Background: The therapeutic potential of EcA is limited due to immunogenicity and short half-life in patients.
Results: Several EcA variants were constructed that showed markedly reduced immunogenicity and cytotoxicity against leukemic lymphoblasts.
Conclusion: Small changes in subunit interface and B-cell epitope significantly reduced immunogenicity and enhanced cytotoxicity.
Significance: These variants have promising potential in the advanced asparaginase therapy of leukemia.
Keywords: Enzyme Mutation, Escherichia coli, Leukemia, Lymphoma, Microbiology, E. coli, Asparaginase, Immunogenicity, Cytotoxicity, Leukemic Cells, Mice
Abstract
l-Asparaginase-II from Escherichia coli (EcA) is a central component in the treatment of acute lymphoblastic leukemia (ALL). However, the therapeutic efficacy of EcA is limited due to immunogenicity and a short half-life in the patient. Here, we performed rational mutagenesis to obtain EcA variants with a potential to improve ALL treatment. Several variants, especially W66Y and Y176F, killed the ALL cells more efficiently than did wild-type EcA (WT-EcA), although nonleukemic peripheral blood monocytes were not affected. Several assays, including Western blotting, annexin-V/propidium iodide binding, comet, and micronuclei assays, showed that the reduction in viability of leukemic cells is due to the increase in caspase-3, cytochrome c release, poly(ADP-ribose) polymerase activation, down-regulation of anti-apoptotic protein Bcl-XL, an arrest of the cell cycle at the G0/G1 phase, and eventually apoptosis. Both W66Y and Y176F induced significantly more apoptosis in lymphocytes derived from ALL patients. In addition, Y176F and Y176S exhibited greatly decreased glutaminase activity, whereas K288S/Y176F, a variant mutated in one of the immunodominant epitopes, showed reduced antigenicity. Further in vivo immunogenicity studies in mice showed that K288S/Y176F was 10-fold less immunogenic as compared with WT-EcA. Moreover, sera obtained from WT-EcA immunized mice and ALL patients who were given asparaginase therapy for several weeks recognized the K288S/Y176F mutant significantly less than the WT-EcA. Further mechanistic studies revealed that W66Y, Y176F, and K288S/Y176F rapidly depleted asparagine and also down-regulated the transcription of asparagine synthetase as compared with WT-EcA. These highly desirable attributes of these variants could significantly advance asparaginase therapy of leukemia in the future.
Introduction
Targeted engineering is an especially promising approach in the field of proteins with therapeutic applications. By applying single or multiple amino acid exchanges, fundamental properties of proteins can be adjusted to meet the requirements of existing biotechnological or medical applications or to tailor them for novel ones. As the majority of protein therapeutics is not of human origin, many of them are recognized as foreign by the human immune system. This often leads to hypersensitivity reactions or the formation of antibodies against the respective protein, the so-called anti-drug antibodies (ADA).2 Moreover, the majority of such proteins have stability as well as systemic toxicity issues. There are several approaches to generate proteins with desired properties such as directed evolution, structure-based targeted mutagenesis, and humanizing the protein by modifying relevant B-cell and helper T-cell epitopes.
In this study, we focused on the engineering of Escherichia coli l-asparaginase isoenzyme II (EcA) that has been used for decades in the treatment of childhood acute lymphoblastic leukemia (ALL), a disease that affects lymphocytes and lymphocyte precursor cells in the bone marrow (1, 2). EcA is also used in the treatment of other leukemias, such as Hodgkin lymphoma, chronic lymphocytic leukemia, and acute myelocytic leukemia (3). EcA is a homotetrameric enzyme that hydrolyzes asparagine to aspartic acid and ammonia (4). Basically, the therapeutic application of EcA is based on the fact that the transformed blasts that cause ALL have lost the ability to synthesize asparagine from aspartate due to very low levels of the enzyme asparagine synthetase. As asparagine is indispensable for protein synthesis, the blast has to take it from the blood plasma to build new proteins. EcA administration leads to the depletion of asparagine in the bloodstream and it therefore interferes with the proliferation of the leukemic blasts, ultimately driving them into apoptosis (5). Moreover, it was shown that l-asparaginase therapy interferes with cellular signaling pathways (6) and inhibits the expression of oncogenic translation factors (7).
The current protocols for the treatment of ALL use a combination of 10 or more drugs, including EcA as an integral part of the chemotherapy. There is evidence that EcA potentiates the antileukemic effect of drugs (8) and markedly improves outcome and overall survival in childhood ALL (9–11). Recent studies have also indicated that novel herbal compounds from different medicinal plant sources also have the potential to reduce leukemic blasts in different lineages in in vivo and in vitro conditions (12, 13). However, the use of EcA in chemotherapy is accompanied by a number of undesired side effects. Because of the bacterial origin, EcA administration may cause strong immunogenic and hypersensitive reactions in the patients, necessitating withdrawal of the drug (14). Sensitive individuals react to repeated EcA administration with formation of ADAs that bind to and thereby inactivate the enzyme (15). This leads to inadequate plasma levels of the EcA, which limit its efficacy. However, it was reported that not all patients with hypersensitivity develop ADAs, and not all patients who develop ADAs exhibit hypersensitivity (16, 17). Another serious drawback of the anti-leukemic drugs is their generalized cytotoxic effects on healthy cells along with leukemic cells. Although a number of attempts have been made to alleviate these problems by rational protein engineering, the optimization of therapy with EcA for ALL patients still remains a challenge.
In previous studies, we have attempted to improve the properties of EcA by amino acid exchanges at dimer-dimer interfaces. These experiments showed that mutations of certain amino acid residues change the enzyme's substrate specificity, the flexibility of an active site loop, and the overall stability of the enzyme protein.3 Moreover, we have shown that the glutaminase side activity of EcA, which is partly responsible for neurotoxicity, can be markedly reduced by site-directed mutagenesis (18). In another study, we have identified several B-cell epitopes on the surface of EcA that are responsible for the immunogenicity (19). These data now provide a sound basis for a knowledge-based engineering of EcA aimed at the reduction of formation of ADA. A study by Jianhua et al. (17) on a single EcA epitope suggests that the antigenicity of EcA, at least in vitro, can indeed be reduced in this way. The aim of this study was to create EcA variants that can enhance the therapy of ALL. In the light of this, we succeeded in creating variants that showed greater cytotoxicity and reduced glutaminase activity in leukemic cell lines and lymphoblasts isolated from patients with ALL. Mice experiments showed that K288S/Y176F variant is significantly less immunogenic than wild-type EcA. Interestingly, sera obtained from mice immunized with wild-type EcA and the ALL patients after several weeks of asparaginase therapy recognized the K288S/Y176F mutant significantly less than the wild-type EcA. We also present evidence indicating that the killing of leukemic cells by EcA occurs through the induction of apoptosis, rapid depletion of asparagine, down-regulating transcript of asparagine synthetase, and arresting the cells at G0/G1 phase rather than by inducing DNA damage and micronuclei formation.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis
The EcA encoding the ansB gene cloned in plasmid pTEW1 (ansB-pTEW1), previously developed in our laboratory (20), was used as template for PCR-based site-directed mutagenesis using the QuikChange XL II kit (Agilent Technologies, Santa Clara, CA). A pTEW1-type plasmid harboring the gene encoding variant W66Y was kindly provided by Dr. C. Derst, Berlin, Germany. The mutagenic primers (all synthesized by MWG Biotec, Germany) were as follows: EcA (Y176F), 5′-TCTGTTAACTTCGGTCCTCTG-3′, template pTEW1; EcA (Y176S), 5′-TCTGTTAACTCCGGTCCTCTG-3′, template pTEW1; EcA (K288S), 5′-GTGGATGATGCGAGTTACGGCTTCGTCGCCTCTGGC-3′, template pTEW1; EcA (K288S/Y176F), 5′-GTGGATGATGCGAGTTACGGCTTCGTCGCCTCTGGC-3′, template pTEW-Y176F; EcA (K288R), 5′-GTGGATGATGCGCGCTACGGCTTCGTCGCCTCTGGC-3′, template pTEW1; EcA (K288R/Y176F), 5′-GTGGATGATGCGCGCTACGGCTTCGTCGCCTCTGGC-3′, template pTEW1-Y176F (mutagenic base changes are shown as underlined boldface). In each case, the mutated codon was flanked by at least 10 bp on each side. DpnI-treated PCR amplicons were transformed into XL10-Gold ultracompetent cells. The mutant gene was excised by EcoRI/HindIII digestion and subcloned into pTWE1. In each case, the mutant genes were sequenced to exclude the presence of additional unwanted base changes. The mutants were propagated using the replica plate method.
Expression and Purification of EcA
Wild-type EcA and mutants were expressed in E. coli BL21Ω, released from the periplasm by osmotic shock and purified by fractional ammonium sulfate precipitation and chromatofocusing as described previously (21, 22). Final purification was achieved by gel filtration on a Sephacryl® S-300 column equilibrated and eluted with 100 mm Tris/HCl, pH 7.0. Protein concentrations were determined by the BCA method (23). With purified EcA preparations, UV spectrometry was employed, using the relationship A280(0.1%) = 7.4 (i.e. a 10 mg/ml solution has an absorption of A280 = 0.74). Molecular masses of the purified proteins were determined by denaturing gel electrophoresis (12% SDS-PAGE) and/or gel filtration.
Enzyme Assays
The asparaginase activity of EcA was routinely measured as described previously (24) using the synthetic substrate l-aspartic β-hydroxamate (AHA) purchased from Sigma. Briefly, after stopping the reaction with trichloroacetic acid, the NH2OH liberated from AHA was quantified by reaction with 8-hydroxyquinoline at alkaline pH. To study the effect of serum on the activity of asparaginase, each enzyme was incubated with (10% human serum) or without (HEPES buffer) serum for 1–6 days (25), and activity was measured by described above. One unit of activity is defined as the amount of enzyme that liberates 1.0 μmol of NH2OH from AHA per min at 25 °C. The glutaminase side activity of EcA was determined by a discontinuous coupled enzymatic assay as described previously (26–28). In the first step, l-glutamine is hydrolyzed to l-glutamate and ammonium ion. In the second step, catalyzed by glutamate dehydrogenase, the ammonium ion formed in step 1 reacts with 2-oxoglutarate and NADH to form l-glutamate and NAD+. In step 1, reaction mixtures (1 ml) containing 10 mm l-glutamine and 5 μg of enzyme in 50 mm phosphate buffer, pH 7.4, were incubated for 10 min at 37 °C followed by addition of 100 μl of 3 m HCl to stop the reaction. In the second step, 600 μl of reaction mixture from step 1 was added to Tris/Cl (100 mm, pH 9.0), NADH (2 mm), and 2-oxoglutarate (3.3 mm) in a total volume of 3 ml. Glutamate dehydrogenase enzyme (22 units) was added to the mixture and incubated for 30 min at 37 °C. NADH consumption was measured by the decrease in absorbance at 340 nm. The enzyme activity was calculated using the molar absorption coefficient of NADH (6220 m−1 cm−1).
