Background: The type III secretion system allows bacteria to inject effectors directly into the host cytoplasm through a translocation pore.
Results: Translocator proteins were characterized in lipid-bound and chaperone-associated forms.
Conclusion: Pseudomonas translocators use different regions to recognize their common chaperone and the eukaryotic membrane.
Significance: Mapping of key regions in translocators provides a starting point for the potential development of novel antibacterials.
Keywords: Crystal Structure, Host-Pathogen Interactions, Membrane Proteins, Pseudomonas aeruginosa, Type III Secretion System
Abstract
The type III secretion system is a widespread apparatus used by pathogenic bacteria to inject effectors directly into the cytoplasm of eukaryotic cells. A key component of this highly conserved system is the translocon, a pore formed in the host membrane that is essential for toxins to bypass this last physical barrier. In Pseudomonas aeruginosa the translocon is composed of PopB and PopD, both of which before secretion are stabilized within the bacterial cytoplasm by a common chaperone, PcrH. In this work we characterize PopB, the major translocator, in both membrane-associated and PcrH-bound forms. By combining sucrose gradient centrifugation experiments, limited proteolysis, one-dimensional NMR, and β-lactamase reporter assays on eukaryotic cells, we show that PopB is stably inserted into bilayers with its flexible N-terminal domain and C-terminal tail exposed to the outside. In addition, we also report the crystal structure of the complex between PcrH and an N-terminal region of PopB (residues 51–59), which reveals that PopB lies within the concave face of PcrH, employing mostly backbone residues for contact. PcrH is thus the first chaperone whose structure has been solved in complex with both type III secretion systems translocators, revealing that both molecules employ the same surface for binding and excluding the possibility of formation of a ternary complex. The characterization of the major type III secretion system translocon component in both membrane-bound and chaperone-bound forms is a key step for the eventual development of antibacterials that block translocon assembly.
Introduction
Type III secretion systems (T3SSs)3 are complex macromolecular nanomachines that play key roles in the infectivity processes of a number of Gram-negative bacteria. These complex systems harbor >20 proteins that associate into a needle-like structure located directly on the bacterial surface. T3SSs encompass both bacterial membranes and serve as conduits to export specific effectors directly from the bacterial cytosol into target eukaryotic cells (1). Because the internal diameter of the needle is on the order of 25 Å, it is believed that effectors must travel in a semi-unfolded state (2, 3) due to energy provided by an ATPase located at the base of the system (4–6). Introduction of point mutations into key proteins of the T3SS have shown to be highly deleterious for the cytotoxicity potential of certain pathogens (7–11), and T3SS-deficient strains display attenuated infectivity in animal models (12–14), indicating that an understanding of T3SS formation and regulation mechanisms could lead to the development of strategies for infection control.
T3SS can be divided into three main sections: 1) a ring-like base structure that encompasses both bacterial membranes and bypasses the periplasm, 2) a hollow needle formed through the polymerization of a single protein through which effectors are thought to travel, and 3) the translocon, a pore formed on the target bilayer by two T3SS-translocated membrane proteins and one hydrophilic partner. The base and the needle are collectively known as the injectisome and have been elegantly characterized through a combination of structural biology, biochemistry, and microbiology techniques (3, 10, 15–21). The translocon, however, remains less well studied despite the fact that it is essential for T3SS functionality (22–24).
All three proteins that compose the translocon are encoded in a single operon that also carries genes that code for chaperones for the different molecules. In Pseudomonas aeruginosa, for example, the translocon is composed of membrane proteins PopB and PopD (chaperoned by PcrH) and the hydrophilic partner PcrV (chaperoned by PcrG), all of which are encoded within the genetic element pcrGVHpopBD (25). PopB is the “major translocator,” a 390-residue protein with two putative transmembrane regions, whereas PopD, the “minor translocator,” carries 295 amino acids and a single predicted transmembrane domain and makes a 1:1 complex with PcrH (26). The Yersinia proteins YopB and YopD display 64 and 60% sequence similarity with their counterparts from Pseudomonas, whereas IpaB and IpaC from Shigella spp. are considerably longer, harboring 580 and 363 amino acids, respectively. All major translocators studied to date display at least one predicted coiled coil region and two transmembrane domains, whereas minor translocators are considerably shorter molecules, mostly with one predicted coiled coil and transmembrane region (22). Understanding how translocon molecules interact with their respective chaperones within the bacterial cytoplasm and associate to the target membrane upon passage through the needle has been the objective of a number of laboratories, as both aspects have been shown to play key roles in bacterial pathogenesis.
P. aeruginosa is a major human pathogen that targets immuno-depressed and cystic fibrosis patients. Its type III secretion system plays an essential role in the initial phases of infection (12, 24, 27). In this work we characterize its major translocator PopB through lipid binding studies, nuclear magnetic resonance, x-ray crystallography, and cellular toxicity assays. The flexible N-terminal region does not become inserted into membranes upon lipid recognition and has characteristics of a molten globule, which could be important for its unfolding and passage through the needle. Notably, the high resolution structure of a complex between a peptide located within the N terminus of PopB and PcrH reveals that central hydrophobic residues of the peptide are important for chaperone recognition and excludes the formation of a 1:1:1 complex between PcrH and the two translocator molecules. These results shed light on common features of many of the characteristics of major translocator proteins in bacterial pathogens.
EXPERIMENTAL PROCEDURES
Liposome Preparation and Sucrose Gradients
Liposomes were prepared by initially preparing a lipidic biofilm using a 2:3 PS:PC (α-Phosphatidyl-l-serine:1,2-dipalmitoyl-sn-glycero-3-phosphocholine) mixture in chloroform through evaporation under nitrogen for 30 min. The lipidic film was dissolved in 3 ml of PBS, pH 7.0, and 0.7% octyl-β-d-glucopyranoside by vortexing followed by repeated freeze/thaw cycles in liquid nitrogen. The mixture was stored at −80 °C. Subsequently, the lipidic mixture was thawed and extruded, producing liposomes with an average diameter of 100 μm. 5 mg of liposomes were incubated overnight at RT with 0.8 mg of PopB or 0.8 mg of PopD. The two different proteoliposome mixtures were then incubated on a rotating wheel at RT with different agents: 1 m KCl, 6 m urea, 0.425 mm n-Dodecyl-β-d-maltopyranoside (DDM) or PBS, pH 7.0, as a control. After 4 h of incubation the solutions were tested on a 0–45% sucrose gradient. Tubes were ultracentrifuged at 230,000 × g for 16 h in a SW41.Ti rotor. 1-ml fractions were recovered from the top to the bottom of the gradient and analyzed by Western blots, which were developed with either anti-PopB or anti-PopD rabbit antibodies.
Trypsin Digestion
PopB and PopD were purified by nickel affinity chromatography and gel filtration at pH 5.0 as described in Schoehn et al. (26). Liposomes were prepared by employing an extruder (Avanti Polar Lipids) using a PS:PC:sphingomyelin for PopD and PS:PC:sphingomyelin:cholesterol for PopB, with the same ratio for all lipids. Proteoliposomes were then prepared by incubation of liposomes at RT with PopD or PopB. Both proteoliposome samples were dialyzed into PBS, pH 7.0, before trypsin digestion. Samples were subsequently proteolyzed using trypsin:protein ratios of 1:500 and 1:100, respectively, at RT. Samples were taken at different times and loaded on SDS-PAGE and subsequently analyzed by Coomassie staining or Western blotting.
One-dimensional NMR
The one-dimensional proton spectrum was recorded at 25 °C with excitation-sculpting water suppression (28). The experiment was performed on an Agilent Direct Drive 800 MHz spectrometer equipped with a cryogenic triple resonance probe. The experimental time was set to 17 min using 1024 scans and a recovery time of 1s.
