide bonds in cells, it is anticipated that their activities are highly regulated by either cofactors, post-translational modifications, or subcellular localizations (5).
Most DUBs are soluble proteins residing in either cytosol or nucleus with the exception of USP19, which contains a C-terminal transmembrane domain (6). It was previously demonstrated that overexpressed USP19 uses its transmembrane domain to localize itself in the membrane of the endoplasmic reticulum (ER), where it was proposed to function in ER-associated protein degradation (ERAD) pathway (6). ERAD is an essential protein quality control mechanism that eliminates misfolded proteins from the ER of eukaryotic cells. In this process, misfolded proteins are selectively retained by chaperones and then retrotranslocated across the ER membrane via a large membrane protein complex containing the ubiquitin ligase Hrd1 and other cofactors such as SEL1L, Derlin1, UbxD8, HERP. Once emerging into the cytosol, the ERAD substrates are ubiquitinated and the ubiquitinated products are then pulled out of the membrane by the p97 ATPase, which hands substrates over to the proteasome for degradation (7). USP19 overexpression was shown to promote deubiquitination of ERAD substrates to stabilize them (6). In addition, USP19 also seems to regulate the stability of many cellular proteins including the cell cycle factor KPC1, the apoptosis regulators c-IAP1 and c-IAP2, and the hypoxia-inducible factor 1α (8–10). However, whether and how USP19 activity is regulated in cells are unknown.
In this study, we characterize the protein interaction network of USP19, which reveals USP19 as the first DUB regulated by the heat shock protein Hsp90. We show that Hsp90 promotes the DUB activity of USP19 via enhancing substrate recognition. Biochemical studies further demonstrate that endogenous USP19 is primarily localized in the cytosol despite carrying a transmembrane domain. The transmembrane domain of USP19 appears to be packed onto its catalytic domain, resulting in auto-inhibition of USP19 deubiquitinating activity. Our data do not support the previously proposed function of USP19 in ERAD, but the study reveals an unexpected mode of DUB regulation and a previously unknown link between a component of the ubiquitin proteasome system and Hsp90, a major anti-cancer drug target.
EXPERIMENTAL PROCEDURES
Cells, Plasmids, and Other Reagents
All cell lines were obtained from ATCC and cultured under the standard conditions using DMEM medium. Plasmids expressing FLAG-tagged USP19, USP7, USP13, USP5 were described previously (11). To express the various USP19 mutant expressing plasmids, we amplified the DNA fragments and cloned these DNA fragment between the SalI and NotI sites of the pRK-FLAG vector. NHK, NHK QQQ, and TTR D18G plasmids were generously provided by John Christianson (University of Oxford). USP19 siRNAs were purchased from Invitrogen. TransIT®-293 (Mirus) was used for plasmid transfection. Antibodies used are FLAG (M2, Sigma), Hsp90 (4F10 and F-8, Santa Cruz Biotechnology), Hsp70/HSC70 (Stressgen), SEL1L (Sigma), UbxD8 (Proteintech), Tom20 (Santa Cruz Biotechnology). The polyclonal anti-USP19 antibody was generated by immunizing rabbits with recombinant protein containing 1 to 494 amino acid residues of human USP19. The USP19 antibody was further purified by affinity chromatography using immobilized GST-USP19 1–494. The USP19 transmembrane peptide (YFVLGTVAALVALVLNVFYPLVSQSRWR) was purchased from Elim Biopharmaceuticals. Geldanamycin was purchased from Sigma.
Immunoblotting
Cells were lysed in the Nonidet P-40 lysis buffer containing 50 mm Tris-HCl pH 7.4, 150 mm sodium chloride, 2 mm magnesium chloride, 0.5% Nonidet P-40, and a protease inhibitor mixture (12). Cell extracts were subject to centrifugation to remove insoluble materials. Immunoblotting was performed according to the standard protocol. Fluorescence-labeled secondary antibodies (Rockland, MD) were used for detection. The fluorescent bands were imaged and quantified by a LI-COR Odyssey infrared imager using the software provided by the manufacture.