Determination of Ammonia Release Using Nessler's Reagent
The hydrolysis of asparagine to aspartate by asparaginase leads to the release of the ammonia group that can be measured by Nessler's reagent. The amount of ammonia released corresponds to the rate of asparagine depletion due to asparaginase activity. Therefore, to determine the activity of the EcA variant, the EcA variants (20 μg/ml) and the substrate l-asparagine were incubated at 37 °C. After 10 min, the reaction was stopped by adding 0.1 ml of the 1.5 m TCA. The amount of ammonia released was determined by adding Nessler's reagent, and the absorbance was measured from 300 to 600 nm.
Isolation of Peripheral Blood Mononuclear Cells (PBMCs)
PBMCs were isolated from buffy coats as described elsewhere (29). Briefly, after sedimentation with 2.5% dextran T-500 (HiMedia, India), 25 ml of supernatant was carefully layered over 15 ml of Ficoll and centrifuged at 1,600 rpm for 30 min with brake and acceleration. After centrifugation, the interphase was collected and centrifuged again at 1,800 rpm for 10 min. Subsequently, the pellet was resuspended in 1× phosphate-buffered saline (PBS) and centrifuged at 1,000 rpm for 10 min. Finally, the pellet was resuspended in RPMI medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS). Approximately 3 × 105 cells were seeded and treated with 0.8 units/ml of purified wild-type EcA and mutants for 96 h. Untreated cells were taken as negative control.
Isolation of Lymphocytes from ALL Patients
All research using patient material was approved by the Institutional Review Boards of Kalinga Institute of Industrial Technology University and Shrirama Chandra Bhanj Medical College. Written informed consent was provided by the study participants. Lymphocytes were isolated from peripheral blood and bone marrow obtained from ALL patients by the LymphoprepTM Axis shield as described previously (29, 30). Briefly, blood samples were collected in a tube containing anticoagulant and diluted by addition of 0.9% NaCl in a 1:1 ratio. 6 ml of the diluted blood was layered over 3 ml of LymphoprepTM and centrifuged at 800 × g for 20 min (∼20 °C) in swing-out rotor. After centrifugation, the mononuclear cells from a distinct band were taken out carefully with the help of a Pasteur pipette. Subsequently, the harvested fraction was diluted with 0.9% NaCl or medium to reduce the density of the solution and centrifuged for 10 min at 250 × g. The cells from the pellet were collected, and ∼3 × 105 cells were treated with 0.8 units/ml of purified wild-type EcA and mutants for 96 h. Untreated cells were taken as negative control. Finally, flow cytometric analysis was performed by analyzing 10,000 gated cells using a FACS Canto II flow cytometer and FACS Diva software (BD Biosciences).
Enzyme-linked Immunosorbent Assay (ELISA)
The antigenicity of EcA and mutants was estimated by indirect ELISA using a modification of the protocol described by Wang et al. (31). Wells of microtiter plates were coated with 100 μl of EcA solution (2–5 μg/ml) in 50 mm carbonate/bicarbonate buffer, pH 9.5, and incubated overnight at 2–8 °C. Then the plates were drained without washing and blocked for at least 90 min at room temperature with 300 μl of 0.1 m PBS, pH 7.2, containing 0.1% BSA and 0.05% Tween 20. The plates were again washed three times with 0.05% Tween 20 in PBS (PBST) before 100 μl per well of 1:8,000 diluted primary antibody (anti-EcA IgG fraction from Abcam) in PBST was added. After incubation for at least 1 h at room temperature, the plates were washed as described above and incubated with 100 μl of HRP-conjugated polyclonal goat anti-human IgG (1:10,000 v/v; Thermo Scientific, Rockford, IL) for 1 h at RT. After another four washes with PBS, the wells were incubated with 100 μl of freshly prepared 2,2′-azino-bis-3-ethylbenzothiazoline-6-sulfonic acid, 0.5 mg/ml, in 0.03% H2O2, 0.1 m Na2HPO4, and 0.08 m citric acid, pH 4.0, for 30 min in the dark. Finally, the absorption was measured at 405 nm using a microplate reader (Epoch, Biotek).
Immunization Schedule of Mice
Six- to 8-week-old female BALB/c mice were used for all immunization experiments. All mice were kept in our animal facilities, in high efficiency particulate air (HEPA)-filtered cages under 12-h light cycles and were given sterile chow and autoclaved water ad libitum. All animal experiments were performed in accordance with national guidelines for the care and handling of laboratory animals and have been approved by the Institutional Animal Ethical Committee. Mice (five mice per group) were first bled prior to immunization to collect pre-bleed and then immunized subcutaneously (31) with 30 μg of purified EcA and K288S/Y176F variant emulsified in an equal volume of Freund's complete adjuvant at a final concentration of 0.5 mg/ml. Three weeks after primary immunization, the mice were again immunized (day 21, boosting immunization) with wild-type and K288S/Y176F variant emulsified in Freund's incomplete adjuvant. Then the mice were rested for 45 days and then immunized for the third time (day 66) following the same protocol used for the second immunization. One week after the second and third immunization, blood samples were drawn. Sera were prepared after centrifugation of coagulated blood at 2,000 rpm for 30 min, and samples were stored at −20 °C until further analysis. Sera obtained before immunization (pre-bleed) were used as control to obtain a reference base line for serum antibody titers.
Antibody Titer Determination by ELISA
The titer of IgG and IgM antibodies was determined in sera obtained from wild-type EcA and K288S/Y176F-immunized mice and ALL patients using indirect ELISA. Antibody titer was expressed as the dilution of serum that gave an absorbance of 0.2. Microtiter plates were coated overnight with wild-type EcA and K288S/Y176F proteins dissolved in 0.05 m Na2CO3/NaHCO3 buffer, pH 9.6, at 4 °C, washed, and blocked using 1% BSA in 0.05 m Na2CO3/NaHCO3 buffer, pH 9.6, for 8 h at room temperature. Then 100 μl of serial dilutions of serum samples (for IgG and IgM) in PBST, pH 7.2, were added to duplicate wells and incubated overnight at 2–8 °C. The plates were then washed three times with PBST and incubated with 100 μl per well of a 1:2,000 dilution of commercially available antibodies (goat anti-mouse IgM or anti-mouse IgG, Sigma) in PBST and incubated for 3 h at RT. The plates were washed three times with PBST and incubated with 100 μl of HRP-conjugated polyclonal rabbit anti-goat Ig (1:2,000 v/v; Sigma) for 2 h at RT. The wells were then washed four times with PBST, and the wells were incubated with 100 μl of freshly prepared 2,2′-azino-bis-3-ethylbenzothiazoline-6-sulfonic acid (0.5 mg/ml) in citrate phosphate buffer, pH 4.0, containing 2 μl of 30% H2O2 for 20 min in the dark, and absorption was measured at 405 nm. The human serum was also used the same way as described above except for using goat anti-human IgG antibody for detection of the titer of human antibodies in the ALL patient sera.
Cytotoxicity Assay
An MTT (MP Biomedicals) viability assay was performed (32) to estimate the cytotoxic activity of EcA variants against Ramos cells (obtained from American Type Culture Collection) and the MV4:11 cell line (kindly provided by Dr. Vaskar Saha, University of Manchester, UK). Ramos cells are immortalized B-lymphocytes, and the MV4:11 cell line is a human acute monocytic leukemia cell line. Cells (5 × 103/ml) of each type were grown in RPMI in 24-well plates (Tarson, India) at 37 °C, 5% CO2, for 24 h. The next day, the cells were centrifuged at 900 rpm for 5 min at room temperature; the supernatant was removed carefully, and the cells were treated with different concentrations of purified wild-type EcA and mutants for 96 h. To determine the cell viability, MTT dye (500 μl from 0.1 mg/ml stock) was added to each well and incubated for 1 h at 37 °C, 5% CO2 in the dark. The cells were then collected by centrifugation at 900 rpm for 5 min. The formazan crystals formed as a result of cellular reduction of MTT were dissolved in dissolving buffer (11 g of SDS in 50 ml of 0.02 m HCl and 50 ml of isopropyl alcohol) and incubated for 1 h at 37 °C; then the absorption was read at 570 nm in an ELISA reader (Biotek, Germany). The percentage of viable cells was calculated as cell viability (%) = average A of wells/average A of control wells × 100.
Apoptosis Assay
The percentage of apoptotic cells was determined by using the annexin V/FITC apoptosis detection kit (Sigma). Briefly, cells (3 × 105/ml) in 2 ml of RPMI medium were grown in culture flasks (Tarson, India) overnight at 37 °C. The medium was then removed, and 3 ml of fresh medium containing 0.8 units/ml of EcA or variants was added, and the flasks were incubated for another 96 h. The cells were collected and washed three times with Dulbecco's PBS (0.1 m, pH 7.4). 500 μl of 1× binding buffer was added, followed by 4 μl of annexin V-FITC and 8 μl of propidium iodide, followed by a 10-min incubation at room temperature in the dark. Untreated cells were taken as negative control. Finally, flow cytometric analysis was performed by analyzing 10,000 gated cells using a FACS Diva software (BD Biosciences).
Western Blotting
MV4:11 cells were seeded on 12-well plates at a density of 3 × 105 cells per well and incubated overnight. The next day, the cells were treated with wild-type EcA and mutants and harvested at the indicated time points. The pellets were washed twice with ice-cold PBS and resuspended in lysis buffer for 45 min at 4 °C. The cells were centrifuged at 14,000 rpm for 25 min to remove the debris. The supernatant was collected, and protein estimation was done by the Bradford method. Approximately 50 μg of protein lysate per well was separated on a 12% SDS gel and transferred to a PVDF membrane. Membranes were blocked overnight in either 5% nonfat milk or 5% BSA in 1× TBS-T (Tris-buffered saline containing 0.05% of Tween 20) at 4 °C and immunoblotted with primary anti-caspase-3, anti-Bcl-XL, anti-PARP, and anti-cytochrome C antibodies (1:1,000). After primary blotting, the membranes were probed either with secondary anti-rabbit or anti-mouse antibodies (1:1,000) for 2 h, and signals were detected using ECL reagent. Chemiluminescence was detected with Kodak X-Omat films. The anti-β-actin antibody was used as the loading control.
Quantitative Real Time PCR
MV4:11 cells were treated with 0.8 units/ml EcA variants for 12, 24, and 48 h. Total RNA was isolated using TRIzol reagent according to the recommendations of the supplier (Invitrogen). The concentration of the RNA was determined spectrophotometrically. cDNA synthesis was performed using cDNA synthesis kit (Thermo). The synthesized cDNA was used as a template for RT-PCR amplification using gene-specific primers. The primer pairs used for asparagine synthetase were forward 5′-TGCTTTCACCTTCAACTGCC-3′ and reverse 5′TTCTGTACTTGGTCGCCTGT-3′; and for GAPDH were forward 5′-AGGGCCCTGACAACTCTTTT-3′ and reverse 5′-AGGGGTCTACATGGCAACTG-3′. The volume of all the reaction mixtures was maintained at 10 μl using SYBR® Green PCR master mix (Applied Biosystems) and carried out in Real Plex master cycler (Eppendorf, Germany) with initial denaturation at 95 °C for 10 min, final denaturation at 95 °C for 30 s, annealing at 52 °C for 30 s, and extension at 72 °C for 30 s. The mRNA levels were normalized to the transcript levels of GAPDH, and the relative fold changes were calculated by calculations based on the real time PCR efficiencies.
