Background: Effects of scyllo-inositol, a modulator of misfolded protein accumulation, were tested in a cellular model of Huntington disease.
Results: scyllo-Inositol lowered mutant huntingtin aggregation and decreased protein abundance through proteasomal and lysosomal degradation.
Conclusion: scyllo-Inositol promotes mutant huntingtin degradation in a model of Huntington disease.
Significance: In contrast to other compounds targeting mutant huntingtin aggregation and accumulation, scyllo-inositol promotes effective degradation.
Keywords: Autophagy, Huntington Disease, Lysosomes, Neurodegenerative Diseases, Proteasome, huntingtin, scyllo-Inositol
Abstract
Huntington disease is characterized by neuronal aggregates and inclusions containing polyglutamine-expanded huntingtin protein and peptide fragments (polyQ-Htt). We have used an established cell-based assay employing a PC12 cell line overexpressing truncated exon 1 of Htt with a 103-residue polyQ expansion that yields polyQ-Htt aggregates to investigate the fate of polyQ-Htt-drug complexes. scyllo-Inositol is an endogenous inositol stereoisomer known to inhibit accumulation and toxicity of the amyloid-β peptide and α-synuclein. In light of these properties, we investigated the effect of scyllo-inositol on polyQ-Htt accumulation. We show that scyllo-inositol lowered the number of visible polyQ-Htt aggregates and robustly decreased polyQ-Htt protein abundance without concomitant cellular toxicity. We found that scyllo-inositol-induced polyQ-Htt reduction was by rescue of degradation pathways mediated by the lysosome and by the proteasome but not autophagosomes. The rescue of degradation pathways was not a direct result of scyllo-inositol on the lysosome or proteasome but due to scyllo-inositol-induced reduction in mutant polyQ-Htt protein levels.
Introduction
Huntington disease is an inherited late-onset neurodegenerative disorder characterized by motor and cognitive decline (1). The disease is caused by an abnormal CAG/polyglutamine (polyQ) repeat expansion located at the N terminus of the huntingtin protein (Htt) (1). Inclusion bodies normally found within neurons of Huntington disease patients and mouse models contain mutant Htt (mt-Htt,2 polyQ-Htt) (2) and have been proposed to be part of a broader cellular coping mechanism (3). Inclusion body formation occurs in concert with the protein degradation pathways, proteasomal and autophagic, to decrease the levels of toxic polyQ-Htt and promote cell survival (3). In aging and in the presence of aggregated proteins, both proteasomal and autophagic functions are decreased, contributing to an increase in toxic species and in neuronal toxicity (3–5). In vivo and in vitro models have demonstrated that inclusion body formation is saturable suggesting that this is a highly regulated process and that targeting polyQ-Htt prior to aggregate formation represents a potential treatment strategy for Huntington disease (3, 6, 7).
A number of compounds have been identified that target the polyQ-Htt aggregation pathway, including antibodies, small molecules, and polyphenols. However, these compounds have no effect on overall mutant Huntingtin protein levels (8–14). These compounds have demonstrated anti-aggregation activity with decreases in the number and/or the size of inclusion bodies. However, they exhibit properties that potentially lead to enhanced cellular toxicity or poor CNS bioavailability (10–14). Similarly, endogenous chaperones have been shown to inhibit polyQ-Htt aggregation also without affecting total polyQ-Htt protein levels (15–17). Recently, overexpression of the endogenous protein, negative regulator of ubiquitin-like protein 1 (NUB1), decreased mt-Htt protein accumulation and rescued toxicity via activation of proteasomal degradation of mt-Htt in both in vitro and in vivo models (18). These latter experiments suggest that compounds that promote reduction of mutant huntingtin protein and resultant toxicity via efficient protein degradation pathways may be sufficient for disease modification.