Plasmid Constructs
The pET15b-PopB/PcrH vector was obtained by first cloning the two genes separately into two distinct pET15b plasmids. popB was cloned using NdeI/BamHI sites, whereas PcrH was cloned in the BamHI site of pET15b. Subsequently, pET15b-PcrH was digested with XbaI-BamHI to maintain the ribosomal binding site and cloned blunt-ended in the BamHI blunt-ended pET15b-PopB. The pETDuet-PopD/PcrH vector was obtained by cloning popD in the NdeI-XhoI sites present in the second multiple cloning site of pETDuet1-PcrH(1–160) vector, described previously (8).
For the in vivo experiments, the section of P. aeruginosa CHA operon containing pcrHpopBpopD was cloned into the NdeI-HindIII restriction sites of the pIApG vector. Two different PopB mutants were generated with the QuikChange II site-directed mutagenesis kit (Stratagene), namely mutants L55D/P58D (named PopB-2D) and V53D/L55D/P57D/P58D (named PopB-4D). The presence of only these mutations was verified by DNA sequencing.
Cytotoxicity toward Macrophages
Macrophage cell line J774 was grown in DMEM/F-12 media with 20% of FCS and antibiotics. 2.5 × 105 cells were separated into aliquots in 24-well plates and washed in DMEM without antibiotics before infection with a culture of P. aeruginosa at A600 = 1.0. A multiplicity of infection (MOI) of 5 was used, and 30-μl aliquots of the supernatant were taken after 1, 2, 3, and 4 h of infection for quantification of LDH release with the LDH cytotoxicity kit (Roche Applied BioScience). The value of LDH release obtained after the addition of 0.5% Triton X-100 was considered as 100% cell death, and uninfected macrophages were used as negative controls.
Protein Stability
Pseudomonas cultures were grown up to an A600 = 1.0 absorbance units; subsequently, induction of the T3SS was performed by the addition of 5 mm EDTA and 20 mm MgCl2, whereas protein synthesis was blocked by the addition of 500 μg/ml chloramphenicol. After 1, 2, and 3 h cells were centrifuged, and the pellet was analyzed by Western blotting.
Construction of a PopB-β-Lac Fusion
A chimeric PopB-β-Lac fusion protein was created by using the overlapping PCR strategy described by Yon and Fried (29). The bla gene from vector pBR322 encoding the β-lactamase (30) was inserted between the popB and popD genes in the pIApG-pcrH-popB-popD expression vector. Sense and reverse primers were designed in such a way that the sequence encoding the β-lactamase was placed in-frame with the end of the popB gene and that the ribosome entry site upstream the start codon of popD was preserved. The final PCR product was cleaved at the unique BsmI restriction site located in the region encoding the C-terminal domain of PopB and the unique FspAI restriction site located in the region encoding the N-terminal region of PopD. The resulting 1.1-kb restriction fragment was ligated into pIApG-pcrH-popB-popD vector yielding the pIApG-pcrH-popB-bla-popD expression vector, which was used to complement the P. aeruginosa CHAΔBD, CHAΔF, and CHAΔV strains.
Cell Infection and CCF2 Measurements
A549 cells were infected with strains harboring either ExoS-β-lactamase or PopB-β-lactamase fusions. Intracellular β-lactamase activity was quantified by using CCF2 substrate (Invitrogen) as described by Verove et al. (30).
Cell Surface-associated β-Lactamase Activity
The measurement of β-lactamase activity of the PopB-β-lactamase fusion relies on the ability of PopB-β-Lac to hydrolyze nitrocefin, a yellow non-cell-permeant β-lactamase substrate that becomes red upon cleavage of the β-lactam ring. Briefly, A549 epithelial cells were seeded into 12-well plates at a density of 2 × 105 cells per well and cultured for 24 h in complete RPMI 1640 medium devoid of antibiotics. Cells were infected with various CHA mutant strains at an MOI of 30. Three hours post-infection, the adherent cell monolayers were washed 4 times with 2 ml of PBS supplemented with calcium and magnesium. Subsequently, β-lactamase activity present on the cell surface was assayed at 486 nm 1 h after the addition of 1 ml of nitrocefin (500 μg/ml of PBS).
Antibodies
Anti-PopB rabbit Abs were purified and used at a 1:3000 dilution, whereas anti-PopD rabbit Abs were used at 1:1000 and anti-PcrH at 1:500 dilutions. Incubation was done at 37 °C for 1 h, and after washing, secondary antibodies (Anti-rabbit HRP-conjugated) were used at a dilution of 1:3000.
Crystallization, Data Collection, and Structure Solution
PcrH (21–160) was expressed and purified as previously described (8). Purified PcrH was mixed with a synthetic peptide corresponding to residues 51–59 of PopB (TGVALTPPS) in molar ratios 1:1, 1:2, and 1:4 and submitted to high throughput crystallization (HTXLab; Partnership for Structural Biology, Grenoble, France) at 4 °C. The best crystals were obtained in 30% PEG 6000, 1.0 m LiCl, 0.1 m Tris-HCl, pH 8.0, with 28 mg/ml of protein and in all three molar ratios of the peptide. Crystals of the 1:4 sample were flash-cooled in liquid nitrogen before data collection at the BM14 beamline in the European Synchrotron Radiation Facility in Grenoble, France. Data were processed and scaled using XDS (31). The structure was solved by molecular replacement using Phaser (32) and the apo structure of PcrH (PDB code 2XCC (8)) as a search model. Cycles of refinement and model building were performed with Refmac 5.6 (33) and Coot (34). The quality and stereochemistry of the final model was checked with SFCHECK (35) and Procheck (36). Figures were generated using PyMOL.
RESULTS AND DISCUSSION
Only PopB Is Stably Inserted into Membranes
We previously determined that PopB-PcrH and PopD-PcrH complexes could be purified in soluble form upon overexpression in Escherichia coli. In addition, we showed that chaperone release could be accomplished by gel filtration of the complexes at acidic pH, leading to oligomerization of both PopB and PopD. However, Pop proteins could only form rings once in the presence of membranes, suggesting that a large conformational change could occur upon lipid binding (26).
To gain insight into the mode of association of PopB and PopD into target membranes, we initially sought to determine if they could be stably inserted into an artificial bilayer or if they remained superficially associated to it. We thus purified PopB-PcrH and PopD-PcrH by overexpressing the bicistronic forms (Fig. 1A) in E. coli and isolated the Pop oligomers by gel filtration at pH 5.0, as previously described (26). These samples were subsequently incubated with artificial liposomes prepared with a 2:3 ratio of α-phosphatidyl-l-serine and 1,2-dipalmitoyl-sn-glycero-3-phosphocholine in the presence/absence of agents known to release peripheral membrane proteins (37, 38). Experiments were performed with (a) PBS, pH 7.0, as a negative control, (b) 1 m KCl, (c) 6 m urea, and (d) 0.425 mm DDM (which corresponds to 2.5 × critical micelle concentration (CMC)). We then separated soluble from membrane-bound proteins on sucrose gradients; fractions were analyzed by Western blotting by employing anti-PopB and anti-PopD antibodies (Fig. 1B).
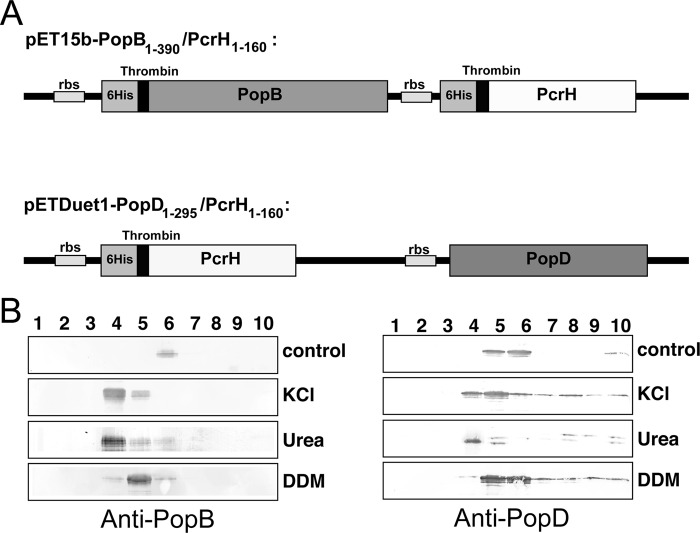
FIGURE 1.