Cell Cycle Analysis
MV4:11 cells (3 × 105/ml) were grown overnight in 12.5-cm2 culture flasks and then treated with 0.8 units/ml of EcA or variants as mentioned above. After 96 h of incubation, cells were harvested, washed with 1× PBS, and fixed in ice-cold 70% ethanol for 1 h at −20 °C. Following incubation, cells were centrifuged at 1,500 rpm for 5 min and washed two times with 1× PBS. Cells were stained with 250 μl of the propidium iodide (50 μg/ml stock) containing 10 μg/ml RNase, incubated at 37 °C for 1 h in the dark, and analyzed by FACS Canto II flow cytometer (San Jose, CA). The acquired data were analyzed using FACS Diva software (BD Biosciences). In each case 10,000 cells were analyzed.
Comet Assay
The effect of EcA variant treatment on DNA damage was determined by alkaline single cell electrophoresis (Comet) assay. MV4:11 cells were treated with 0.8 units/ml of W66Y for 96 h. Untreated cells were used as control. Cells were trypsinized, and cell suspension was prepared in 1× PBS. 10 μl of cell suspension was mixed with 60 μl of 0.5% low melting agarose. A thin smear of cell suspension was prepared in glass cavity slides (Blue Label Scientifics, Mumbai, India). The agarose was allowed to solidify in the dark at 4 °C for 45 min, and then the slides were submerged in lysis solution (10 mm Tris, 100 mm EDTA, 2.5 m NaCl, 1% Triton X-100 and 10% DMSO) for 30 min in the dark at 4 °C. After lysis, the slides were washed with distilled H2O, transferred to an electrophoresis unit containing freshly prepared electrophoresis buffer (500 mm EDTA, 200 mm NaOH, (pH >13), and left for unwinding of DNA for 45 min, and the cells were electrophoresed for 15 min at 15 V. The cells were washed twice with distilled H2O, fixed with 70% chilled ethanol, and stained with 0.5 μg/ml propidium iodide for 15 min in the dark. The slides were dried and observed using a fluorescence microscope (Nikon, Japan).
Micronuclei Assay
MV4:11 cells (3 × 105/well) were grown overnight in 6-well plates and treated with 0.8 units/ml EcA or variants for 48 h. Cells grown in media were used as control. The cells were then processed for micronuclei assay as described previously (33). Briefly, the cells were arrested at the cytokinesis stage by the addition of cytochalasin-B (0.5 mg/ml) in 1× PBS at a concentration of 10 μg/ml, and the cells were allowed to grow for 12 h. Then the cells were trypsinized and centrifuged at 1,000 rpm for 8 min at room temperature. Cells were fixed by addition of chilled methanol and acetic acid (3:1), vortexed simultaneously, and incubated at 4 °C for 1 h. The above process was repeated twice until the cell suspension appeared clear. Finally, a few drops of cell suspension were added onto the slide, air-dried, and stained with propidium iodide stain. The images were captured by Olympus BX61 fluorescent microscope using appropriate filters and analyzed by Cytovision 7.2 software.
Statistical Analysis
Significant differences between the groups were determined by one-way analysis of variance. All the statistical calculations were performed with the help of GraphPad prism version 5.0. Significance was indicated as *** for p < 0.001, ** for p < 0.01, and * for p < 0.05.
RESULTS
Selection of Amino Acid Residues for Mutagenesis
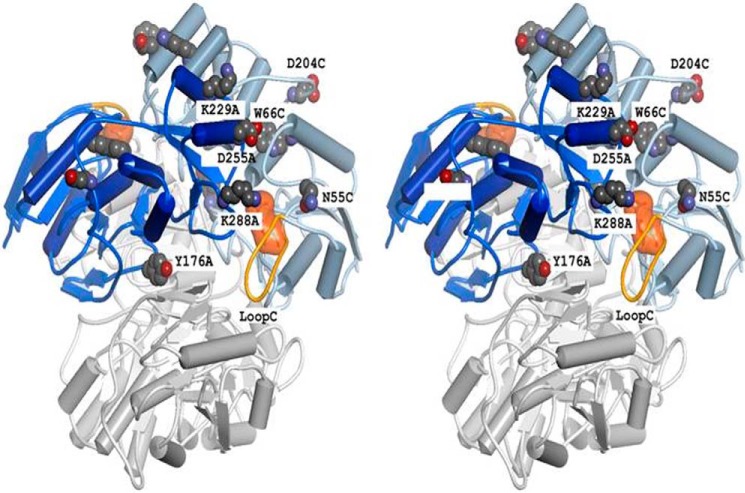
EcA is a homotetramer made up from two so-called “intimate” dimers (one consisting of subunits A and C and the other of B and D, respectively, see Fig. 1). In previous studies, a number of residues with crucial importance for EcA activity and stability have already been identified. Among these are residues at dimer-dimer interfaces, such as Ser-122 (34) and Tyr-181 (35). In this work, we have augmented and expanded these observations by mutating additional amino acids, including Tyr-176, a residue in close vicinity to Tyr-181, and Lys-288, which is the central residue of a strong B-cell epitope on the surface of EcA (17). The role of Trp-66, the only tryptophan residue of EcA, was examined in connection with a study in which the movement of a mobile loop at the enzyme's active site was followed by using rapid reaction techniques (36). It turned out that variant W66Y was only slightly impaired with regard to both stability and catalytic properties. As shown in Fig. 1, Trp-66 is inserted between two α-helices near the active site of each monomer, whereas residue Lys-288 is at close proximity to the active site of the adjacent intimate subunit (i.e. in the intimate dimer AC, Lys-288 of subunit A is near the active site of subunit C).
FIGURE 1.
Structure of EcA (stereo representation). The figure at the top consists of subunits A (dark blue) and C (light blue), and the BD dimer (bottom) is colored grey. An aspartate residue bound to the active site of subunit C and the mobile loop of monomer C are shown in orange. Individual residues are labeled according to position and subunit, e.g. K288A stands for lysine 288 in subunit A.
Construction, Expression, and Purification of EcA Mutants
Mutations in the EcA-encoding ansB gene were created by site-directed mutagenesis and confirmed by DNA sequencing. All enzyme variants were readily expressed and purified as described for wild-type EcA (21). The resulting proteins were at least 95% pure as judged by SDS-PAGE (data not shown). Yields varied between 5 and 20 mg of purified enzyme per liter of E. coli culture. Gel filtration studies showed that all EcA variants retained their native tetrameric structure (data not shown).
Mutations in EcA Affect Both Asparaginase and Glutaminase Activity
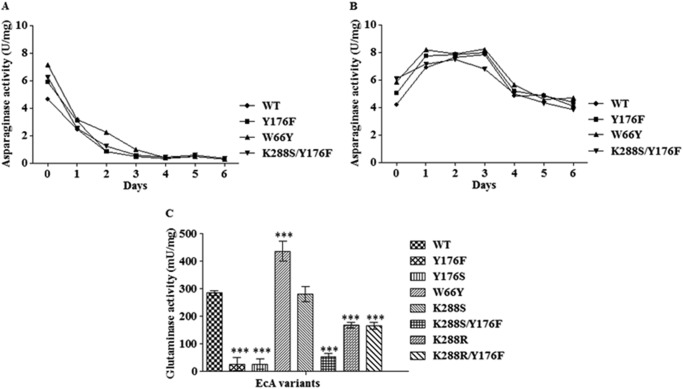
First the effects of the mutations on the enzyme's activity were studied. The asparaginase activity of all EcA variants (assayed with AHA as the substrate) and its glutaminase side activity were assayed under physiological conditions (37 °C, pH 7.4). As shown in Fig. 2, the asparaginase activities of all variants examined were comparable with each other. Interestingly, the asparaginase activity increased and was stable for few days when the enzyme samples were preincubated with human serum (Fig. 2, A and B). By contrast, the glutaminase activities were markedly different. All variants harboring mutations at position 176 (Y176F, Y176S, and K288S/Y176F) had much lower glutaminase activities than the wild-type EcA (p < 0.001), whereas the glutaminase activity of variant W66Y was significantly higher (Fig. 2C). With the exception of K288S, all variants with amino acid changes at position 288 also exhibited significantly reduced glutaminase activities (p < 0.001).
FIGURE 2.
Asparaginase and glutaminase activities of wild-type EcA and variants. A, asparaginase activity of wild-type EcA and variants in the absence of human serum (incubated with HEPES buffer only) at 37 °C using AHA as the substrate. B, asparaginase activity of wild-type EcA and variants in the presence of 10% human serum. C, glutaminase activity of wild-type EcA and variants (5 mg/ml) after 10 min of incubation. Each experiment was performed three times; means ± S.D. are shown. ***, p < 0.001.
Variants Y176F and W66Y Exhibit Higher Cytotoxicity against Leukemic Cell Lines
We further investigated the effect of EcA variants on cytotoxicity against leukemic cell lines MV4:11, a B-myelomonocytic ALL cell line, and Ramos, a B-lymphocyte cell line. Nonleukemic PBMCs were used as control. Leukemic cell lines were used for the in vitro cytotoxicity assays instead of primary patient samples because cell lines are homogeneous and readily available for experimental replications. Moreover, this approach minimizes potential artifacts due to differences in the in vivo to in vitro conditions.
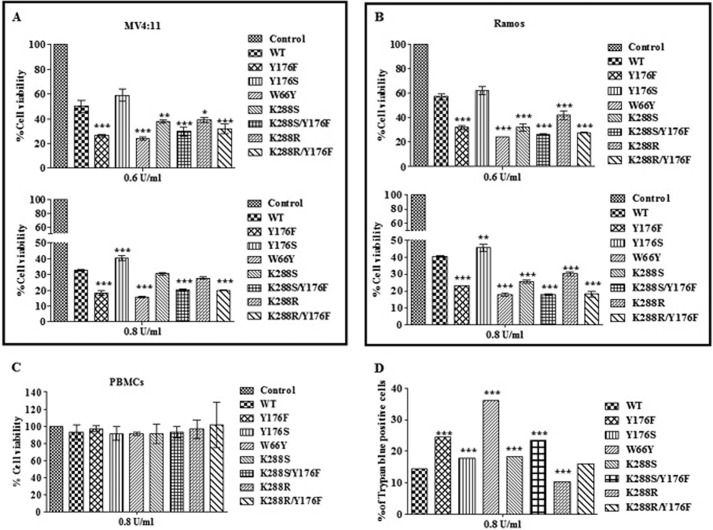
First the viability of MV4:11 cells was determined following treatment with different doses of wild-type EcA. The survival profiles were determined by the MTT assay, which relies on the fact that only metabolically active cells reduce MTT to a purple formazan dye. As expected, this resulted in a dose-dependent, significant decrease in the number of viable cells. For instance, treatment with 0.6 and 1.0 units/ml wild-type EcA led to a 50 and 75% reduction in cell viability, respectively (data not shown). Fig. 3 summarizes the viability of MV4:11, Ramos, and PBMC cells after treatment with 0.6 and 0.8 units/ml of EcA variants for 96 h. Interestingly, MV4:11 cells treated with 0.6 units/ml of Y176F and W66Y showed a much greater reduction in viability as compared with wild-type EcA and other variants (p < 0.001; Fig. 3A, upper panel). Variants W66Y and Y176F decreased the number of viable cells by 78 and 76%, respectively (p < 0.001), as compared with 49% after treatment with wild-type enzyme (Fig. 3A, upper panel). The single mutants K288S and K288R led to an approximate 60% reduction in cell viability (p < 0.01 and p < 0.05), whereas treatment with the double mutants K288S/Y176F and K288R/Y176F resulted in a 67% reduction in viability as compared with wild-type enzyme (p < 0.001). Mutant Y176S showed no cytotoxic activity at all.