scyllo-Inositol (SI) has been shown to inhibit toxicity and accumulation of amyloid-β peptide and α-synuclein and promote degradation (19–21). SI is also actively transported into the brain as well as neurons by the sodium myo-inositol transporters (22). An interesting parallel between neurodegenerative diseases is the lack of correlation between the abundance of visible inclusions or extracellular deposits and the extent of neurotoxicity (1, 3, 23, 24) suggesting that other species with specific conformations are the primary propagators of disease (1, 3, 23, 24). Importantly, recent work by Lu et al. (18) that demonstrated the modulation of mt-Htt protein abundance by targeting proteasomal degradation was sufficient to rescue cellular toxicity in vitro, and neurodegeneration in vivo supports the rationale for examining compounds that promote degradation of misfolded proteins.
The aim of this study was to determine whether SI can modulate polyQ-Htt accumulation and examine the downstream cellular degradation pathways. We used an established PC12 cell line that, upon treatment with ponasterone, overexpresses polyQ-Htt resulting in mutant Htt accumulation (10). This model system has the benefit of examining Htt accumulation in the context of basic cellular processes and bears similarities to in vivo Htt aggregation in deposition of both cytosolic and nuclear aggregates (10). We show that after induction of polyQ-Htt expression and aggregate formation, PC12 cells exhibit impaired lysosomal and proteasomal activity, although autophagy is unaffected. Treatment with SI significantly but modestly decreased the number of polyQ-Htt aggregates and robustly decreased polyQ-Htt protein abundance. The reduction in polyQ-Htt abundance was accompanied by a rescue of the lysosomal and the proteasomal deficits. The rescue of protein degradation pathways was not a direct result of SI on these pathways, as SI treatment of PC-12 cells before induction of polyQ-Htt was indistinguishable from untreated cells. Our combined results demonstrate that SI is an effective promoter of polyQ-Htt protein clearance.
EXPERIMENTAL PROCEDURES
Reagents
SI was purchased from Wako (catalog no. 096-04163, Saitama, Japan). Cystamine dihydrochloride (catalog no. C121509), MG132 (catalog no. C2211), 3-methyladenine (3-MA, catalog no. M9281), bafilomycin (catalog no. B1793), and ammonium chloride (catalog no. A9434) were obtained from Sigma. The following antibodies were used: mouse anti-polyQ (1:1000, catalog no. MAB1574, Millipore, Billerica, MA); mouse anti-Htt (1:1000, catalog no. MAB5374, Millipore); rabbit anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH, 1:50,000, catalog no. G9545, Sigma); rabbit anti-cathepsin-B (1:500, catalog no. sc-13985, Santa Cruz Biotechnology, Santa Cruz, CA); rabbit anti-LC3 (1:3000, catalog no. NB600–1384, Novus Biologicals, Littleton, CO); mouse anti-p62 (1:500, catalog no. H00008878-M01, Novus Biologicals, Littleton, CO); rabbit anti-Lys-48-ubiquitin (1:1000, catalog no. 05–1307, Millipore); rat anti-Hsc70 (1:10,000, catalog no. ab19136, Abcam, Cambridge, UK); and horseradish-peroxidase (HRP)-conjugated secondary antibodies (1:2000, Santa Cruz Biotechnology).
Cell Line
The Htt14A2.5 line was a generous gift from Dr. Leslie M. Thompson (University of California, Irvine, CA). Cells were plated at a density of 500,000 cells per well of 24-well plates and maintained in DMEM/F-12 (catalog no. 11330107, Invitrogen) containing 10% fetal bovine serum, 5% horse serum, 250 μg/ml Zeocin (catalog no. 460509, Invitrogen), and 100 μg/ml geneticin (catalog no. 10131035, Invitrogen). After 24 h in culture, the cells were treated with 5 μm ponasterone (catalog no. PON123, BioShop, Burlington, Ontario, Canada) to induce polyQ-Htt protein production. For experiments measuring protein or mRNA expression of polyQ-Htt, 50 ng/ml nerve growth factor (NGF, catalog no. 5221LC, Cell Signaling, Danvers, MA) was added to the plating media to differentiate the cells.