Protein Purification
To purify deubiquitinating enzymes, HEK293 cells grown in 10-cm culture dishes were transfected with the individual DUB expressing plasmid. Cells were propagated for 72 h prior to lysis in a buffer containing 1% CHAPS, 1% Triton X-100, 50 mm Tris-HCl, pH 7.4, 150 mm sodium chloride, 2 mm magnesium chloride, 1 mm EDTA, and a protease inhibitor mixture. The cleared cell extract was incubated with FLAG-agarose beads (Sigma) and the bound materials were extensively washed with a buffer containing 0.1% CHAPS, 0.1% Triton X-100, 50 mm Tris-HCl, pH 7.4, 150 mm sodium chloride, 2 mm magnesium chloride, 1 mm EDTA. The bound materials were eluted using 0.2 mg/ml FLAG peptide (Sigma) in 25 mm Hepes pH 7.2, 115 mm potassium acetate, 5 mm sodium acetate, 2.5 mm magnesium chloride. For purifications under the reducing condition, the lysis and wash buffers also contain 1 mm DTT and the elution buffer contains 0.5 mm DTT. Quantitative immunoblotting was performed using known concentrations of FLAG-tagged ubiquitin or bovine serum albumin (BSA) as a reference to determine the concentrations for the purified proteins.
In Vitro DUB Assays
The activity of purified deubiquitinases was determined by detecting the increase in fluorescence upon cleavage of Ubiquitin-AFC, as described previously (13). Briefly, purified DUBs (∼200 nm) were added individually to 200 μl of deubiquitinating assay buffer (50 mm Tris-HCl pH 7.4, 20 mm potassium chloride, 5 mm magnesium chloride including 0.5 μm Ub-AFC), and incubated at 37 °C. The fluorescence intensity was measured using an Aminco Bowman Luminescence spectrometer with excitation and emission wavelengths set at 400 and 505 nm, respectively. Data obtained were plotted and analyzed using Kaleidagraph v4.0 (Synergy Software). In some experiments, we used K48-linked di-ubiquitin as a model substrate to determine DUB activity. In this case, 1 μg of di-ubiquitin was incubated with ∼500 nm deubiquitinase in 20 μl of DUB assay buffer. The cleavage of di-ubiquitin was monitored by SDS-PAGE gel followed by Coomassie blue staining.
Biochemical Fractionation
To fractionate cells, cells washed with an ice cold phosphate saline buffer were treated for 10 min on ice with a hypotonic buffer containing 20 mm Hepes pH 7.2, 10 mm potassium chloride, 1 mm magnesium chloride, 1 mm DTT. After incubation, cells were passed through a Dounce homogenizer equipped with a tight pestle. Cell lysis was confirmed by staining with Trypan blue dye. The cells were then centrifuged at 1000 × g for 10 min to remove the nuclei. The supernatant fractions were further fractionated by centrifugation at 16,200 × g for 10 min at 4 °C. The resulting supernatant cytosol and membrane pellet fractions were analyzed by immunoblotting.
RESULTS
Interaction of USP19 with Hsp90
To study the function of USP19, we wished to identify its interaction partner(s). We transiently expressed FLAG-tagged wild type USP19 in HEK293 cells and prepared cell extract. As a control, cells transfected with an empty vector were used. Proteins immunoprecipitated with FLAG beads were analyzed by two approaches. First, the samples were analyzed by SDS-PAGE gel followed by silver staining (Fig. 1A). Two abundant USP19-interacting proteins were identified by mass spectrometry and later confirmed by immunoblotting as Hsp90 and Hsp70/Hsc70 (Fig. 1B). Alternatively, the immunoprecipitated materials were analyzed by mass spectrometry using a shotgun approach, which also identified these chaperones together with some less abundant proteins as potential USP19 interactors (supplemental Table S1). Given that Hsp90 and Hsp70 are often present in precipitated protein samples due to their high expression levels, we performed immunoprecipitation to confirm that these chaperones interacted with USP19 at endogenous levels. Immunoprecipitation using a USP19 specific antibody but not a control antibody pulled down Hsp90 from untransfected cell extract. By contrast, Hsp70 was precipitated by both antibodies (Fig. 1C). These results suggest a specific interaction between Hsp90 and USP19. Furthermore, when purified FLAG-USP19-Hsp90 complex was subject to a second round of immunoprecipitation using Hsp90 antibodies, the precipitated material still contained a large amount of USP19. Thus, USP19 and Hsp90 can form a stable complex in cells (Fig. 1D).