FIGURE 3.
Cytotoxic activity of EcA and variants on MV4:11 (A), Ramos (B), and PBMCs (C) are shown. MV4:11, Ramos, and PBMCs were treated with different concentrations of enzymes for 96 h. Cells viability was determined by the MTT assay. D, trypan blue assay. MV4:11 cells were treated with 0.8 units/ml of enzymes for 96 h. Cell viability was determined by trypan blue exclusion assay. Each experiment was performed three times; mean ± S.D. are shown. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Differences in cell viability were also noted at higher enzyme concentrations (Fig. 3A, lower panel). Incubation with 0.8 units/ml of W66Y and Y176F reduced the number of viable MV4:11 cells by about 85 and 82% (p < 0.001), respectively, whereas in case of wild-type EcA about 66% reduction in cell viability was observed. Similar killing patterns were observed when Ramos cells were treated with 0.6 and 0.8 units/ml of wild-type EcA and the variants (Fig. 3B). To address the question whether the cytotoxicity of EcA variants is preferential for leukemic cells relative to nonmalignant cells, we also studied their effect on PBMCs. No significant changes in the viability of PBMCs were observed under similar experimental conditions (Fig. 3C).
The cytotoxicity data described above was further corroborated by scoring the number of trypan blue-positive MV4:11 cells. Because of the presence of intact cell membrane, viable cells exclude this stain, whereas dead cells appear blue in color. As shown in Fig. 3D, treatment with 0.8 units/ml EcA variants increased the number of trypan blue-positive cells as compared with wild-type EcA treatment. These results also indicate that variants Y176F and W66Y exhibit a significantly higher cytotoxic effect on leukemic cells (p < 0.001) but had no cytotoxic activity on nonleukemic cells.
K288S/Y176F Double Mutation Reduces Antigenicity of EcA
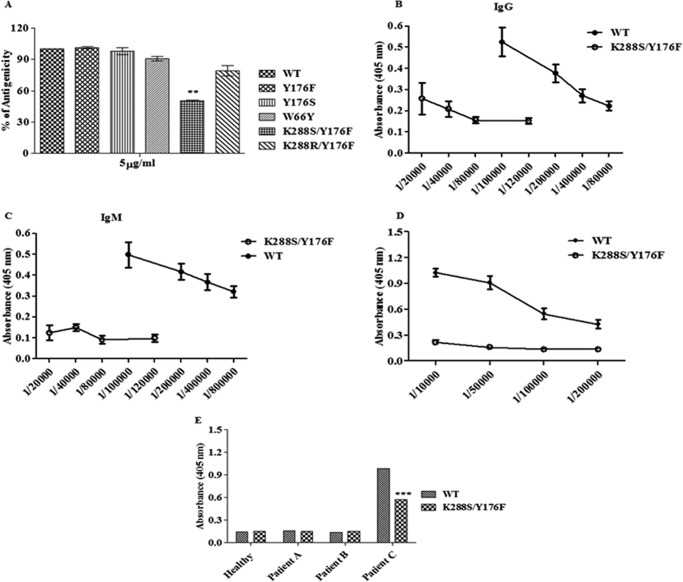
We previously identified several Bcell epitopes spanning the entire EcA sequence (19). In this study, we selected five amino acid residues (Asn-55, Asp-204, Asp-255, Lys-229, and Lys-288, cf. Fig. 1), which occur in different strong epitopes of EcA, and replaced them by site-directed mutagenesis. About 2,000 mutants resulting from each mutated residue were screened for reduced antigenicity using commercially available rabbit anti wild-type EcA antibody by ELISA. The variants with mutations in four epitopes, viz. Asn-55, Asp-204, Asp-255, and Lys-229 (data not shown) as well as variants Y176F, Y176S, W66Y, and K288R/Y176F, did not exhibit any significant differences in binding to rabbit anti-EcA antibody. However, binding of antibodies to the double mutant K288S/Y176F was reduced to less than 40% as compared with that of the wild-type EcA (p < 0.001; Fig. 4A). However, the single mutation Lys-288 to Ser did not show altered binding ability to rabbit anti-EcA antibodies.
FIGURE 4.
Indirect ELISA of wild-type EcA and variants. A, microtiter plates were coated with wild-type EcA and variants (5 μg/ml), probed with primary (rabbit anti-EcA), and developed by secondary (HRP-conjugated polyclonal goat anti-rabbit IgG) antibodies by the indirect ELISA method. The experiment was performed three times. B and C, immunogenicity study in mice. Microtiter plates were coated with wild-type EcA and K288S/Y176F variant (5 μg/ml). Different dilutions of serum (obtained on day 74 after immunization) from mice immunized with wild-type EcA and K288S/Y176F was prepared and added to the wells in duplicate. Primary antibodies (goat anti-mouse IgM or anti-mouse IgG) and secondary antibody (HRP-conjugated polyclonal rabbit anti-goat Ig) were used. The titer of IgG (B) and IgM (C) antibodies were determined by the indirect ELISA method. D, microtiter plates were coated with wild-type EcA and K288S/Y176F (5 μg/ml). Different dilutions of serum obtained from mice immunized with wild-type EcA were added to the wells. Binding of serum IgG antibody to wild-type EcA and K288S/Y176F was determined as described above. Immunization studies were repeated two times. E, microtiter plates were coated with wild-type EcA and K288S/Y176F. Appropriate dilution of serum obtained from a healthy individual, an ALL patient without asparaginase therapy (patient A), and ALL patients who were given asparaginase therapy for 5 days (patient B) and 45 days (patient C) were added to the wells. The data shown here are representative of three (n = 3) patients from each group. The binding of serum IgG with wild-type EcA and K288S/Y176F was determined as described above. Mean ± S.D. are shown. ***, p < 0.001.
K288S/Y176F Mutant Is Less Immunogenic in Mice
To evaluate the in vivo immunogenicity, mice (five mice in each group) were first bled to prepare prebleed serum and then immunized subcutaneously with wild-type EcA and K288S/Y176F variant emulsified in Freund's complete adjuvant. The same group of mice was reimmunized (boosting immunization) 3 weeks later (second immunization) and again after 6 weeks (third immunization) using the same immunogen emulsified in Freund's incomplete adjuvant through the same route. Then the sera from mice immunized with wild-type EcA and the K288S/Y176F variant were prepared 8 days after the second and third immunization and were used to determine the titer of IgG and IgM antibodies against the respective immunogen. All mice showed a very similar response, i.e. low antibody response to K288S/Y176F as compared with wild-type EcA, and the dose of the immunogen did not have any effect on this pattern. As shown in Fig. 4, B and C, mice immunized with K288S/Y176F variant showed ∼10-fold lower IgG and IgM titers as compared with IgG and IgM antibody responses induced by wild-type EcA. The antibody titers against the wild-type EcA was 1:400,000 after the second immunization, which increased to 1:800,000 after the third immunization; in the case of K288S/Y176F, the antibody titer was 1:60,000 after the second immunization, and it did not increase further after the third immunization.
K288S/Y176F Is Recognized Significantly Less by Serum Antibody from Mice Immunized with Wild-type EcA and Asparaginase-treated ALL Patient
Remission of ALL due to premature inactivation of EcA by antibodies remains a major problem in patients receiving EcA therapy. Moreover, the survival rate is significantly less in ALL relapse patients due to circulating anti-asparaginase antibodies that neutralize EcA and reduce its efficacy. As shown above, K288S/Y176F variant showed less immunogenicity, and we therefore determined whether sera obtained from mice after the third immunization with wild-type EcA and the ALL patients (n = 3) who were given asparaginase therapy for 45 days (patient C) could recognize the K288S/Y176F mutant or not. For this, 1:10,000 to 1:200,000 dilutions of antisera prepared from mice immunized with wild-type EcA after the third immunization was reacted with the wild-type EcA and the K288S/Y176F proteins. The bound antibodies were detected as described under “Experimental Procedures.” For detecting human antibodies bound to wild-type EcA and the K288S/Y176F mutant, human sera (1:100 dilution) from ALL patients were reacted and the antibodies bound to wild-type EcA and K288S/Y176F were detected as described under “Experimental Procedures.” It is assumed that due to the immunogenic nature, administration of EcA will lead to antibody production in mice and in the ALL patients. Sera obtained from healthy individuals and confirmed ALL patients who received no asparaginase therapy (patient A) or received therapy for less than a week (patient B) were used as controls. In agreement with in vitro ELISA results (Fig. 4A), we observed ∼5- and 2-fold less binding of the K288S/Y176F mutant to the sera IgG of mice immunized with wild-type EcA and from an ALL patient (patient C; p < 0.001), respectively, as compared with wild-type EcA (Fig. 4, D and E).