Aggregation Assay
After experimental treatments, PC12 cells were fixed with 4% paraformaldehyde (in phosphate-buffered saline) for 15 min. The number of aggregates within the fixed cells was analyzed under the TE200 fluorescent microscope (Nikon, Melville, NY). Visible enhanced green fluorescent protein (EGFP) punctum was counted as an aggregate. For each well, five fields were randomly counted at a ×20 objective totaling at least 100 cells. Counts by two blinded investigators were compared with ensure reproducibility of the results. The level of aggregation is expressed as the total number of aggregate-containing cells per total number of EGFP-positive cells.
Cell Viability Assay
Thiazolyl blue (catalog no. M2128, Sigma) was added to the PC12 cells post-treatment at a final concentration of 0.5 mg/ml and incubated for 30 min. The PC12 cells were then lysed with dimethyl sulfoxide, and thiazolyl blue absorbance was read on a ThermoMax spectrometric plate reader (Molecular Devices, Sunnyvale, CA) at 540 nm.
Subcellular Fractionation and Immunoblotting
Post-experiment PC12 cells analyzed by immunoblotting were either solubilized directly or fractionated sequentially using buffers of increasing solubilization strengths. Samples solubilized directly were homogenized in Lysis Buffer T/S (50 mm Tris, 1 mm EDTA, 1 mm EGTA, 150 mm NaCl, 0.8% Triton, 0.2% SDS, and 1 mm phenylmethanesulfonyl fluoride). Samples that underwent four-step fractionations were first solubilized in High Salt Buffer (1% protease inhibitor mixture (catalog no. 539134, Billerica, MA), 50 mm Tris, 1 mm EDTA and EGTA, and 500 mm NaCl), followed by solubilization in Triton Buffer (High Salt Buffer plus 1% Triton), solubilization in SDS Buffer (High Salt Buffer plus 1% SDS), and finally solubilization in urea buffer (1% protease inhibitor mixture, 7 m urea, 2 m thiourea, 4% CHAPS, and 30 mm Tris). Nonsoluble components from each buffer were pelleted by ultracentrifugation at 100,000 × g for 1 h and solubilized by the next buffer in sequence. Samples that underwent a two-step fractionation were first solubilized in Lysis Buffer T/S followed by Urea Buffer.
Prior to electrophoresis, samples were boiled in lithium dodecyl sulfate sample buffer (catalog no. NP0007, Invitrogen) containing 1% β-mercaptoethanol. Boiled samples were subjected to electrophoresis in 12% bis-Tris gels (Invitrogen) and transferred to polyvinylidene fluoride membranes. The membranes were blocked in Tris-buffered saline, 0.1% Tween (TBS-T) containing 10% milk for 1 h and probed with a primary antibody solution (in TBS-T containing 5% milk) overnight in the cold room followed by an HRP-conjugated secondary antibody solution (in TBS-T containing 5% milk) for 1 h. The membranes were developed with ECL prime (catalog no. RPN2232, GE Healthcare) and visualized upon film exposure. The scanned protein bands were quantified using ImageJ and normalized to either GAPDH or Ponceau S. Staining of membranes with 0.5% Ponceau S (in 5% acetic acid) was carried out for 15 min followed by two 5-min washes with 5% acetic acid.
Filter Trap Assay
To detect SDS-insoluble aggregates, PC12 cells were solubilized in Lysis Buffer T/S. Both soluble and insoluble components of the lysates constituting 50 μg of protein were diluted in TBS containing 1% SDS, boiled for 5 min, and passed through a cellulose acetate membrane (Whatman 0.2-μm pore), using a dot-blot apparatus (catalog no. 170-6545, Bio-Rad). The membranes were washed twice with TBS containing 1% SDS before immunoblot detection of Htt aggregates as described above.