PopB and PopD interact with artificial membranes. A, scheme of expression clones used in this work. Both Pop proteins are co-expressed with their common chaperone PcrH, which carries a thrombin-cleavable 6His tag. rbs, ribosomal binding site. B, liposomes containing PopB (left) or PopD (right) were treated with different agents that destabilize the association of proteins to membranes (1 m KCl, 6 m urea, 2.5 × CMC DDM) to characterize their association with membranes. After 4 h of incubation and sucrose gradient centrifugation, fractions were recovered from the top to the bottom of the gradient and were analyzed by Western blotting using anti-PopB and/or anti-PopD rabbit antibodies.
Our results show that PopB remains bound to liposomes in all conditions, i.e. in control experiments as well as in the presence of high levels of urea, KCl, and DDM, indicating that it is stably inserted into membranes. PopD, however, although remaining associated to lipids in the control experiment, could be partly detached from liposomes in the presence of all three agents (Fig. 1B), suggesting that it is not stably/integrally associated to the membrane but could be superficially associated to it. This is in agreement with ex vivo observations made on the translocon-forming proteins of Shigella in which the major translocator IpaB is strongly associated to RBC membranes, whereas the minor translocator IpaC is associated to a lesser extent (39, 40). In addition, this observation also supports in vitro studies performed on other translocon molecules of Salmonella (40), Yersinia (41, 43), and Shigella (42). These results suggest that, in Pseudomonas, PopB could be the major translocon component that binds most stably to target membranes upon infection, whereas PopD remains superficially associated to the bilayer, a suggestion that is in agreement with model membrane studies which showed that PopB plays the central role in pore formation (44, 45).
The Exposed N Terminus of PopB Is a Molten Globule
To gain an understanding into the orientation of the molecules on target membranes, we isolated Pop-containing fractions after sucrose gradient centrifugation and submitted them to trypsinolysis. We reasoned that trypsin would only target the exposed regions of the molecules, allowing us to map the sequences that were inserted into the bilayer. Pop proteins purified by gel filtration but not submitted to interaction with membranes were used as a control. After cleavage, samples were loaded onto an SDS-PAGE gel and blotted onto PVDF membranes to allow for N-terminal sequencing of the resulting bands.
For PopD, trypsin cleaved at several sites within the protein sequence irrespective of whether PopD was bound to liposomes or not (Fig. 2, A and C). N-terminal sequencing of the bands produced identified that residues Gln-56, Met-170, Thr-156, Ile-183, Ala-205, and Asn-210 (Fig. 2, A and D) were immediately juxtaposed to the trypsin cleavage site, indicating that PopD was proteolyzed in its entirety both in the lipid-free and liposome-bound forms, potentially due to the fact that it is only superficially associated to the membrane (which would confirm the results presented in Fig. 1B). In the case of the lipid-free PopD, however, a cleavage site juxtaposed to Asp-96 could also be identified (diamond in Fig. 2, C and D), suggesting that there is a potential structural rearrangement of PopD upon lipid binding.
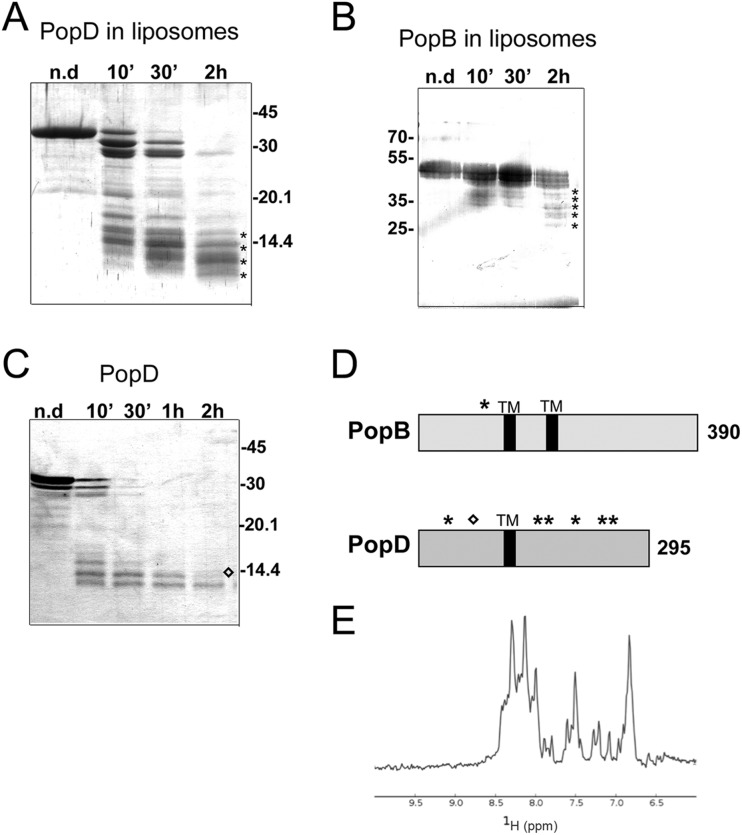
FIGURE 2.
Trypsin digestion of PopB and PopD inserted into liposomes. A and C, purified PopD (in solution or inserted into liposomes) was digested with trypsin at RT at a molar ratio of (1:500), and samples were loaded on SDS-PAGE after different digestion times. n.d, non-digested sample. B, PopB proteoliposomes were digested with trypsin at the same ratio as described above; aliquots were loaded on SDS-PAGE, and the gel was developed by Western blotting using purified anti-PopB antibodies. D, representation of PopB and PopD with the positions of the residues recognized by trypsin and identified by N-terminal sequence are highlighted. The black rectangles represent the two transmembrane (TM) regions. Asterisks indicate bands analyzed by N-terminal sequencing for proteins in liposomes, and the diamond indicates a band seen for PopD only in solution. E, one-dimensional NMR spectrum of PopB (1–149) recorded at 25 °C on an 800 MHz spectrometer.
Although extensive proteolysis was also observed for a PopB sample trypsinized in the absence of liposomes (not shown), as expected for a molten globule protein (46), for the PopB sample in the lipid-bound form proteolysis was restricted to the N-terminal region (Fig. 2, B and D). After trypsin treatment, most of the protein still ran on SDS-PAGE with a molecular weight similar to that of the non-digested sample (Fig. 2B); N-terminal sequencing of the five minor bands in the sample treated with trypsin for 2 h revealed the same sequence for all of them (ESEEAAKE). The first Glu corresponds to residue 153, which is located in a position that is immediately N-terminal to the beginning of the first predicted transmembrane region (residues 170–192; Fig. 2D; Ref. 26). This result suggests that in the membrane-bound form the 152 N-terminal residues of PopB are trypsin-accessible and thus could be located toward the outside of the membrane, whereas sequences that immediately follow this region are stably imbedded in the bilayer. These results suggest that as opposed to PopD, which only associates to bilayers superficially, PopB is stably inserted through its predicted transmembrane region, and its N terminus is exposed.
To gain insight into the fold of the exposed N terminus of PopB, we cloned residues 1–149 into a plasmid harboring an N-terminal His6 coding sequence (pET15b) and expressed the protein in E. coli BL21 (DE3) cells. PopB (1–149) was purified by nickel chelation chromatography and eluted as a soluble monomer in gel filtration. Liposome flotation assays indicated that the protein does not have affinity for lipids, as it migrated at the bottom of the sucrose gradient (not shown). We subsequently performed a one-dimensional NMR spectrum of the sample (Fig. 2E), which shows poor dispersion of the amide resonances and broad line widths, suggesting that there may be exchange between the different conformations adopted by the protein. Notably, molten globule forms are folded proteins with extensive native-like secondary structures but that lack tertiary structures and are often characterized by spectra with extremely broad peaks with poor chemical shift dispersions. Thus, this is in agreement with studies that suggested that Pop proteins are molten globules (46), which could be a requirement for their passage through the interior of the T3SS needle and subsequent oligomerization and membrane interaction steps (26). These results collectively indicate that, upon activation of the T3SS and cellular recognition, PopB could be initially inserted into the membrane through its two transmembrane regions, whereas its N-terminal domain remains positioned toward the outside of the cell, where it could be recognized (and stabilized) by PopD, which is superficially positioned, and/or PcrV, which is potentially organized into a pentamer at the tip of the T3SS needle (47, 48). Notably, the presence of two predicted two transmembrane regions in PopB suggest that its C-terminal residues are also exposed to the outside of the cell (see below).