Treatment with EcA Variants Induces Apoptosis in Leukemic Cells
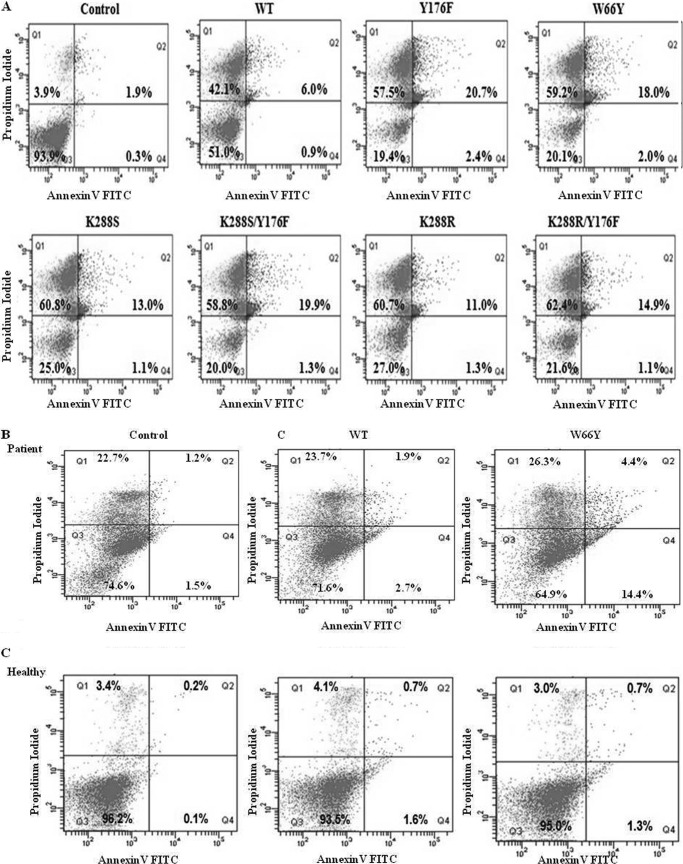
To further understand the mechanism of enhanced cytotoxicity of EcA variants, we examined their effects on apoptosis, PARP activation and cytochrome c release in cell line MV4:11. We first investigated whether the observed cytotoxicity was due to apoptosis. Because variant Y176S did not show cytotoxic activity, it was not included in these experiments. MV4:11 cells were treated with 0.8 units/ml of wild-type EcA and variants for 96 h and then analyzed for apoptosis by annexin V/FITC and propidium iodide (PI) staining (Fig. 5A). Living and apoptotic cells are impermeable to PI, whereas dead cells take up PI and then show red fluorescence. Annexin V binds to phosphatidylserine residues that appear on the surface of apoptotic cells. Thus, after staining a cell population with FITC annexin V and PI, apoptotic cells show green fluorescence, although dead cells show red and green fluorescence, and live cells show little or no fluorescence. After treatment with Y176F and W66Y, 23 and 20% of the cells, respectively, were positive for apoptosis (annexin V-FITC+; PI−, quadrants Q2 and Q4), whereas only 6.9% were annexin V-FITC+ in wild-type EcA-treated cells. No significant difference in the percentage of apoptotic cells was observed after treatment with K288S/Y176F relative to Y176F-treated cells; however, the percentage of apoptosis in K288S/Y176F-treated cells was significantly higher as compared with wild-type EcA. In case of K288S, K288R, and K288R/Y176F, the percentage of positive cells was in the range of 12–16%. In unstimulated controls, only 2% of the cells were positive. Moreover, the fraction of dead cells (upper left quadrant) was significantly higher in cells treated with EcA variants as compared with wild-type EcA (Fig. 5A).
FIGURE 5.
Analysis of apoptosis of MV4:11 and ALL patient lymphoblast cells after treatment with EcA and variants. MV4:11 cells (A), primary lymphoblasts isolated from ALL patients (B), and lymphocytes from healthy individuals (C) were cultured in complete medium for 24 h and then treated with 0.8 units/ml of wild-type EcA and variants for 96 h and analyzed for apoptosis by annexin V binding in flow cytometry. PI stains dead cells with red fluorescence, FITC-annexin V and PI stain apoptotic cells and show green fluorescence, and live cells show little or no fluorescence. Annexin V binding of untreated (control) cells is depicted in upper panel. D, Western blotting of caspase-3, Bcl-XL, cytochrome c, and PARP in MV4:11 cells treated with wild-type EcA, W66Y, and K288S/Y176F (0.8 units/ml) for different periods of time. E, quantification of ammonia production. Wild-type EcA, W66Y, Y176F, and K288S/Y176F variants (20 μg/ml) were incubated with the substrate l-asparagine at 37 °C. After 10 min, the reaction was stopped by adding 1.5 m TCA. The amount of ammonia released was determined by adding Nessler's reagent, and the absorbance was measured from 300 to 600 nm. F, expression of asparagine synthetase transcripts was measured by quantitative real time-PCR. MV4:11 cells were treated with 0.8 units/ml wild-type EcA and variants. RNA was isolated from the treated and untreated cells at different time points. cDNA was synthesized, and the expression of asparagine synthetase was determined using quantitative real time-PCR. Transcript levels are represented relative to mRNA levels of untreated (control) cells. The expression values were normalized with GAPDH. Each experiment was performed three times; means ± S.D. are shown. Significant differences are indicated as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
W66Y Induces Apoptosis in Leukemic Blasts from ALL Patients
To determine the in vivo efficacy of the EcA variants constructed here, leukemic blast cells were isolated from confirmed ALL patients (n = 5), and induction of apoptosis was determined after treatment with variant W66Y. Because of the limited cell numbers obtained from patient samples, we performed this experiment only with W66Y, which showed similar cytotoxic and apoptosis patterns as Y176F and K288S/Y176F. Similar results were obtained with cells obtained from all ALL patients; therefore, Fig. 5B represents the data obtained from one of the confirmed ALL patients. Consistent with the cytotoxicity and apoptosis effects observed in MV4:11 cell line, W66Y showed ∼4-fold higher induction of apoptosis in leukemic blast cells as compared with cells treated with wild-type EcA (Fig. 5B). Importantly, the percentage of apoptosis was significantly higher in W66Y-treated ALL lymphoblasts (18.8%) than in cells from healthy individuals (4.6%) (Fig. 5B), suggesting that W66Y may also kill leukemic cells in patient blood more efficiently. However, in cells from healthy individuals the percentage of apoptosis was 2.3 and 2.0%, respectively (Fig. 5C). Table 1 shows the expression of ALL CD markers in lymphoblasts isolated from same ALL patient (cf. Fig. 5B).
TABLE 1.
Expression of ALL CD markers in the lymphocyte blast cells isolated from patient sample (cf. Fig. 5)
Flow cytometric analysis was performed by analyzing 10,000 gated cells using FACS Diva software (BD Biosciences).
| Patient Serial No. | CD markers | % of expression |
|---|---|---|
| 1 | CD 45 | 64.89 |
| 2 | CD 10 | 53.66 |
| 3 | CD 19 | 59.8 |
| 4 | CD 34 | 0.01 |
| 5 | CD 7 | 0.46 |
| 6 | CD 5 | 0.02 |
| 7 | HLDR | 8.81 |
| 8 | CD 13 | 50.35 |
| 9 | CD 33 | 0.09 |
| 10 | CD 117 | 0.01 |
| 11 | CD 3 | 50.35 |
| 12 | CD 79a | 0.09 |
| 13 | G Ampo | 0.01 |
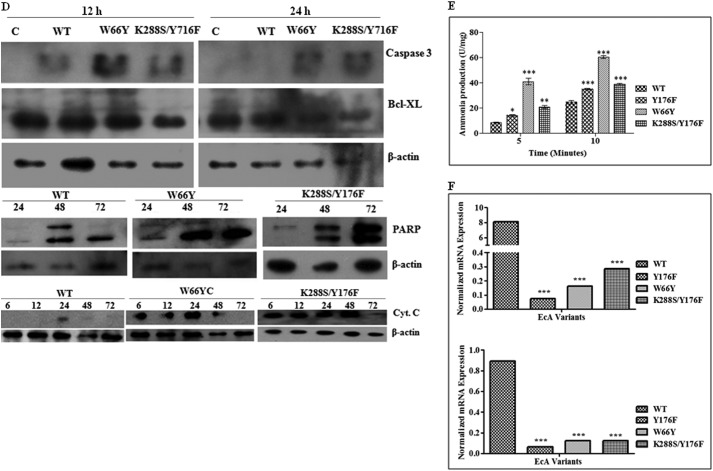
EcA Variants Induce the Formation of Apoptotic Proteins
We next investigated the effects of EcA variants on apoptotic proteins and cytochrome c release from mitochondria in the MV4:11 cell line by Western blotting. Because variants Y176F and W66Y showed similar killing and apoptosis patterns and because K288S/Y176F was found to be less immunogenic and also induced more apoptosis than the other EcA variants, we used W66Y and K288S/Y176F variants only. Both W66Y and K288S/Y176F induced expression of caspase-3, a member of the cysteine aspartic acid protease that plays a central role in induction of apoptosis, as compared with wild-type EcA-treated cells. In contrast, expression of anti-apoptotic protein Bcl-XL was significantly down-regulated in W66Y and K288S/Y176F-treated cells (Fig. 5D). Moreover, W66Y and K288S/Y176F also stimulated cytochrome c release more efficiently than wild-type EcA (Fig. 5D).
We also determined the effects of variants W66Y and K288S/Y176F on the expression of PARP by Western blot analysis of MV4:11 cell extracts. Apoptosis was induced by exposing cells to W66Y and K288S/Y176F variants for increasing lengths of time and was confirmed by monitoring the degradation of PARP to the characteristic 89-kDa C-terminal fragment. As shown in Fig. 5D, the level of the 89-kDa fragment significantly increased after treatment with 0.8 units/ml W66Y and K288S/Y176F. These results suggest that several EcA variants, in particular W66Y and K288S/Y176F, strongly induced PARP-mediated apoptosis, leading to cytochrome c release.
EcA Variants Cause Leukemic Cell Death by Depletion of Asparagine and Down-regulate the Transcription of Asparagine Synthase
To obtain more mechanistic insights of leukemic cell death by EcA variants, we determined the production of ammonia using Nessler's reagent. EcA hydrolyzes asparagine to aspartic acid and ammonia. Asparagine is indispensable for protein synthesis. As shown in Fig. 5E, significantly more (p < 0.001) ammonia production was observed in the presence of Y176F, W66Y, and K288S/Y176F variants as compared with wild-type EcA, indicating that EcA variants deplete asparagine more rapidly than wild-type EcA. Moreover, we also determined the expression of asparagine synthetase in EcA variant-treated MV4:11 cells. As mentioned above, ALL have lost the ability to synthesize asparagine from aspartate due to very low levels of asparagine synthetase. Treatment with Y176F, W66Y, and K288S/Y176F significantly down-regulated the expression of asparagine synthetase as compared with wild-type EcA (Fig. 5F).
Treatment with EcA Variants Arrests the Cells at the G0/G1 Control Point
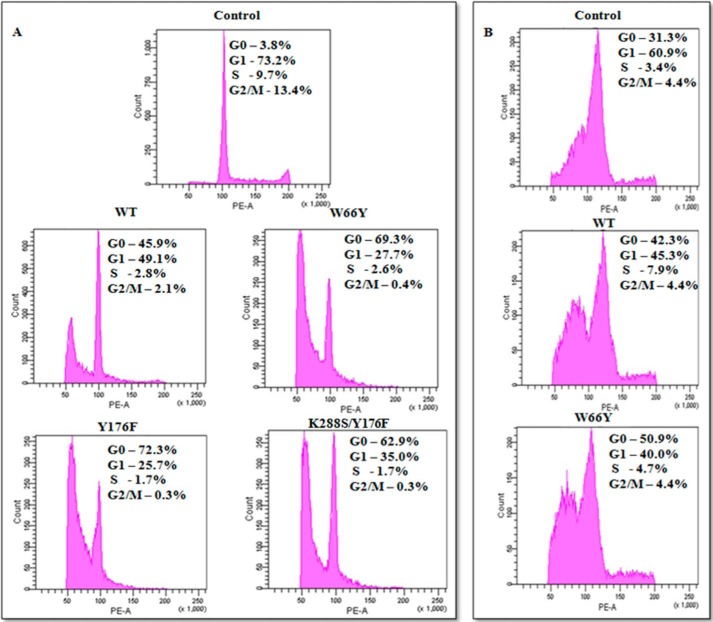
Previous studies have shown that EcA treatment inhibits protein synthesis and arrests the cell cycle in G1 phase (37, 38). We therefore studied the growth behavior of MV4:11 cells after treatment with EcA variants by flow cytometry. In agreement with previous reports, we observed cell cycle arrest at the G0/G1 transition. In addition, we now report that the number of arrested cells was significantly higher after treatment with W66Y (69%), Y176F (72%), and K288S/Y176F (62.9%) in G0 phase as compared with wild-type-treated cells (46%, Fig. 6A). The percentage of cells arrested at different cell cycle stages in response to EcA variant treatment is summarized in Table 2. These data demonstrate that certain EcA variants, particularly Y176F, W66Y, and K288S/Y176F, are more efficient in inducing cell cycle arrest at the G0/G1 control point.