FIGURE 1.

Interaction of USP19 with Hsp90. A, cells transfected with either a control or FLAG-USP19-expressing vector were lysed. USP19 was purified using beads conjugated with FLAG antibodies. The purified samples were analyzed by SDS-PAGE and silver staining. B, fraction of the purified samples in A was analyzed by immunoblotting (IB). C, endogenous interaction of USP19 with Hsp90. D, FLAG-USP19 purified from mammalian cells in A was subject to another round of immunoprecipitation with either control IgG or Hsp90 specific antibodies.
We mapped the domain in USP19 responsible for Hsp90 binding using a set of USP19 constructs expressing various USP19 segments as indicated in Fig. 2A. The result showed that the catalytic domain of USP19 was sufficient for binding Hsp90 (Fig. 2B). Because USP family members share significant sequence homology, particularly in the catalytic domains, we tested whether Hsp90 interacted with other USPs. Extracts from cells transiently expressing FLAG-tagged USP19, USP7, USP13, or USP5 were subject to immunoprecipitation with FLAG beads. Immunoblotting showed that only USP19, but not other DUBs co-precipitated with Hsp90 (Fig. 2C). These results suggest that despite similarity in sequence and structure, only USP19 binds Hsp90 specifically via its catalytic domain.
FIGURE 2.
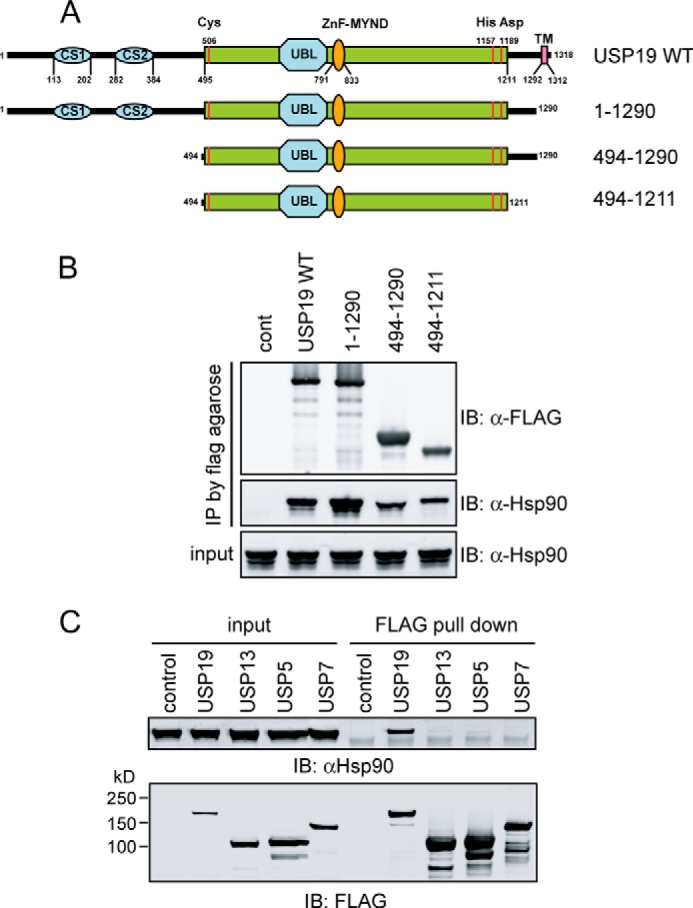
Hsp90 binds USP19 specifically via its catalytic domain. A, schematic representation of the USP19 mutant constructs used in the interaction studies. B, Hsp90 binds USP19 via the catalytic domain. Cells expressing the indicated FLAG-tagged USP19 variants were lysed, and the extracts were subject to immunoprecipitation by FLAG antibodies. cont, control. C, as in B, except that cells expressing the indicated DUBs were used.