Both N and C Termini of PopB Are Exposed to the outside of the bilayer
To further explore the topology of PopB in an in vivo (infectious) setting, we generated a construct that expressed a fusion protein encompassing PopB followed by a β-lactamase reporter (β-Lac) at its C terminus (Fig. 3). The β-Lac reporter assay has been in the past successfully used to measure the level of injection and secretion of the T3SS toxins ExoS and ExoY by P. aeruginosa with two different molecules, CCF2 (30) and nitrocefin. In the first assay, if the C terminus of PopB were to be located within the target cell, the fluorescent substrate CCF2 would be recognized and cleaved by β-Lac, emitting a signal at 460 nm (blue, Fig. 3A). Conversely, location of the C terminus toward the outside of the cell would not generate CCF2 cleavage, causing a detection of a signal at 535 nm (green, Fig. 3B). Alternatively, β-Lac activity was also measured using the non-permeable, chromogenic β-lactamase substrate nitrocefin whose color was shifted from yellow to red (abs 486 nm) when it was cleaved by β-Lac (Fig. 3B).
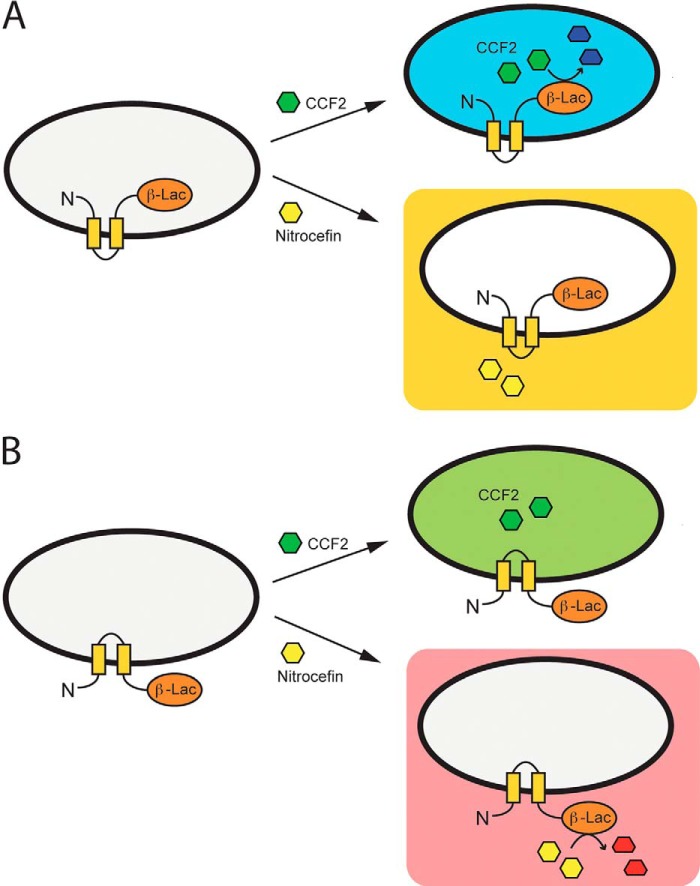
FIGURE 3.
CCF2 and nitrocefin assays used to determine the orientation of the C terminus of PopB on membranes. PopB could potentially be inserted in host cell membranes with its N and C termini pointing toward the inside (panel A) or the outside (panel B) of the infected cell. To discriminate between these two orientations, a β-lactamase reporter gene was fused to the C terminus of PopB, and its activity was followed using two β-lactamase substrates: CCF2 (upper parts), that penetrates eukaryotic cells and turns from green to blue upon cleavage, and nitrocefin (bottom parts), a non-permeable substrate that turns from yellow to red upon hydrolysis of the β-lactam ring. The two transmembrane regions of PopB are shown as yellow boxes.
We initially verified that the presence of the β-Lac located C-terminally to PopB did not interfere with its function in the P. aeruginosa clinical strain CHA. The PopB-β-Lac fusion proved to be functional, as it was secreted upon induction of T3SS in vitro (Fig. 4A) and was able to complement the P. aeruginosa PopB knock-out mutant (CHAΔBD (49)) in a cytotoxicity assay performed with J774 macrophages (Fig. 4B). Subsequently, to investigate whether the β-Lac reporter was found within the eukaryotic cell cytoplasm, A549 epithelial cells were infected with the CHAΔBD strain supplemented with PopB-β-Lac and PopD. The cells were then incubated with CCF2, and the level of substrate cleavage was assessed by measuring the ratio of emission of light at 460 and 535 nm, as previously described (30). As a positive control we used a P. aeruginosa CHA strain expressing an ExoS-β-Lac fusion that displayed high levels of CCF2 cleavage due to the fact that ExoS is translocated into the eukaryotic cytoplasm, and thus, CCF2 is recognized by the β-lactamase (Fig. 4C, fourth bar). Notably, infection of A549 cells with the same strain expressing PopB-β-Lac instead of ExoS-β-Lac generated CCF2 cleavage levels that were comparable to background (Fig. 4C, fifth bar), strongly suggesting that in the case of PopB the C terminus is not located within the cytoplasm of the eukaryotic cells.
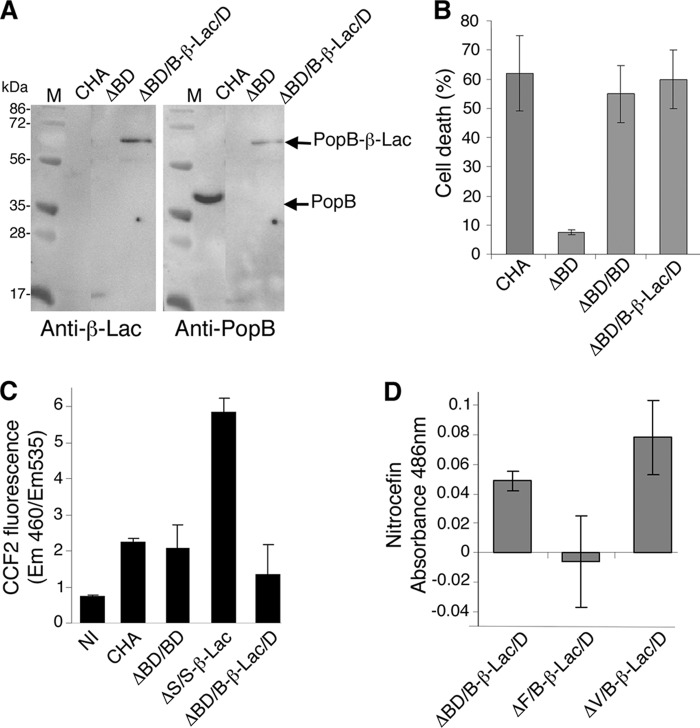
FIGURE 4.