FIGURE 6.
Cell cycle analysis of MV4:11 and primary lymphoblast cells isolated from ALL patient after treatment with wild-type EcA and variants. MV4:11 cells (A) and primary lymphoblasts from ALL patients (B) were cultured in complete medium for 24 h and then treated with 0.8 units/ml EcA and variants for 96 h. The cells were stained with PI and analyzed by flow cytometry. Untreated (control) cells were taken as negative control. Each experiment was performed three times.
TABLE 2.
Cell cycle studies of MV4:11 cells after treatment with EcA variants
The cells were treated with 0.8 units/ml EcA variants for 96 h. Flow cytometric analysis was performed by analyzing 10,000 gated cells using FACS Diva software (BD Biosciences).
| EcA variants | Percentage of cell arrested |
|||
|---|---|---|---|---|
| G0 | G1 | S | G2/M | |
| Control | 3.8 | 73.2 | 9.7 | 13.4 |
| WT | 45.9 | 49.1 | 2.8 | 2.1 |
| K288S | 60.5 | 37.4 | 1.5 | 0.6 |
| K288R | 63.3 | 34.2 | 2.1 | 0.4 |
| K288R/Y176F | 58.2 | 38.9 | 2.2 | 0.6 |
We also monitored the cell cycle progression of primary lymphocytes isolated from ALL patients. Because of limited cell numbers obtained from ALL patient samples, this experiment was performed in EcA wild-type- and W66Y-treated ALL lymphoblasts only. As shown in Fig. 6B, treatment with 0.8 units/ml W66Y arrested significantly more cells (51%) in the G0 phase than wild-type EcA (42%).
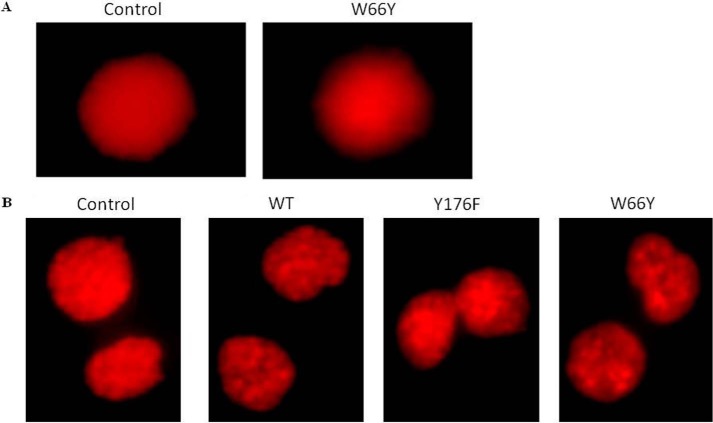
Genotoxic Effects of EcA Variants in an ALL Cell Line
Most drugs that kill cells are causing either cytotoxicity or genotoxicity or both. To further understand the mechanisms of EcA toxicity, we used the so-called comet assay to examine DNA damage in MV4:11 cells treated with W66Y, Y176F, and K288S/Y176F. The comet assay is a sensitive technique for measuring DNA damage, detecting single strand breaks in DNA. In this assay, the length of the tail increases with the extent of DNA damage. Treatment of MV4:11 cells with 0.8 units/ml of W66Y showed no increase in tail length as compared with control cells (Fig. 7A). Similar results were obtained with K288S/Y176F (data not shown). To corroborate the DNA damage studies, micronucleus formation was also studied. 1,000 binucleated cells were analyzed for detecting micronuclei frequency in treated samples. No micronuclei formation was observed in cells treated with EcA-WT, Y176F, and W66Y (Fig. 7B). These results indicate that EcA variants decrease leukemic cell viability predominantly by cytotoxic mechanisms.
FIGURE 7.
Genotoxic effect of EcA and variants in MV4:11 cells. A, comet assay, and B, micronuclei formation of wild-type EcA- and W66Y-treated MV4:11 cells. MV4:11 cells were cultured in complete medium for 24 h, then treated with 0.8 units/ml EcA and variants for 96 h, and analyzed for DNA damage by alkaline Comet assay. The cells were stained with 0.5 μg/ml propidium iodide and observed using a fluorescence microscope (Nikon, Japan). Micronuclei formation was checked by PI staining. Cells were arrested at the cytokinesis stage by cytochalasin-B (0.5 mg/ml) and stained with propidium iodide stain. 1,000 cells were analyzed to check the frequency of micronuclei formation. Images were captured by Olympus BX61 fluorescent microscope and analyzed by Cytovision7.2 software. Untreated (control) cells were taken as negative control. Each experiment was performed three times.
DISCUSSION
EcA is an important component of most protocols for the treatment of primary (39, 40) and relapsing (9) ALL. Despite significant improvements in the clinical efficacy of EcA, relapses due to premature inactivation of EcA by circulating antibodies and the resulting shorter half-life of the enzyme in vivo remains a major problem in ALL therapy. One of the key factors to avoid remissions is the effective depletion of serum asparagine, which will inhibit leukemic cell growth (41). In the context of multiagent therapy, EcA contributes crucially to the dynamic depletion of asparagine (34, 42). Thus, there is a critical need to improve the relevant therapeutic properties of EcA to improve the outcome of ALL chemotherapy. One strategy to augment the therapeutic efficacy is to identify amino acid residues in the EcA that are responsible for its stability and immunogenicity in vivo as well as the efficacy of leukemic cell killing. In this study, we used a rational protein engineering approach to create EcA variants with these desired properties. In the process, we identified amino acid residues present at the dimer interface and within immunogenic epitope regions whose substitution with another amino acid residue reduced immunogenicity and glutaminase activity and increased cytotoxicity against leukemic cells, while preserving their asparaginase activity. Interestingly, one EcA variant K288S/Y176F was recognized significantly less by serum antibodies from mice immunized with wild-type EcA and an ALL patient who has been given asparaginase therapy for 45 days.
In the EcA tetramer, the “intimate dimers” (dimers AC and BD, respectively) have two areas of contact with the nonintimate ones that mainly involve residues 115–125 as well as 175–195 (see Fig. 1). In previous studies, several EcA variants with amino acid substitutions in region 115–125 have already been created and examined in previous studies (24). As shown here, substitutions at position 176 did not impair asparaginase activity, although the glutaminase activity was markedly reduced. These results indicate that, with the exception of Y176S, the mutations made here probably did not change the overall conformation of the enzyme. It has been shown that human serum increases anti-tumor activity of EcA (43). Addition of human serum maintained asparaginase activity of all EcA variants, indicating that serum components may cause stabilization of the asparaginase protein and thus extend its half-life.
Some side effects of EcA treatment have been attributed to its glutaminase activity (44–47). Therefore, attempts were made to reduce glutaminase activity of the variants and thereby toxicity (18). In this study, no clear correlation between asparaginase and glutaminase activity of different mutants was observed. So residue Tyr-176 for which a mutation to Phe and double mutation K288S/Y176F caused significant reduction in glutaminase activity nevertheless had a slightly higher asparaginase activity and significantly improved cytotoxicity as compared with wild-type EcA (discussed below). The mutation Tyr-176 to Phe impairs turnover of the weaker glutamine substrate to a higher degree than that of asparagine. This may result from small conformational changes at a subunit interface that would affect binding or be due to an impact of mutation on the relatively solid and rigid three-dimensional structure of the active site of the enzyme, which limits the accommodation of extra CH2 group of glutamine. Variant Y176S, although it was devoid of glutaminase activity, had a comparable asparaginase activity but significantly reduced cytotoxicity as compared with wild-type EcA. In addition to Y176F, W66Y with both high glutaminase activity and higher cytotoxicity also appears to be an effective drug. Similar observations were reported by others for an EcA variant N24A mutation that showed stable glutaminase activity and also improved cell-killing properties (48).
Glutamine is essential for many cell types as a major source of energy, nitrogen, and carbon (49, 50). It is possible that glutamine metabolism links with the tricarboxylic acid cycle to maintain the availability of metabolic intermediates critical for cancer cell survival and proliferation. Indeed, glutamine addiction in cancer cells represents a potential therapeutic target (51). This may explain why, at least in vitro, depletion of exogenous glutamine by EcA enhances cytotoxicity. Therefore, an enzyme with lower glutaminase activity but comparable cytotoxicity has potential therapeutic applications. Both asparaginase as well as glutaminase side activities are responsible for the production of ammonia. Although Y176F and K288S/Y176F mutants exhibited low glutaminase activity, high ammonia production was observed. We have seen that Y176F and K288S/Y176F are significantly more stable, show high catalytic activity, and also exhibit more asparaginase activity as compared with wild-type EcA.3 Therefore, the increased ammonia production could be due to higher stability and asparaginase and catalytic activities.
As a bacterial protein, EcA causes undesired effects in many children, including serious allergic reactions. Therefore, we attempted to create EcA variants with reduced antigenicity relative to wild-type EcA. For this purpose, we created several thousand mutants by substituting amino acid residues by the 19 other naturally occurring amino acids in the B-cell epitope regions that we have identified previously (19). The mutants were created by random mutagenesis of a specific amino acid residue present in different epitopic regions and selected by a replica plate method. At least 2,000 colonies resulting from each mutation were selected and tested for asparaginase activity, stability, and antigenicity. The activities of variants with mutations at Lys-288 were only slightly altered. Combining the substitution at the dimer interface (Y176F) with a second mutation in the B-cell epitope (K288S) led to the development of a less immunogenic mutant and may therefore form a basis for further studies aimed at evaluating the use of antigenically modified enzymes for therapy. In this study, we demonstrated that immunization of mice with a double mutant K288S/Y176F induced significantly less antibodies as compared with the wild-type EcA even after three immunizations. However, the antibody titers against the wild-type EcA in mice increased from 1:400,000 to 1:800,000 (almost 2-fold) between the second and third immunization, whereas in case of K288S/Y176F there was no significant increase (1:60,000) under the same condition. Interestingly, K288S/Y176F also showed significantly less binding to serum antibodies from mice immunized with wild-type EcA and ALL patients who were given asparaginase therapy for several weeks. In therapeutic perspective, this is an important finding. We have found that K288S/Y176F has reduced ability to induce antibodies in vivo in mice and binds less to existing anti-EcA antibodies present in the serum of mice immunized with wild-type EcA and in the serum of ALL patients taking asparaginase therapy. Therefore, it may prove very useful in the treatment of relapse in ALL cases. Many sensitive patients react to repeated EcA administration with the rapid formation of antibodies that bind to and thereby neutralize the activity of administered EcA, which renders the therapy ineffective. It is worth noting that Y176F and K288S as single mutants did not show reduced antigenicity. It suggests that a substitution of Tyr-176 to Phe affects the conformation of the Lys-288 epitope and thus its immunogenic properties. We have found that the single mutant Y176F is extremely stable as compared with wild-type EcA.3 Similarly, in Erwinia chrysanthemi l-asparaginase, a mutation of Glu-288, a residue occupying a similar position as Lys-288 in EcA reduced its antigenicity by 1.5-fold (52). Examination of the structures show that this immunodominant region near the C terminus is located at the surface of the enzyme, with a high relative flexibility, and therefore it would be expected to be more accessible for interaction with antibodies.