Hsp90 Promotes USP19 Deubiquitinating Activity
Because recombinant USP19 purified from Escherichia coli had no activity, but full-length USP19 purified from mammalian cells was active (data not shown), we hypothesized that the activity of USP19 might be regulated by its associated proteins. Because USP19 associates with Hsp90 via the catalytic domain, and because Hsp90 is a ubiquitous molecular chaperone required for folding, activation and function of many client proteins in cells (14), we tested whether Hsp90 was required for USP19 DUB activity. To this end, we treated the FLAG-USP19-Hsp90 complex purified from mammalian cells in vitro with the Hsp90 specific inhibitor geldanamycin (GA) and then measured the deubiquitinating activity using ubiquitin-AFC as the substrate. The cleavage of the AFC group from ubiquitin by wild type USP19 generated fluorescence. By contrast, a catalytically inactive USP19 mutant purified under the same condition did not contain any DUB activity (13). When treated with GA, the activity of USP19 was reduced by ∼90% (Fig. 3A). This phenotype could be recapitulated using a polyubiquitinated protein as the substrate (Fig. 3B). Because treatment of USP7, a DUB that did not bind Hsp90 with GA did not affect its activity (Fig. 3C), the effect of GA appears to be specific to USP19, a Hsp90-interacting DUB.
FIGURE 3.
Hsp90 regulates USP19 activity. A, deubiquitinating activity of USP19 was measured using 0.4 μm Ub-AFC as the substrate in the absence or presence of the indicated inhibitor. GA, geldanamycin. B, in vitro ubiquitinated Ub-V-GFP was incubated with FLAG-USP19 purified from mammalian cells in the absence or presence of 1 mm DTT (−/+). Where indicated, USP19 was pre-incubated with GA for 1 h before the DUB activity was measured. C, GA did not affect USP7 activity. As in A, except that purified USP7 was used. D, cells treated with GA (2.5 or 10 μm) for 16 h were lysed. USP19 immunoprecipitated from cell extracts was analyzed by immunoblotting. E, GA inhibits USP19 activity at low substrate concentrations. USP19 activity was measured as in A using Ub-AFC at the indicated concentrations. The plot shows the relative inhibition of the USP19 activity.
To understand how Hsp90 affects USP19 activity, we first tested whether GA treatment disrupted the Hsp90-USP19 interaction. Co-immunoprecipitation using cells treated with GA showed that the interaction between Hsp90 and USP19 was moderately reduced after cells were exposed to GA for 16 h (Fig. 3D). However, the interaction was not affected if the cells were treated for 1 h (data not shown). Since 1 h exposure to GA is sufficient to inhibit USP19 activity, we concluded that GA inhibited USP19 without disrupting its interaction with Hsp90. Because GST-tagged USP19 purified from E. coli could not be activated by purified recombinant Hsp90 (data not shown), these results also implied that the association of USP19 with Hsp90 alone is insufficient to activate USP19. Moreover, since USP19 remained stable in cells treated with the Hsp90 inhibitor, Hsp90 inhibition does not seem to alter the global conformation of USP19. Otherwise, misfolded USP19 would be turned over by a cellular quality control pathway.
To further understand the mechanism by which Hsp90 activates USP19, we tested the inhibition of FLAG-USP19 by GA in the presence of different concentrations of ubiquitin-AFC. Interestingly, GA significantly inhibits USP19 activities when ubiquitin-AFC concentration was low, but increasing Ub-AFC concentration rendered USP19 resistant to GA (Fig. 3E). Thus, Hsp90 is only required for USP19 function when the substrate level is low, suggesting that it may promote USP19 binding to ubiquitin. In this regard, the Hsp90 chaperoning activity may maintain structural integrity of a cryptic ubiquitin binding motif in USP19 to promote deubiquitination at low substrate concentrations. This interpretation is consistent with the fact that Hsp90 inhibition does not significantly affect USP19 folding and stability.
The Complex of USP19-Hsp90 Is Dispensable for ERAD
Given the previously established link between USP19 and ERAD, we asked whether the interaction of USP19 with Hsp90 is involved in ER protein quality control. We first treated cells expressing several misfolded ER proteins with GA and tested whether inhibition of Hsp90 would lead to accumulation of misfolded ER proteins, a phenotype usually associated with ERAD inhibition. Immunoblotting showed that GA treatment (16 h) only caused small accumulation of several ERAD substrates including NHK, TTR, and TCRα in cells (Fig. 4, A and B). When we treated the cells for a shorter period time and performed pulse-chase experiments to examine the degradation kinetics for NHK and TCRα, we found that inhibition of Hsp90 by GA did not lead to significant stabilization of these substrates (data not shown). We conclude from these experiments that longer treatment with GA may cause accumulation of some ERAD substrates by indirectly affecting their biogenesis and/or turnover, but acute Hsp90 inhibition did not affect ERAD. Given the role of Hsp90 in USP19 regulation, it appears that USP19 may not play a significant role in ERAD.