Characterization of the PopB-β-Lac fusion expressed in P. aeruginosa. A, P. aeruginosa CHA wild type or mutant strains were grown in T3SS-inducing conditions. When the cultures reached an A600 nm value of 1.0 absorbance units, 100 μl of culture were centrifuged, and the supernatants were analyzed by Western blotting with rabbit anti-β-Lac and anti-PopB antibodies. B, cytotoxicity of PopB-β-Lac mutants. Macrophages were infected with P. aeruginosa CHA wild type, popBpopD mutant (ΔBD) or mutant complemented with either PopBwt/PopDwt (ΔBD/BD) or PopB-β-Lac/PopDwt (ΔBD/B-β-Lac/D). Non-infected (NI) cells were used as negative control. After 3 h of infection, LDH released was measured to determine the % of cell death. C, intracellular β-Lac activity was determined after infection of A549 cells with the indicated strains using CCF2. A P. aeruginosa strain expressing the fusion protein ExoS-β-Lac (ΔS-S-β-Lac) was used as a positive control. D, the complemented cytotoxic P. aeruginosa strain CHAΔBD/B-β-Lac/D, the non-secreting strain CHAΔF/B-β-Lac/D, and the constitutively secreting strain CHAΔV/B-β-Lac/D were used to infect A549 epithelial cells in triplicate to an MOI of 30. β-Lactamase activity was assayed by measuring the absorbance at 486 nm using nitrocefin as a substrate. M, molecular mass standards.
To confirm the outer membrane exposure of the C terminus of PopB, we again infected A549 epithelial cells with P. aeruginosa mutant strains carrying PopB-β-Lac and measured the β-lactamase activity by adding nitrocefin to infected cells (Fig. 4D). Here, we employed two strains that insert PopB into the membrane: P. aeruginosa ΔBD complemented with both PopB-β-Lac and PopD (ΔBD/B-β-Lac/D) and ΔPcrV complemented with the same constructs (ΔV/B-β-Lac/D). This latter strain, despite not being able to express PcrV, is able to form the translocon, although it is non-infectious (49). β-Lactamase cleavage of nitrocefin could be measured for both transfected constructs. Subsequently as the negative control, we complemented a strain that cannot form the secretion needle with the aforementioned constructs (ΔF/B-β-Lac/D). The observation that little activity was associated to cells infected with this strain is consistent with its inability to secrete or insert the translocators into the host cell plasma membrane. Collectively, these data suggest that the PopB translocator is inserted within the eukaryotic membrane with its flexible, N-terminal domain and its C-terminal tail exposed to the outside of the bilayer.
The N Terminus of PopB Binds to the Concave Region of PcrH
After having mapped the architecture of PopB on bilayers, we thus set out to investigate its cytoplasmic form; that is, in a complex with its chaperone PcrH. We had previously shown through limited proteolysis that the 95 N-terminal residues of the minor translocator PopD could associate to PcrH with 1:1 stoichiometry (26, 46). By solving the high resolution crystal structure of PcrH, we had also shown that it folds into seven TPR motifs that generate a concave region that is capable of binding PopD residues 49–56 (8). Notably, IpgC, the type II chaperone of the type III secretion system of Shigella, also recognizes a short peptide of the N terminus of the major translocator molecule (50, 51), and SycD from Yersinia enterocolitica was also structurally characterized in complex with a peptide from minor translocator YopD (52).
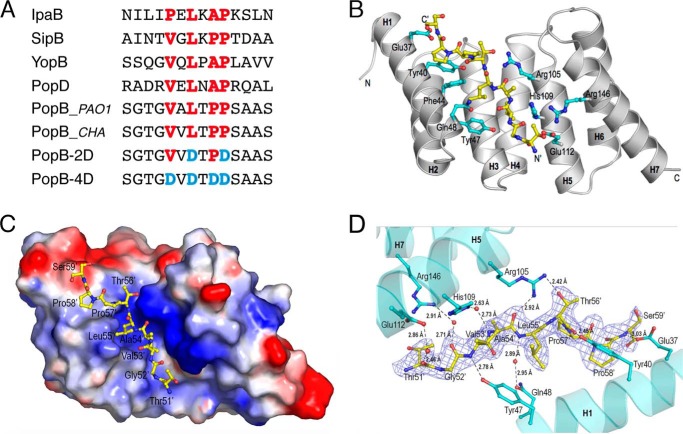
By performing sequence alignments between PopB and PopD, we identified a short peptide in PopB (residues 51–59) within the N-terminal flexible region that displayed sequence similarities with the minimal region in PopD that interacts with PcrH (residues 49–56, Fig. 5A; Ref. 8) and harbors the six-amino acid chaperone-binding consensus sequence initially identified by Kolbe and co-workers (50). This PopB peptide, 51TGVALTPPS59, was thus synthesized and incubated with PcrH before crystallization trials at 4 °C. Diffraction data were collected to a resolution of 2.25 Å at the European Synchrotron Radiation Facility in Grenoble, and crystals of the complex belonged to space group P21221 with two molecules per asymmetric unit. The structure was solved by molecular replacement using the structure of the apo form of PcrH as a search model (PDB 2XCC). Data collection and refinement statistics are shown in Table 1. Calculation of a difference Fourier map (Fo − Fc) revealed density corresponding to the peptide in both monomers, but only the peptide associated to monomer A could be traced unambiguously in its entire length (residues 51–59). We will thus describe the analysis of this monomer.
FIGURE 5.
PopB 51TGVALTPPS59 binds within the concave region of PcrH. A, sequences of N-terminal peptides of translocon proteins that interact with type II chaperones as well as of the mutants tested in Fig. 7. B, schematic representation of the structure of PcrH (in gray) displaying most residues that interact with PopB 51TGVALTPPS59. D, a close-up view of the established hydrogen bonds. The omit density map is contoured at 1.0 σ. C, electrostatic surface representation of PcrH in complex with the PopB peptide. Peptide residues are labeled, and carbon atoms are represented as yellow sticks (and in cyan for PcrH), with oxygen atoms in red and nitrogen atoms in blue.
TABLE 1.
Data collection, molecular replacement and structure refinement statistics
Values in parentheses correspond to the last resolution shell. r.m.s.d., root mean square deviation.
| Data collection | |
| Wavelength (Å) | 0.977 |
| Space group | P21221 |
| Unit cell parameters (Å) | a = 45.10, b = 73.29, c = 98.94 |
| Resolution (Å) | 2.25 (2.25–2.39) |
| Number of observed/unique reflections | 113,610/16,247 |
| Completeness (%) | 99.4 (96.8) |
| Rmeasured | 5.1 (44.7) |
| Rmrgd-Fa | 6.6 (39.8) |
| I/σ (I) | 29.7 (4.8) |
| Refinement | |
| Rwork | 0.214 |
| Rfree | 0.264 |
| r.m.s.d.bond | 0.008 |
| r.m.s.d.angle | 1.069 |
| Mean B factor (Å2) | 43.242 |
| No. of protein atoms | 2,028 |
| No. of solvent atoms | 117 |
| Ramachandran analysis (%) | |
| Residues in favored regions | 98.1 |
| Residues in allowed regions | 1.9 |
| Residues in outlier regions | 0 |
a Rmrgd-F is defined in Ref. 55.
51TGVALTPPS59 binds in an extended conformation within a cleft in the concave region of PcrH, and it presents an antiparallel orientation in respect to the first helix of the chaperone (Fig. 5, B and D). Notably, residues Val-53′, Leu-55′, and Pro-58′ (the prime indicates PopB peptide residues), which form the conserved translocator-chaperone binding motif P′/V′XL′XXP′ (50–52), fit into PcrH surface cleft and interact mostly with conserved residues (Tyr-40, Ala-41, Phe-44, Phe-59, Leu-74, Gly-75, Ala-78, Tyr-93, His-109). Additionally, the main chain atoms of Thr-51′, Gly-52′, Val-53′, and Leu-55′ establish hydrogen bonds with the side chain atoms of Glu-112, Arg-146, His-109, Gln-48, and Tyr-40 from PcrH intermediated by water molecules, whereas the main chain atoms of Val-53′, Ala-54′, Thr-56′, and Ser-59′ establish direct hydrogen bonds with the Tyr-47, Arg-105, Tyr-40, and Glu-37 side chains of PcrH (Fig. 5, B–D). Only one peptide side chain, the hydroxyl group of Thr-56′, makes a direct hydrogen bond with PcrH (with the side chain of Arg-105). Last, the side chains of residues Thr-51′, Ala-54′, and Pro-57′ point out, to the solvent. These observations indicate that the concave region of PcrH acts as the palm of a hand, “grasping” a common region of the Pop molecules (Fig. 5C), with the rest of the protein potentially wrapping around the chaperone and also interacting with the convex region of PcrH.