In the second part of this study, we explored whether mutations Y176F and W66Y as single changes or in combination show a preference in killing leukemic cells relative to normal cells. For this, we compared the cytotoxicity of these two variants in leukemia cell lines and PBMCs. In ALL cell lines, Y176F and W66Y killed significantly more cells as compared with wild-type EcA, although PBMCs were minimally affected by the same dose. The increased cytotoxicity of W66Y may be associated with high glutaminase activity, whereas the enhanced cytotoxicity due to the substitution Y176F remains to be explained. Although previous studies indicate that EcA causes DNA fragmentation at higher doses, our study shows that EcA causes cell death by inducing cytotoxicity and not by genotoxicity under these experimental conditions. Thus, the data presented here suggest that both Y176F and W66Y are superior antileukemic agents as compared with wild-type EcA. Moreover, our study confirms the previously reported lack of significant effect of EcA on nonmalignant cells (53).
A significant induction in the apoptosis of ALL cells was observed in cells treated with Y176F, W66Y, and K288S/Y176F. W66Y and K288S/Y176F induced cell death through the mitochondrion-dependent apoptotic pathway, mediated by cytochrome c release. Previous studies have shown that after release into the cytoplasm cytochrome c initiates apoptosis by binding to apoptotic protease activating factor-1 (Apaf-1) (54). Caspase-mediated apoptotic cell death is accomplished through the cleavage of several key proteins required for cellular functioning and survival. Among these, the induction of caspases and cleavage of PARP into 89- and 24-kDa fragments by caspases is considered a hallmark of apoptosis. This occurs during drug-induced apoptosis in a variety of cells (55) by depleting the cell of NAD and ATP (56). We found a significant increase in the level of caspase-3 and the cleaved 89-kDa fragment in W66Y- and K288S/Y176F-treated cells indicating that PARP-dependent apoptosis indeed takes place. In contrast, the expression of anti-apoptotic protein Bcl-XL was down-regulated in EcA variant-treated cells. Moreover, W66Y and Y176F also induced more apoptosis in primary ALL lymphoblasts. The above results indicate that W66Y, Y176F, and K288S/Y176F have good potential as therapeutic agents. Further mechanistic studies revealed that W66Y and K288S/Y176F variants caused rapid depletion of asparagine, which is essential for protein synthesis. Moreover, both enzymes down-regulated the expression of asparagine synthetase enzyme. It is well established that ALL lymphoblasts have lost the ability to synthesize asparagine from aspartate due to low levels of asparagine synthetase. Therefore, the blast must take asparagine from blood plasma to build new proteins. Our study suggests that EcA variants cause rapid depletion of asparagine and also down-regulate the expression of asparagine synthetase, which therefore interferes with the proliferation of the leukemic blasts and drive them to apoptosis. However, it remains to be clarified how EcA leads to down-regulation of asparagine synthetase.
As mentioned above, the clinical use of EcA in the treatment of ALL is thought to be based on the degradation of l-asparagine, which is required for cell proliferation. It has been suggested that protein and DNA synthesis are delayed following EcA treatment and that the protein synthesis required to initiate or complete a phase of the cell cycle was inhibited due to l-asparagine deprivation (35, 57). In our study, we observed that treatment of MV4:11 cells and primary lymphoblasts from ALL patients with EcA variants, in particular Y176F, K288S/Y176F, and W66Y, blocked the cell cycle at G0 phase, although in the case of wild-type EcA, this block occurred in the G1 phase. It appears that Y176F, K288S/Y176F, and W66Y deplete l-asparagine more rapidly as compared with wild-type EcA and thereby block protein synthesis at the early phase of the cell cycle. It could also be due to an increase in internal protein turnover because of absence of net synthesis of proteins or blocking of de novo l-asparagine biosynthesis. Blocking the leukemic cell cycle progression at an early phase may prove crucial for ALL therapy.
In summary, we altered the properties of EcA to allow a different treatment scenario. EcA was engineered to enhance cytotoxicity against leukemic cells and to abate “silent inactivation” by decreasing immunogenicity. We further show that reduced glutaminase activity is not necessarily correlated with higher cytotoxicity (Y176F). Three variants (W66Y, Y176F, and K288S/Y176F) appear especially promising as substitutes for wild-type EcA in ALL therapy, showing a potential for therapeutic use and forming a sound basis for future clinical trials.
Acknowledgments
We thank Soumitra Mohanty and Prajna Jena for technical support, critical reading of the manuscript, and for fruitful discussions.
This work was supported by Grant SR/FT/LS-041/2009 from the Department of Science and Technology, Government of India (to A. S.).
S. Verma, R. Mehta, P. Maiti, K. H. Roehm, and A. Sonawane, unpublished data.
- ADA
- anti-drug antibody
- ALL
- acute lymphoblastic leukemia
- PI
- propidium iodide
- PARP
- poly(ADP-ribose) polymerase
- PBMC
- peripheral blood mononuclear cell
- AHA
- l-aspartic β-hydroxamate
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
REFERENCES
- 1. Cantor J. R., Yoo T. H., Dixit A., Iverson B. L., Forsthuber T. G., Georgiou G. (2011) Therapeutic enzyme deimmunization by combinatorial T-cell epitope removal using neutral drift. Proc. Natl. Acad. Sci. U.S.A. 108, 1272–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wriston J. C., Jr., Yellin T. O. (1973) l-Asparaginase: a review. Adv. Enzymol. Relat. Areas Mol. Biol. 39, 185–248 [DOI] [PubMed] [Google Scholar]
- 3. Ravindranath Y., Abella E., Krischer J. P., Wiley J., Inoue S., Harris M., Chauvenet A., Alvarado C. S., Dubowy R., Ritchey A. K., et al. (1992) Acute myeloid leukemia (AML) in Down's syndrome is highly responsive to chemotherapy: experience on Pediatric Oncology Group AML Study 8498. Blood 80, 2210–2214 [PubMed] [Google Scholar]
- 4. Cooney D. A., Milman H. A. (1972) A radiometric technique for measuring l-asparagine in picomole quantities. Biochem. J. 129, 953–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Müller H. J., Boos J. (1998) Use of l-asparaginase in childhood ALL. Crit. Rev. Oncol. Hematol. 28, 97–113 [DOI] [PubMed] [Google Scholar]
- 6. Iiboshi Y., Papst P. J., Hunger S. P., Terada N. (1999) l-Asparaginase inhibits the rapamycin-targeted signaling pathway. Biochem. Biophys. Res. Commun. 260, 534–539 [DOI] [PubMed] [Google Scholar]
- 7. Suto H., Yasuda H., Isobe Y., Sasaki M., Imai H., Tsutsui M., Oshimi K., Komatsu N., Sugimoto K. (2010) Suppression of eIF4E expression by l-asparaginase. Acta Haematol. 123, 215–219 [DOI] [PubMed] [Google Scholar]
- 8. Yang L., Panetta J. C., Cai X., Yang W., Pei D., Cheng C., Kornegay N., Pui C. H., Relling M. V. (2008) Asparaginase may influence dexamethasone pharmacokinetics in acute lymphoblastic leukemia. J. Clin. Oncol. 26, 1932–1939 [DOI] [PubMed] [Google Scholar]
- 9. Abshire T. C., Pollock B. H., Billett A. L., Bradley P., Buchanan G. R. (2000) Weekly polyethylene glycol conjugated l-asparaginase compared with biweekly dosing produces superior induction remission rates in childhood relapsed acute lymphoblastic leukemia: a Pediatric Oncology Group Study. Blood 96, 1709–1715 [PubMed] [Google Scholar]
- 10. Pession A., Valsecchi M. G., Masera G., Kamps W. A., Magyarosy E., Rizzari C., van Wering E. R., Lo Nigro L., van der Does A., Locatelli F., Basso G., Aricò M. (2005) Long-term results of a randomized trial on extended use of high dose l-asparaginase for standard risk childhood acute lymphoblastic leukemia. J. Clin. Oncol. 23, 7161–7167 [DOI] [PubMed] [Google Scholar]
- 11. Moghrabi A., Levy D. E., Asselin B., Barr R., Clavell L., Hurwitz C., Samson Y., Schorin M., Dalton V. K., Lipshultz S. E., Neuberg D. S., Gelber R. D., Cohen H. J., Sallan S. E., Silverman L. B. (2007) Results of the Dana-Farber Cancer Institute ALL Consortium Protocol 95-01 for children with acute lymphoblastic leukemia. Blood 109, 896–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bhattacharya K., Samanta S. K., Tripathi R., Mallick A., Chandra S., Pal B. C., Shaha C., Mandal C. (2010) Apoptotic effects of mahanine on human leukemic cells are mediated through crosstalk between Apo-1/Fas signaling and the Bid protein and via mitochondrial pathways. Biochem. Pharmacol. 79, 361–372 [DOI] [PubMed] [Google Scholar]
- 13. Mondal S., Mandal C., Sangwan R., Chandra S., Mandal C. (2010) Withanolide D induces apoptosis in leukemia by targeting the activation of neutral sphingomyelinase-ceramide cascade mediated by synergistic activation of c-Jun N-terminal kinase and p38 mitogen-activated protein kinase. Mol. Cancer 9, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holleman A., den Boer M. L., Kazemier K. M., Janka-Schaub G. E., Pieters R. (2003) Resistance to different classes of drugs is associated with impaired apoptosis in childhood acute lymphoblastic leukemia. Blood 102, 4541–4546 [DOI] [PubMed] [Google Scholar]
- 15. Hak L. J., Relling M. V., Cheng C., Pei D., Wang B., Sandlund J. T., Rubnitz J., Pui C. H. (2004) Asparaginase pharmacodynamics differ by formulation among children with newly diagnosed acute lymphoblastic leukemia. Leukemia 18, 1072–1077 [DOI] [PubMed] [Google Scholar]
- 16. Albertsen B. K., Schrøder H., Ingerslev J., Jakobsen P., Avramis V. I., Müller H. J., Carlsen N. T., Schmiegelow K. (2001) Comparison of intramuscular therapy with Erwinia asparaginase and asparaginase Medac: pharmacokinetics, pharmacodynamics, formation of antibodies and influence on the coagulation system. Br. J. Haematol. 115, 983–990 [DOI] [PubMed] [Google Scholar]