FIGURE 4.
Hsp90 and USP19 are not involved in ERAD. A, cells transiently expressing the ERAD substrate NHK-GFP were treated with GA (10 μm) or MG132 (10 μm) for 15 h. Whole cell extracts (WCE) were analyzed by immunoblotting. B, cells expressing the ERAD substrates TTR D18G-GFP or TCRα-YFP were treated with the indicated inhibitors for 15 h. Shown are the relative fluorescence intensities measured by flow cytometry. C and D, USP19 knockdown does not affect ERAD of NHK-GFP and TTR D18G-GFP. E, interaction of overexpressed USP19 with a retrotranslocation complex. ER membrane fractions isolated from either control or USP19-expressing cells were solubilized. Protein extracts were subject to immunoprecipitation with FLAG beads. A fraction of the input and the precipitated samples were analyzed by immunoblotting. F, As in E, except that protein extracts from both the ER membrane (memb) and the cytosol (cyto) fractions were subject to immunoprecipitation with anti-USP19 antibodies. # indicates IgG.
To directly test the role of USP19 in ERAD, we used siRNA to knock down USP19 in HEK293 cells. Knockdown of USP19 by ∼90% did not lead to significant stabilization of the ERAD substrates NHK, TTR, and TCRα (Fig. 4, C and D, data not shown), indicating that USP19 does not have an essential function in ER quality control.
Our conclusion is contrary to a previous study that suggested direct involvement of USP19 in ERAD based in part on the observation that when overexpressed, USP19 interacts with the ERAD machinery protein Derlin1(6). We re-investigated the interaction of USP19 with known ERAD components using co-immunoprecipitation. To increase the possibility of detecting weak or transient interactions, we first isolated an ER-enriched membrane fraction from HEK293 cells transiently expressing FLAG-tagged USP19 and then solubilized the membranes using a mild detergent. USP19 was immunoprecipitated from the membrane extract. Immunoblotting showed that FLAG-USP19 co-immunoprecipitated with many ERAD factors with various efficiencies. These included Derlin1, UbxD8, Hrd1, and SEL1L (Fig. 4E). We then tested whether these interactions could be detected for endogenous USP19, but our experiments failed to detect these interactions under the same immunoprecipitation condition (Fig. 4F; data not shown). Surprisingly, we noticed that little endogenous USP19 was present in the membrane fraction. On the other hand, the same antibody detected a significant amount of USP19 signal from a whole cell extract and also from a cytosolic fraction. Thus, it seems that unlike overexpressed USP19, endogenous USP19 is predominantly localized in the cytosol (Fig. 4F).
Endogenous USP19 Is Primarily Localized to the Cytosol Despite Carrying a Transmembrane Domain
To resolve the controversy regarding the subcellular distribution of USP19, we re-investigated the localization of overexpressed USP19 by both biochemical fractionation and immunostaining. Consistent with the previous report (6), biochemical fractionation showed that FLAG-USP19 was predominantly localized to the ER-enriched membrane fraction purified from HEK293 cells expressing FLAG-USP19 (Fig. 5A). Immunostaining with FLAG antibodies revealed a reticulum-like pattern in a perinuclear region, indicative of ER localization (Fig. 5B). However, when we performed a similar fractionation experiment to examine endogenous USP19, we found that more than 80% of the protein was localized in the cytosol where it interacted with Hsp90 (Fig. 5C). Similar cytosolic localization of USP19 was observed in other cell types including U2OS, NCI-H226, NCI-H460, HCT-15, and HT29 (Fig. 5D). Thus, while overexpressed USP19 is predominantly membrane bound, endogenous USP19 is largely cytosolic.
FIGURE 5.