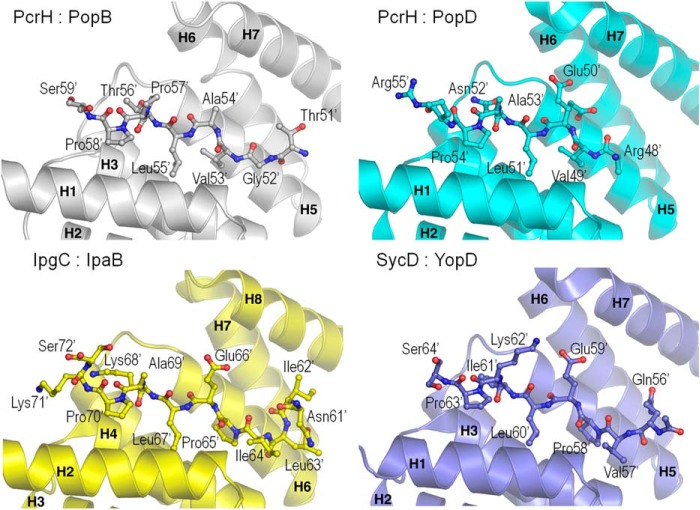
Other structures of T3SS type II chaperones have already been solved in complex with peptides corresponding to the chaperone binding region of their respective translocators, as mentioned above. The comparison of these structures with our present model reveals that all peptides bind in a very similar fashion to the concave region of their respective chaperones, with their N termini toward helix 1 and their C termini toward helix 5 (Fig. 6). Notably, within the region that encompasses the common motif P′/V′XL′XXP′, the conformation of the main chain peptide atoms is very similar, and the side chains of the conserved residues Pro/Val, Leu, and Pro fit deeply in the cleft of the chaperones. The most dissimilar conformations are observed for the residues located at the extremities of the peptides, not included within the conserved motif (Fig. 6).
FIGURE 6.
Comparison of structures of chaperone-translocator peptide complexes: PcrH-PopB (gray), PcrH-PopD (cyan; Ref. 8), IpgC-IpaB (yellow; Ref. 50), and SycD-YopD (blue; Ref. 52). The seven (eight for IpgC) helices of the different chaperones are shown. The structures were aligned with Coot. The translocator peptides bind in a very similar fashion to the concave region of their respective chaperones, especially within the region corresponding to the conserved P′/V′XL′XXP′ motif.
PcrH is now the only type II chaperone that has been structurally characterized in complex with peptides from both major and minor translocator partners, unequivocally showing that the concave region of the chaperone serves as the binding site for both proteins. It is of note that PopD and PopB share <15% sequence identity levels, and yet PcrH recognizes both molecules stably. In addition PcrH is a dimer in the asymmetric unit and has exactly the same orientation as observed previously for the apo- and PopD-bound forms, with a back-to-back interaction surface of 546 Å2. Interestingly, in our structure both PcrH monomers harbor the major translocator peptide. This result is distinct from previous structures of type II chaperones (8, 51), in which the dimer in the asymmetric unit only recognized the translocator peptide in one of the binding sites, suggesting that the other site could be occupied by the “other” translocator molecule. Our present result could thus suggest that the binding pattern is so similar among different peptides, even from divergent bacterial species, with hydrophobic residues binding deep into the cleft and playing the key roles in chaperone recognition that the binding sites are actually interchangeable. Thus, it seems unlikely that PcrH could recognize both PopB and PopD concomitantly, although a complex between a PcrH dimer and each one of the two translocators could still be a possibility. However, Tomalka et al. (53) recently showed that an apoPcrH exists in Pseudomonas as a dimer with a head-to-head conformation; however, dimerization is not required for substrate binding and targeting, which suggests that, within the cytoplasm, 1:1 PcrH-Pop complexes are the most likely forms.
To test if the interactions between PcrH and the N-terminal peptide of PopB involving residues 51–59 are important for functionality of the T3SS, we modified the highly cytotoxic P. aeruginosa CHA strain (originally isolated from a cystic fibrosis patient) to carry mutations within this motif in PopB (Fig. 5A). We tested these mutants for (i) expression and secretion of PopB mutants and (ii) cytotoxicity toward macrophages. A mutant strain carrying a PopB variant with point mutations in the 51TGVALTPPS59 region, in which two (L55D/P58D; PopB-2D) or four (V53D/L55D/P57D/P58D; PopB-4D) of the central hydrophobic residues were replaced by Asp allowed us to verify the importance of hydrophobic interactions within the PopB-PcrH complex. Plasmids expressing the mutant pcrHpopBpopD section of the operon were introduced individually into the P. aeruginosa CHA strain carrying deletions of popB and popD (ΔpopBD) before testing.
P. aeruginosa strains expressing the two PopB mutants were able to express and secrete proteins at comparable levels (Fig. 7A); in addition, the PopB mutants were stable for up to 3 h in T3SS-induced conditions, suggesting that PcrH binding is maintained (Fig. 7C). Finally the two mutant strains displayed levels of cytotoxicity toward macrophages that were similar to that displayed by the wild type strain (Fig. 7B), suggesting that the mutations within the PopB region that interacts with the PcrH “palm” are not sufficient to disrupt protein-protein interactions. This is distinct from cytotoxicity profile of mutants of the comparable region in PopD, which displayed no cytotoxicity toward macrophages (8), and indicates that Pop proteins display distinct affinities for their common chaperone. This could be explained by the fact that PopB (being a bigger molecule) may remain partly bound to PcrH despite the mutations, suggesting that other regions of the translocator protein may also contribute to chaperone binding.
FIGURE 7.
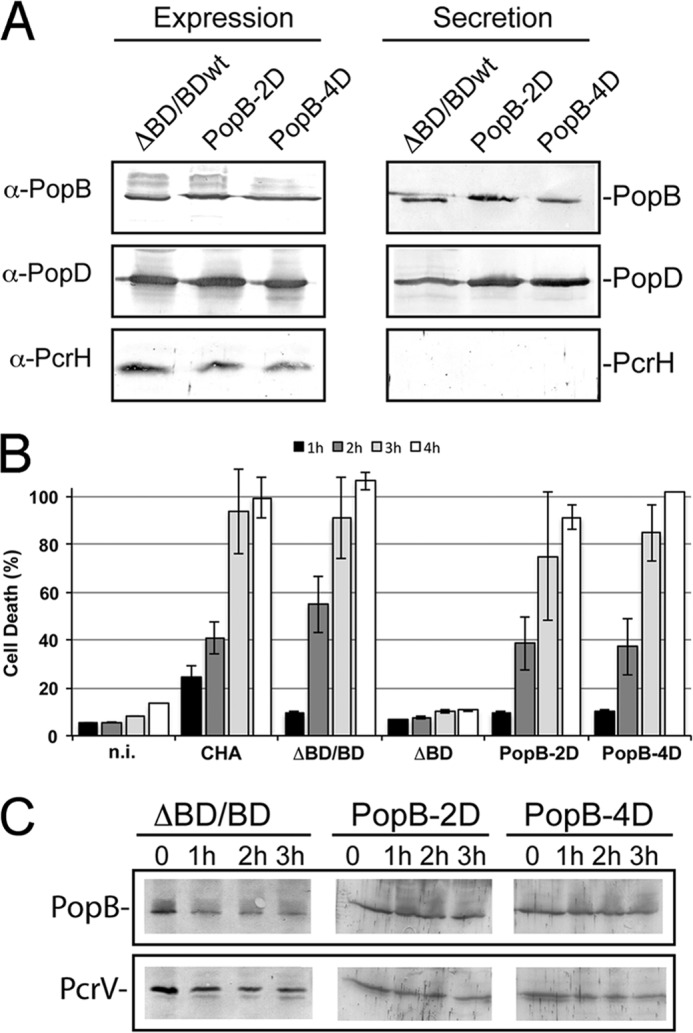
Characterization of PopB mutants in P. aeruginosa. A, P. aeruginosa strains carrying the two different sets of mutations in PopB, L55D/P58D (PopB-2D) and V53D/L55D/P57D/P58D (PopB-4D), were grown in T3SS-inducing conditions. When the cultures reached an A600 nm value of 1.0 absorbance units, 100 μl of cultures were centrifuged, and the pellet (Expression) and the supernatant (Secretion) were analyzed by Western blotting with anti-PopB, anti-PopD, and anti-PcrH antibodies. B, cytotoxicity of PopB mutants. Macrophages were infected with cultures of P. aeruginosa CHA depleted for popB and popD genes (ΔBD) or complemented with PopDwt and PopBwt, PopB-2D, or PopB-4D using a multiplicity of infection of 5. A non-infected culture was used as negative control. After 1–4 h of infection, LDH release into the supernatant was measured to determine the % of cell death. C, stability of PopB mutants was followed for up to 3 h after the addition of chloramphenicol, a protein synthesis inhibitor. The cell contents were analyzed by Western blotting with anti-PopB antibodies and anti-PcrV as a control.