- 17. Jianhua C., Yujun W., Ruibo J., Min W., Wutong W. (2006) Probing the antigenicity of E. coli l-asparaginase by mutational analysis. Mol. Biotechnol. 33, 57–65 [DOI] [PubMed] [Google Scholar]
- 18. Derst C., Henseling J., Röhm K. H. (2000) Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase activity by amino acid replacements at position 248. Protein Sci. 9, 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Werner A., Röhm K. H., Müller H. J. (2005) Mapping of B-cell epitopes in E. coli asparaginase II, an enzyme used in leukemia treatment. Biol. Chem. 386, 535–540 [DOI] [PubMed] [Google Scholar]
- 20. Derst C., Wehner A., Specht V., Röhm K. H. (1994) States and functions of tyrosine residues in Escherichia coli asparaginase II. Eur. J. Biochem. 224, 533–540 [DOI] [PubMed] [Google Scholar]
- 21. Harms E., Wehner A., Jennings M. P., Pugh K. J., Beacham I. R., Röhm K. H. (1991) Construction of expression systems for Escherichia coli asparaginase II and two-step purification of the recombinant enzyme from periplasmic extracts. Protein Expr. Purif. 2, 144–150 [DOI] [PubMed] [Google Scholar]
- 22. Wehner A., Harms E., Jennings M. P., Beacham I. R., Derst C., Bast P., Röhm K. H. (1992) Site-specific mutagenesis of Escherichia coli asparaginase II. None of the three histidine residues is required for catalysis. Eur. J. Biochem. 208, 475–480 [DOI] [PubMed] [Google Scholar]
- 23. Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 24. Derst C., Henseling J., Röhm K. H. (1992) Probing the role of threonine and serine residues of E. coli asparaginase II by site-specific mutagenesis. Protein Eng. 5, 785–789 [DOI] [PubMed] [Google Scholar]
- 25. Rizzari C., Citterio M., Zucchetti M., Conter V., Chiesa R., Colombini A., Malguzzi S., Silvestri D., D'Incalci M. (2006) A pharmacological study on pegylated asparaginase used in front-line treatment of children with acute lymphoblastic leukemia. Haematologica 91, 24–31 [PubMed] [Google Scholar]
- 26. Miller K. E., Balbas J. C., Benton R. L., Lam T. S., Edwards K. M., Kriebel R. M., Schechter R. (2012) Glutaminase immunoreactivity and enzyme activity is increased in the rat dorsal root ganglion following peripheral inflammation. Pain Res. Treat. 10.1155/2012/414697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kenny J., Bao Y., Hamm B., Taylor L., Toth A., Wagers B., Curthoys N. P. (2003) Bacterial expression, purification, and characterization of rat kidney-type mitochondrial glutaminase. Protein Expr. Purif. 31, 140–148 [DOI] [PubMed] [Google Scholar]
- 28. Jeon J. M., Lee H. I., So J. S. (2009) Glutaminase activity of Lactobacillus reuteri KCTC3594 and expression of the activity in other Lactobacillus spp. by introduction of the glutaminase gene. Afr. J. Microbiol. Res. 3, 605–609 [Google Scholar]
- 29. Böyum A. (1968) Isolation of leucocytes from human blood. Further observations. Methylcellulose, dextran, and Ficoll as erythrocyte aggregating agents. Scand. J. Clin. Lab. Invest. Suppl. 97, 31–50 [PubMed] [Google Scholar]
- 30. Böyum A. (1968) Separation of leukocytes from blood and bone marrow. Introduction. Scand. J. Clin. Lab. Invest. Suppl. 97, 7. [PubMed] [Google Scholar]
- 31. Wang B., Hak L. J., Relling M. V., Pui C. H., Woo M. H., Storm M. C. (2000) ELISA to evaluate plasma anti-asparaginase IgG concentrations in patients with acute lymphoblastic leukemia. J. Immunol. Methods 239, 75–83 [DOI] [PubMed] [Google Scholar]
- 32. Fotakis G., Timbrell J. A. (2006) In vitro cytotoxicity assays: comparison of LDH, neutral red, MTT and protein assay in hepatoma cell lines following exposure to cadmium chloride. Toxicol. Lett. 160, 171–177 [DOI] [PubMed] [Google Scholar]
- 33. Fenech M., Bolognesi C., Kirsch-Volders M., Bonassi S., Zeiger E., Knasmüller S., Holland N. (2007) Harmonisation of the micronucleus assay in human buccal cells–a Human Micronucleus (HUMN) project (www.humn.org) initiative commencing in 2007. Mutagenesis 22, 3–4 [DOI] [PubMed] [Google Scholar]
- 34. Kwok C. S., Kham S. K., Ariffin H., Lin H. P., Quah T. C., Yeoh A. E. (2006) Minimal residual disease (MRD) measurement as a tool to compare the efficacy of chemotherapeutic drug regimens using Escherichia coli-asparaginase or Erwinia-asparaginase in childhood acute lymphoblastic leukemia (ALL). Pediatr. Blood Cancer 47, 299–304 [DOI] [PubMed] [Google Scholar]
- 35. Mashburn L. T., Gordon C. S. (1968) The effects of l-asparaginase on the amino acid incorporation of mouse lymphoid tumors. Cancer Res. 28, 961–967 [PubMed] [Google Scholar]
- 36. Benjwal S., Verma S., Röhm K. H., Gursky O. (2006) Monitoring protein aggregation during thermal unfolding in circular dichroism experiments. Protein Sci. 15, 635–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ueno T., Ohtawa K., Mitsui K., Kodera Y., Hiroto M., Matsushima A., Inada Y., Nishimura H. (1997) Cell cycle arrest and apoptosis of leukemia cells induced by l-asparaginase. Leukemia 11, 1858–1861 [DOI] [PubMed] [Google Scholar]
- 38. Krejci O., Starkova J., Otova B., Madzo J., Kalinova M., Hrusak O., Trka J. (2004) Upregulation of asparagine synthetase fails to avert cell cycle arrest induced by l-asparaginase in TEL/AML1-positive leukaemic cells. Leukemia 18, 434–441 [DOI] [PubMed] [Google Scholar]
- 39. Sallan S. E., Hitchcock-Bryan S., Gelber R., Cassady J. R., Frei E., 3rd, Nathan D. G. (1983) Influence of intensive asparaginase in the treatment of childhood non-T-cell acute lymphoblastic leukemia. Cancer Res. 43, 5601–5607 [PubMed] [Google Scholar]
- 40. Otten J., Philippe N., Suciu S., Béhar C., Babin-Boilletot A., Thyss A., Ferster A., Vilmer E., and EORTC Children Leukemia Group (2002) The Children Leukemia Group: 30 years of research and achievements. Eur. J. Cancer 38, S44–S49 [DOI] [PubMed] [Google Scholar]
- 41. Jarrar M., Gaynon P. S., Periclou A. P., Fu C., Harris R. E., Stram D., Altman A., Bostrom B., Breneman J., Steele D., Trigg M., Zipf T., Avramis V. I. (2006) Asparagine depletion after pegylated E. coli asparaginase treatment and induction outcome in children with acute lymphoblastic leukemia in first bone marrow relapse: a Children's Oncology Group study (CCG-1941). Pediatr. Blood Cancer 47, 141–146 [DOI] [PubMed] [Google Scholar]
- 42. zur Stadt U., Harms D. O., Schlüter S., Jorch N., Spaar H. J., Nürnberger W., Völpel S., Gutjahr P., Schrappe M., Janka G., Kabisch H. (2000) Minimal residual disease analysis in acute lymphoblastic leukemia of childhood within the framework of COALL study: results of an induction therapy without asparaginase. Klin. Padiatr. 212, 169–173 [DOI] [PubMed] [Google Scholar]
- 43. Lee M. B., Niblock R. M., Bridges J. M. (1969) The influence of human serum on the anti-tumour activity of two l-asparaginases. Br. J. Cancer 23, 369–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van den Berg H. (2011) Asparaginase revisited. Leuk. Lymphoma 52, 168–178 [DOI] [PubMed] [Google Scholar]
- 45. Davidson L., Brear D. R., Wingard P., Hawkins J., Kitto G. B. (1977) Purification and properties of l-glutaminase-l-asparaginase from Pseudomonas acidovorans. J. Bacteriol. 129, 1379–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Avramis V. I., Tiwari P. N. (2006) Asparaginase (native ASNase or pegylated ASNase) in the treatment of acute lymphoblastic leukemia. Int. J. Nanomedicine 1, 241–254 [PMC free article] [PubMed] [Google Scholar]
- 47. Kotzia G. A., Labrou N. E. (2005) Cloning, expression and characterisation of Erwinia carotovora l-asparaginase. J. Biotechnol. 119, 309–323 [DOI] [PubMed] [Google Scholar]
- 48. Offman M. N., Krol M., Patel N., Krishnan S., Liu J., Saha V., Bates P. A. (2011) Rational engineering of l-asparaginase reveals importance of dual activity for cancer cell toxicity. Blood 117, 1614–1621 [DOI] [PubMed] [Google Scholar]
- 49. DeBerardinis R. J., Mancuso A., Daikhin E., Nissim I., Yudkoff M., Wehrli S., Thompson C. B. (2007) Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. U.S.A. 104, 19345–19350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reitzer L. J., Wice B. M., Kennell D. (1979) Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J. Biol. Chem. 254, 2669–2676 [PubMed] [Google Scholar]
- 51. Wise D. R., Thompson C. B. (2010) Glutamine addiction: a new therapeutic target in cancer. Trends Biochem. Sci. 35, 427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Moola Z. B., Scawen M. D., Atkinson T., Nicholls D. J. (1994) Erwinia chrysanthemi l-asparaginase: epitope mapping and production of antigenically modified enzymes. Biochem. J. 302, 921–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Apostolidou E., Swords R., Alvarado Y., Giles F. J. (2007) Treatment of acute lymphoblastic leukaemia: a new era. Drugs 67, 2153–2171 [DOI] [PubMed] [Google Scholar]
- 54. Mohr A., Zwacka R. M., Debatin K. M., Stahnke K. (2004) A novel method for the combined flow cytometric analysis of cell cycle and cytochrome c release. Cell Death Differ. 11, 1153–1154 [DOI] [PubMed] [Google Scholar]
- 55. Suppipat K., Park C. S., Shen Y., Zhu X., Lacorazza H. D. (2012) Sulforaphane induces cell cycle arrest and apoptosis in acute lymphoblastic leukemia cells. PLoS One 7, e51251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yang B., Fan L., Fang L., He Q. (2006) Hypoxia-mediated fenretinide (4-HPR) resistance in childhood acute lymphoblastic leukemia cells. Cancer Chemother. Pharmacol. 58, 540–546 [DOI] [PubMed] [Google Scholar]
- 57. Becker F. F., Broome J. D. (1967) l-Asparaginase: inhibition of early mitosis in regenerating rat liver. Science 156, 1602–1603 [DOI] [PubMed] [Google Scholar]