Endogenous USP19 is primarily localized in the cytosol. A, subcellular fractionation shows that overexpressed USP19 is mainly localized in the membrane. B, shown is a representative HeLa cell expressing FLAG-USP19 stained with FLAG antibody in green. DAPI stains the nucleus in blue. Scale bar, 10 μm. C, endogenous USP19 is primarily localized in the cytosol. D, cell fractionation shows that USP19 is primarily localized in the cytosol in different kinds of cells. E, As in D, except that HEK293 cells treated at 37 °C or 42 °C for 16 h were used. F, as in E, except that cells treated with an ER stress inducer tunicamycin (Tm) or expressing the non-glycosylated ERAD substrate NHK-QQQ were used. cont, control. G, as in E, except that cells treated with the indicated inhibitors were used. H, as in E, except that cells treated with GA as indicated were used.
Our observation raised the question of whether the localization of endogenous USP19 could be regulated. We explored the possibility that certain stress signals might result in more endogenous USP19 being incorporated into the ER membrane. So far, we have performed fractionation experiments using cells treated with various stressors including the ER stress inducers tunicamycin, overexpression of a misfolded ER protein (NHK QQQ), heat shock incubation at 42 °C, and chemicals that induce either mitochondria or DNA damages, or proteasome inhibition (Bortezomib). Immunoblotting showed that the ratio of cytosolic and membrane-bound USP19 was unaffected by these treatments (Fig. 5, E–G). Although these results have not revealed any conditions that can change the localization of USP19, the results do not exclude the possibility that the two pools of USP19 in cells could be exchanged under other conditions or in specialized cell types not tested here.
Our study also raised the question of how the transmembrane domain of USP19 is stabilized in the cytosol. Because prolonged inhibition of Hsp90 did not alter the subcellular localization of USP19 (Fig. 5H), Hsp90 did not seem to play a major role in this process. This result is not entirely surprising given that the transmembrane domain of USP19 is dispensable for interaction with Hsp90. Perhaps, the transmembrane domain of endogenous USP19 was shielded in the cytosol either directly or indirectly by a cellular factor, but under the USP19 overexpression condition, this factor became insufficient. This model provides a plausible explanation for the differential localization of overexpressed and endogenous USP19.
The Activity of USP19 Is Inhibited by Its Transmembrane Domain
The dual localization of a transmembrane domain-containing DUB also suggested a possibility that the USP19 activity may be regulated by subcellular localization. In accord with this idea, we consistently observed that the USP19 catalytic domain purified from mammalian cells was more active than full length USP19 (data not shown). We therefore tested whether a synthetic USP19 transmembrane peptide could inhibit the DUB activity of a USP19 mutant lacking the transmembrane domain using di-ubiquitin as the substrate. Addition of the peptide resulted in a concentration dependent inhibition of USP19 activity (Fig. 6A). Similar observation was obtained when the peptide was added to just the USP19 catalytic domain (Fig. 6B). These results suggest a functional interaction between the transmembrane segment and the USP19 catalytic domain, which inhibits USP19 deubiquitinating activity.
FIGURE 6.
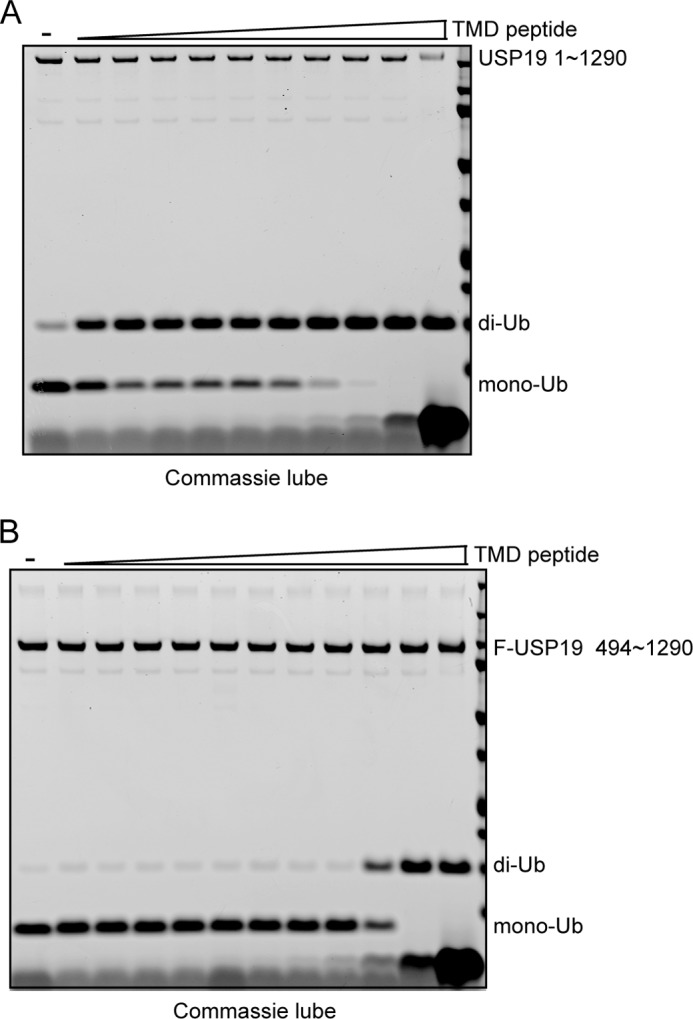