The translocon is a key element of the pathogenicity arsenal of T3SS-carrying bacteria, and understanding the details of the individual steps required for its formation, including how proteins associate both within the cytoplasm and on the target bilayer, is essential for its characterization. The collective results presented here indicate that (i) within the cytoplasm both major and minor translocators use the concave region of their common chaperone, PcrH, for binding. This excludes the possibility of the formation of a 1:1:1 complex and suggests that the functional units are most likely 1:1 associations. (ii) Upon translocation, presumably through the T3SS needle, PopB becomes stably associated into the target bilayer with both N- and C-terminal regions directed toward the outside of the cell. These regions could potentially be the recognition sites for PopD, which is only superficially associated to the membrane, and could subsequently play the role of “bridge” with the tip of the T3SS needle (as suggested for the Yersinia spp YopD homolog; Ref. 54)). Detailed information regarding the distinct steps involved in translocon formation will be essential for the exploitation of such molecules as potential targets for the development of novel antibacterials.
Acknowledgments
We thank J. Marquez and the HTX Laboratory team for access to and help with high throughput crystallization, Eric Faudry and François Cretin for help with β-Lac measurements, J.-P. Andrieu (Institut de Biologie Structurale-Grenoble Partnership for Structural Biology Platform) for N-terminal sequencing, and the European Synchrotron Radiation Facility for access to beamlines. This work used the platforms of the Grenoble Instruct center (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL) with support from FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology.
This work was supported by the joint ministerial R&D program against CBRNE threats and the Commissariat à l'Energie Atomique.
The atomic coordinates and structure factors (code 4JL0) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- T3SS
- type III secretion system
- PS
- α-phosphatidyl-l-serine
- PC
- 1,2-dipalmitoyl-sn-glycero-3-phosphocholine
- β-Lac
- β-lactamase reporter
- RT
- room temperature
- DDM
- n-Dodecyl-β-d-maltopyranoside
- CMC
- critical micelle concentration
- MOI
- multiplicity of infection.
REFERENCES
- 1. Cornelis G. R. (2006) The type III secretion injectisome. Nat. Rev. Microbiol. 4, 811–825 [DOI] [PubMed] [Google Scholar]
- 2. Blocker A., Jouihri N., Larquet E., Gounon P., Ebel F., Parsot C., Sansonetti P., Allaoui A. (2001) Structure and composition of the Shigella flexneri “needle complex,” a part of its type III secreton. Mol. Microbiol. 39, 652–663 [DOI] [PubMed] [Google Scholar]
- 3. Loquet A., Sgourakis N. G., Gupta R., Giller K., Riedel D., Goosmann C., Griesinger C., Kolbe M., Baker D., Becker S., Lange A. (2012) Atomic model of the type III secretion system needle. Nature 486, 276–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zarivach R., Vuckovic M., Deng W., Finlay B. B., Strynadka N. C. (2007) Structural analysis of a prototypical ATPase from the type III secretion system. Nat. Struct. Mol. Biol. 14, 131–137 [DOI] [PubMed] [Google Scholar]
- 5. Thomas J., Stafford G. P., Hughes C. (2004) Docking of cytosolic chaperone-substrate complexes at the membrane ATPase during flagellar type III protein export. Proc. Natl. Acad. Sci. U.S.A. 101, 3945–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Akeda Y., Galán J. E. (2005) Chaperone release and unfolding of substrates in type III secretion. Nature 437, 911–915 [DOI] [PubMed] [Google Scholar]
- 7. Chen L., Ai X., Portaliou A. G., Minetti C. A., Remeta D. P., Economou A., Kalodimos C. G. (2013) Substrate-activated conformational switch on chaperones encodes a targeting signal in type III secretion. Cell Rep. 3, 709–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Job V., Matteï P.-J., Lemaire D., Attree I., Dessen A. (2010) Structural basis of chaperone recognition by type III secretion system minor translocator proteins, J. Biol. Chem. 285, 23224–23232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miki T., Shibagaki Y., Danbara H., Okada N. (2009) Functional characterization of SsaE, a novel chaperone protein of the type III secretion system encoded by Salmonella pathogenicity island 2. J. Bacteriol. 191, 6843–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Quinaud M., Plé S., Job V., Contreras-Martel C., Simorre J.-P., Attree I., Dessen A. (2007) Structure of the heterotrimeric complex that regulates type III secretion needle formation. Proc. Natl. Acad. Sci. U.S.A. 104, 7803–7808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sato H., Frank D. W., Hillard C. J., Feix J. B., Pankhaniya R. R., Moriyama K., Finck-Barbançon V., Buchaklian A., Lei M., Long R. M., Wiener-Kronish J., Sawa T. (2003) The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 22, 2959–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hauser A. R. (2009) The type III secretion system of Pseudomonas aeruginosa. Infection by injection., Nat. Rev. Microbiol. 7, 654–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Silver A. C., Kikuchi Y., Fadl A. A., Sha J., Chopra A. K., Graf J. (2007) Interaction between innate immune cells and a bacterial type III secretion system in mutualistic and pathogenic associations. Proc. Natl. Acad. Sci. U.S.A. 104, 9481–9486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miki T., Iguchi M., Akiba K., Hosono M., Sobue T., Danbara H., Okada N. (2010) Chromobacterium pathogenicity island 1 type III secretion system is a major virulence determinant for Chromobacterium violaceum-induced cell death in hepatocytes. Mol. Microbiol. 77, 855–872 [DOI] [PubMed] [Google Scholar]
- 15. Schraidt O., Lefebre M. D., Brunner M. J., Schmied W. H., Schmidt A., Radics J., Mechtler K., Galán J. E., Marlovits T. C. (2010) Topology and organization of the Salmonella typhimurium type III secretion needle complex components. PLoS Pathog. 6, e1000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hodgkinson J. L., Horsley A., Stabat D., Simon M., Johnson S., da Fonseca P. C. A., Morris E. P., Wall J. S., Lea S. M., Blocker A. J. (2009) Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout. Nat. Struct. Mol. Biol. 5, 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Veenendaal A. K., Hodgkinson J. L., Schwarzer L., Stabat D., Zenk S. F., Blocker A. J. (2007) The type III secretion system needle tip complex mediates host cell sensing and translocon insertion. Mol. Microbiol. 63, 1719–1730 [DOI] [PubMed] [Google Scholar]
- 18. Marlovits T. C., Kubori T., Lara-Tejero M., Thomas D., Unger V. M., Galán J. E. (2006) Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature 441, 637–640 [DOI] [PubMed] [Google Scholar]
- 19. Marlovits T. C., Kubori T., Sukhan A., Thomas D. R., Galán J. E., Unger V. M. (2004) Structural insights into the assembly of the type III secretion needle complex. Science 306, 1040–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wagner S., Königsmaier L., Lara-Tejero M., Lefebre M., Marlovits T. C., Galán J. E. (2010) Organization and coordinated assembly of the type III secretion export apparatus. Proc. Natl. Acad. Sci. U.S.A. 107, 17745–17750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yip C. K., Kimbrough T. G., Felise H. B., Vuckovic M., Thomas N. A., Pfuetzner R. A., Frey E. A., Finlay B. B., Miller S. I., Strynadka N. C. (2005) Structural characterization of the molecular platform for type III secretion system assembly. Nature 435, 702–707 [DOI] [PubMed] [Google Scholar]