Real Time Quantitative PCR
Total mRNA was extracted using the Aurum RNA isolation kit (catalog no. 732-6830, Bio-Rad) according to the manufacturer's protocol. cDNA was generated by reverse transcription using the iScript kit (catalog no. 1708841, Bio-Rad) according to the manufacturer's protocol. Quantitative PCR was carried out by a Bio-Rad CFX384 system employing the SsoAdvanced SYBR Green Supermix dye (catalog no. 1725261). The following primer sequences were used: EGFP forward 5′-ACAACTACAACAGCCACAAC-3′ and EGFP reverse 5′-GACTGGGTGCTCAGGTAG-3′; β-actin forward 5′-CTGACAGGATGCAGAAGG-3′ and β-actin reverse 5′-CTGACAGGATGCAGAAGG-3′; and TATA box forward 5′-GCCTTCCACCTTATGCTCAG-3′ and TATA box reverse 5′-GAGTAAGTCCTGTGCCGTAAG-3′. Bio-Rad CFX Manager was used to perform data analysis. mRNA expression of polyQ-Htt was expressed as total EGFP mRNA normalized to the combined mRNA of β-actin and TATA box.
Assays for Proteolytic Activities
Assays for proteasome activity were carried out as described previously (25, 26). Fluorogenic proteasome substrates tert-butyloxycarbonyl-Leu-Arg-Arg-aminomethylcoumarin (AMC, catalog no. BML-BW8515) and succinyl-Leu-Leu-Val-Tyr-AMC (catalog no. BML-P802) were purchased from Enzo (Farmingdale, NY). Cells were lysed with 50 mm Tris, 1 mm EDTA, 2 mm ATP, and 1% Triton. Each assay well contained 30 μg of cell lysates and 50 μm proteasome substrate and was incubated at 37 °C for 30 min. The assay wells were then measured with an Infinite M200 fluorescent plate reader (Tecan, Männedorf, Switzerland) at excitation 380 nm and emission 460 nm. Proteasome activity was expressed as change in fluorescence per total protein concentration. Protein concentration was measured using the bicinchoninic acid assay kit (catalog no. PI23225, Thermo, Rockford, IL) according to the manufacturer's protocol.
Cathepsin B activity was measured as described previously (20) by proteolysis of the specific substrate carbobenzoxy-l-arginyl-arginine-aminomethylcoumarin (carbobenzoxy-RR-AMC, Sigma). Cells were lysed with 50 mm Tris, 1 mm EDTA, and 1% Triton, and 50 μl of lysates were preincubated with l-cysteine (final concentration 4 mm) for 10 min at 37 °C. An equal volume of the reaction buffer (final concentrations 50 μm carbobenzoxy-RR-AMC, 400 μm sodium acetate, pH 5.5) was then added. After 30 min of incubation at 37 °C, AMC release indicating substrate cleavage was measured with TECAN Infinite 200 at 380 nm excitation and 460 nm emission. Cathepsin B activity was expressed as change in fluorescence per total protein concentration.
Lysosomal activity was measured by the fluorescent lysosomal indicator LysoTracker (catalog no. L7528, Invitrogen). PC12 cells were labeled in culture with 1 μm LysoTracker for 30 min. After two washes with media, total fluorescence indicating lysosomal activity was measured with TECAN Infinite 200 at 570 nm excitation and 590 nm emission. Lysosomal activity was expressed as fluorescence (relative to nonlabeled controls) per total protein concentration.
Statistical Analysis
Statistical significance levels were measured by either analysis of variance followed by Newman-Keuls post hoc or Student's t test in the case of two experimental groups. Each n represents one culture well. * denotes p < 0.05.