The USP19 transmembrane domain inhibits its catalytic activity. A, purified USP19 mutant lacking transmembrane domain was incubated with Lys48-linked di-Ub in the presence of increased concentration of a transmembrane (TMD) peptide at 37 °C for 1 h. B, as in A, except that the purified USP19 catalytic domain was used.
DISCUSSION
In this study, we characterize the regulation and function of USP19, a deubiquitinase previously implicated in ERAD. Unlike the previous study (6), our results do not support an essential function of USP19 in ERAD. The proposed ERAD function of USP19 was based on the observation that overexpression of USP19 inhibits the degradation of several model ERAD substrates including TCRα in a manner dependent on its catalytic activity (6). However, cellular phenotypes induced by DUB overexpression are difficult to interpret because wild type DUB when overexpressed can frequently inhibit cellular processes in a dominant negative manner (15, 16). We found that knockdown of USP19 does not affect the turnover of several ERAD substrates. Importantly, despite having a transmembrane domain, endogenous USP19 is primarily localized to the cytosol, and the reported interactions of USP19 with ERAD machinery factors are only detected when USP19 is overexpressed. These findings suggest that USP19 does not play a major role in degradation of misfolded ER proteins.
The transmembrane domain of USP19 is localized to its carboxyl terminus, making it a member of the so called tail-anchored (TA) proteins. Membrane targeting of TA proteins are mediated by a Sec61-independent mechanism. In vitro, the insertion of ER-bound TA proteins requires a pre-targeting complex consisting of Bag6, Trc35, and Ubl4A (17). In this complex, Bag6, or its yeast ortholog Sgt2 captures newly synthesized TA proteins to deliver them to a downstream ATPase Trc40 for subsequent targeting to the ER membrane (18, 19). The interaction of Bag6 with tail anchors in these proteins shields these hydrophobic segments, facilitating their traverse through the cytoplasm en route to the ER membrane. Thus, our finding that USP19 is predominantly localized to the cytosol despite carrying a tail anchor is quite unexpected. However, this is not without precedent. The pro-apoptotic protein BAX was found to be localized in the cytosol, although it carries a similar C-terminal transmembrane domain. A structural study showed that this hydrophobic segment is sequestered in the cytosol by an interaction with the BH3 domain in BAX until apoptotic signals activate it (20). Likewise, the USP19 transmembrane domain may be somehow stabilized in the cytosol by the observed interactions between this hydrophobic segment and the USP19 catalytic domain. However, because overexpressed USP19 is predominantly localized in the ER membrane, intramolecular interactions obviously cannot be the only factor that stabilizes USP19 in the cytosol. We therefore postulate that the localization of USP19 in the cytosol may require additional cofactor(s) and/or post-translational modification(s), which further shield the hydrophobic transmembrane domain. This safeguarding mechanism apparently becomes inadequate when USP19 is massively overproduced in cells, resulting in membrane translocation of overexpressed USP19. Although we haven't identified a physiological condition under which endogenous USP19 is translocated to the membrane, our results suggest the possibility that the localization of some TA proteins can be regulated in cells.