- 22. Matteï P.-J., Faudry E., Job V., Izoré T., Attree I., Dessen A. (2011) Membrane targeting and pore formation by the type III secretion system translocon. FEBS J. 278, 414–426 [DOI] [PubMed] [Google Scholar]
- 23. Galle M., Jin S., Bogaert P., Haegman M., Vandenabeele P., Beyaert R. (2012) The Pseudomonas aeruginosa type III secretion system has an exotoxin S/T/Y independent pathogenic role during acute lung infection. PLoS ONE 7, e41547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dacheux D., Goure J., Chabert J., Usson Y., Attree I. (2001) Pore-forming activity of type III system-secreted proteins leads to oncosis of Pseudomonas aeruginosa-infected macrophages. Mol. Microbiol. 40, 76–85 [DOI] [PubMed] [Google Scholar]
- 25. Allmond L. R., Karaca T. J., Nguyen V. N., Nguyen T., Wiener-Kronish J. P., Sawa T. (2003) Protein binding between PcrG-PcrV and PcrH-PopB/PopD encoded by the pcrGVH-popBD operon of the Pseudomonas aeruginosa type III secretion system. Infect Immun 71, 2230–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schoehn G., Di Guilmi A. M., Lemaire D., Attree I., Weissenhorn W., Dessen A. (2003) Oligomerization of type III secretion proteins PopB and PopD precedes pore formation in Pseudomonas. EMBO J. 22, 4957–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Engel J., Balachandran P. (2009) Role of Pseudomonas aeruginosa type III effectors in disease. Curr. Opin. Microbiol. 12, 61–66 [DOI] [PubMed] [Google Scholar]
- 28. Van Q. N., Shaka A. J. (1998) Improved cross-peak detection in two-dimensional proton NMR spectra using excitation sculpting. J. Magn. Reson. 132, 154–158 [DOI] [PubMed] [Google Scholar]
- 29. Yon J., Fried M. (1989) Precise gene fusion by PCR. Nucleic Acids Res. 17, 4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Verove J., Bernarde C., Bohn Y. S., Boulay F., Rabiet M. J., Attree I., Cretin F. (2012) Injection of Pseudomonas aeruginosa Exo toxins into host cells can be modulated by host factors at the level of translocon assembly and/or activity. PLoS ONE 7, e30488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murshudov G. N., Skubák P., Lebedev A. A, Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vaguine A. A., Richelle J., Wodak S. J. (1999) SFCHECK. A unified set of procedure for evaluating the quality of macromolecular structure-factor data and their agreement with atomic model. Acta Crystallogr. D Biol. Crystallogr. 55, 191–205 [DOI] [PubMed] [Google Scholar]
- 36. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Damico R. L., Crane J., Bates P. (1998) Receptor-triggered membrane association of model retroviral glycoprotein. Proc. Natl. Acad. Sci. U.S.A. 95, 2580–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Travier L., Mondragon R., Dubremetz J. F., Musset K., Mondragon M., Gonzalez S., Cesbron-Delauw M. F., Mercier C. (2008) Functional domains of the Toxoplasma GRA2 protein in the formation of the membranous nanotubular network of the parasitophorous vacuole. Int. J. Parasitol. 38, 757–773 [DOI] [PubMed] [Google Scholar]
- 39. Blocker A., Gounon P., Larquet E., Niebuhr K., Cabiaux V., Parsot C., Sansonetti P. (1999) The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J. Cell Biol. 147, 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McGhie E. J., Hume P. J., Hayward R. D., Torres J., Koronakis V. (2002) Topology of the Salmonella invasion protein SipB in a model bilayer. Mol. Microbiol. 44, 1309–1321 [DOI] [PubMed] [Google Scholar]
- 41. Ryndak M. B., Chung H., London E., Bliska J. B. (2005) Role of predicted transmembrane domains for type III translocation, pore formation, and signaling by the Yersinia pseudotuberculosis YopB protein. Infect. Immun. 73, 2433–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hume P. J., McGhie E. J., Hayward R. D., Koronakis V. (2003) The purified Shigella IpaB and Salmonella SipB translocators share biochemical properties and membrane topology. Mol. Microbiol. 49, 425–439 [DOI] [PubMed] [Google Scholar]
- 43. Costa T. R., Amer A. A., Farag S. I., Wolf-Watz H., Fällman M., Fahlgren A., Edgren T., Francis M. S. (2013) Type III secretion translocon assemblies that attenuate Yersinia virulence. Cell. Microbiol. 15, 1088–1110 [DOI] [PubMed] [Google Scholar]
- 44. Romano F. B., Rossi K. C., Savva C. G., Holzenburg A., Clerico E. M., Heuck A. P. (2011) Efficient isolation of Pseudomonas aeruginosa type III secretion translocators and assembly of heteromeric transmembrane pores in model membranes. Biochemistry 50, 7117–7131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Faudry E., Vernier G., Neumann E., Forge V., Attree I. (2006) Synergistic pore formation by type III toxin translocators of Pseudomonas aeruginosa. Biochemistry 45, 8117–8123 [DOI] [PubMed] [Google Scholar]
- 46. Faudry E., Job V., Dessen A., Attree I., Forge V. (2007) Type III secretion system translocator has a molten globule conformation both in its free and chaperone-bound forms. FEBS J. 274, 3601–3610 [DOI] [PubMed] [Google Scholar]
- 47. Mueller C. A., Broz P., Müller S. A., Ringler P., Erne-Brand F., Sorg I., Kuhn M., Engel A., Cornelis G. R. (2005) The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science 310, 674–676 [DOI] [PubMed] [Google Scholar]
- 48. Blocker A. J., Deane J. E., Veenendaal A. K., Roversi P., Hodgkinson J. L., Johnson S., Lea S. M. (2008) What's the point of the type III secretion system needle? Proc. Natl. Acad. Sci. U.S.A. 105, 6507–6513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Goure J., Pastor A., Faudry E., Chabert J., Dessen A., Attree I. (2004) The V antigen of Pseudomonas aeruginosa is required for assembly of the functional PopB/PopD translocation pore in host cell membranes. Infect. Immun. 72, 4741–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lunelli M., Lokareddy R. K., Zychlinsky A., Kolbe M. (2009) IpaB-IpgC interaction defines binding motif for type III secretion translocator. Proc. Natl. Acad. Sci. U.S.A. 106, 9661–9666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lokareddy R. K., Lunelli M., Eilers B., Wolter V., Kolbe M. (2010) Combination of two separate binding domains defines stoichiometry between type III secretion system chaperone IpgC and translocator protein IpaB. J. Biol. Chem. 285, 39965–39975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schreiner M., Niemann H. H. (2012) Crystal structure of the Yersinia enterocolitica type III secretion chaperone SycD in complex with a peptide of the minor translocator YopD. BMC Struct. Biol. 12, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tomalka A. G., Zmina S. E., Stopford C. M., Rietsch A. (2013) Dimerization of the P. aeruginosa translocator chaperone PcrH is required for stability not function. J. Bacteriol. 195, 4836–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Costa T. R., Edqvist P. J., Bröms J. E., Ahlund M. K., Forsberg A., Francis M. S. (2010) YopD self-assembly and binding to LcrV facilitate type III secretion activity by Yersinia pseudotuberculosis. J. Biol. Chem. 285, 25269–25284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Diederichs K., Karplus P. A. (1997) Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat. Struct. Biol. 4, 269–275 [DOI] [PubMed] [Google Scholar]