Our study also suggests a dual regulatory mechanism that governs the USP19 activity in cells. The interaction of the transmembrane segment with its own catalytic domain either in cis or in trans (if USP19 forms a homo-oligomer) can inhibit USP19 activity when this enzyme is present in the cytosol. The integration of this enzyme into the ER membrane would in principle relieve this auto-inhibition, but full activation of USP19 also requires its association with Hsp90. The latter appears to promote ubiquitin binding by the USP19 catalytic domain and thus enhance its activity. These findings establish USP19 as a new substrate of Hsp90, which may prove to be a useful system to study how Hsp90 activates its client proteins.
Acknowledgments
We thank K. Lindsten (Karolinska Institutet) and S. Wing (McGill University) for USP19 cDNAs.
This research was supported by the Intramural Research Program of the NIDDK of the National Institutes of Health.

This article contains supplemental Table S1.
- DUB
- deubiquitinating enzyme
- ERAD
- endoplasmic reticulum-associated degradation
- TA
- tail-anchored
- Hsp
- heat shock protein
- GA
- geldanamycin.
REFERENCES
- 1. Pickart C. M., Eddins M. J. (2004) Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695, 55–72 [DOI] [PubMed] [Google Scholar]
- 2. Li W., Ye Y. (2008) Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol. Life Sci. 65, 2397–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ye Y., Rape M. (2009) Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10, 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Komander D., Clague M. J., Urbé S. (2009) Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 [DOI] [PubMed] [Google Scholar]
- 5. Reyes-Turcu F. E., Ventii K. H., Wilkinson K. D. (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78, 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hassink G. C., Zhao B., Sompallae R., Altun M., Gastaldello S., Zinin N. V., Masucci M. G., Lindsten K. (2009) The ER-resident ubiquitin-specific protease 19 participates in the UPR and rescues ERAD substrates. EMBO Rep. 10, 755–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vembar S. S., Brodsky J. L. (2008) One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu Y., Adegoke O. A., Nepveu A., Nakayama K. I., Bedard N., Cheng D., Peng J., Wing S. S. (2009) USP19 deubiquitinating enzyme supports cell proliferation by stabilizing KPC1, a ubiquitin ligase for p27Kip1. Mol. Cell Biol. 29, 547–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mei Y., Hahn A. A., Hu S., Yang X. (2011) The USP19 deubiquitinase regulates the stability of c-IAP1 and c-IAP2. J. Biol. Chem. 286, 35380–35387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Altun M., Zhao B., Velasco K., Liu H., Hassink G., Paschke J., Pereira T., Lindsten K. (2012) Ubiquitin-specific protease 19 (USP19) regulates hypoxia-inducible factor 1alpha (HIF-1alpha) during hypoxia. J. Biol. Chem. 287, 1962–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Q., Liu Y., Soetandyo N., Baek K., Hegde R., Ye Y. (2011) A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble States for proteasome degradation. Mol. Cell 42, 758–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee J. G., Baek K., Soetandyo N., Ye Y. (2013) Reversible inactivation of deubiquitinases by reactive oxygen species in vitro and in cells. Nature Commun. 4, 1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Young J. C., Moarefi I., Hartl F. U. (2001) Hsp90: a specialized but essential protein-folding tool. J. Cell Biol. 154, 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clague M. J., Barsukov I., Coulson J. M., Liu H., Rigden D. J., Urbé S. (2013) Deubiquitylases from genes to organism. Physiol. Rev. 93, 1289–1315 [DOI] [PubMed] [Google Scholar]
- 16. Wang Q., Li L., Ye Y. (2006) Regulation of retrotranslocation by p97-associated deubiquitinating enzyme ataxin-3. J. Cell Biol. 174, 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mariappan M., Li X., Stefanovic S., Sharma A., Mateja A., Keenan R. J., Hegde R. S. (2010) A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature 466, 1120–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang F., Brown E. C., Mak G., Zhuang J., Denic V. (2010) A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol. Cell 40, 159–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stefanovic S., Hegde R. S. (2007) Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell 128, 1147–1159 [DOI] [PubMed] [Google Scholar]
- 20. Suzuki M., Youle R. J., Tjandra N. (2000) Structure of Bax: coregulation of dimer formation and intracellular localization. Cell 103, 645–654 [DOI] [PubMed] [Google Scholar]