RESULTS
SI Lowers the Number of Visible PolyQ-Htt Inclusions
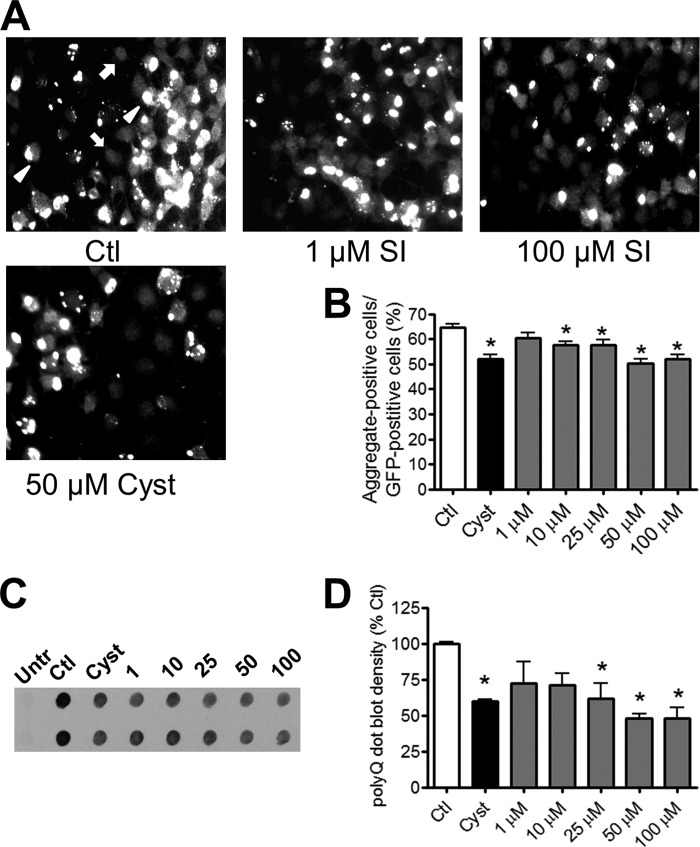
The Htt14A2.5 line was a subclone of the Htt103Q-EGFP PC12 cell line that contained a high percentage of cells with visible polyQ-Htt aggregates after induction of protein expression with ponasterone (10). In control cultures, 5 μm ponasterone resulted in ∼65% of EGFP-positive cells containing visible polyQ-Htt aggregates after 24 h (Fig. 1, A and B). Cells containing visible EGFP puncta representing polyQ-Htt aggregates (Fig. 1A, arrowheads) are present along with cells containing only diffuse EGFP fluorescence (Fig. 1A, arrows). In agreement with previous studies showing that inclusion body formation is a saturable process (3, 6, 7, 10), ponasterone induction of polyQ-Htt expression for 48 h did not yield significantly higher levels of aggregation than at 24 h (data not shown). We thus conducted all experiments using the 24-h induction paradigm.
FIGURE 1.
SI dose-dependently decreases accumulation of Htt inclusions in PC12 cells. Expression and accumulation of Htt containing expanded polyglutamine (polyQ) were induced by 5 μm ponasterone for 24 h. Induced cultures were treated with either vehicle (Ctl), 1–100 μm SI, or 50 μm cystamine. A, representative images of induced cultures treated with either vehicle (Ctl, top left), 1 μm SI (top middle), 100 μm SI (top right), or 50 μm cystamine (Cyst, bottom left). White arrowheads are examples of Htt aggregates, and white arrows are examples of EGFP-positive cells without aggregates. Magnification = ×20. B, quantification of aggregation levels in control, cystamine-treated, and SI-treated cultures (n = 45). C, representative dot blots; D, quantification of SDS-insoluble polyQ-Htt from untreated/uninduced (Untr) cells and induced cells treated with vehicle, SI, or cystamine (n = 3). * denotes p < 0.05 relative to control.
To examine the effects of SI on polyQ-Htt accumulation, we co-treated the Htt14A2.5 PC12 cells with SI and ponasterone for induction of polyQ-Htt protein expression. This prevention paradigm resulted in a dose-dependent decrease in the number of visible polyQ-Htt aggregates as demonstrated by a reduction in cells containing EGFP puncta (Fig. 1, A and B). A significant reduction in visible aggregates was observed from 10 to 100 μm SI. At 100 μm SI, cells containing aggregates decreased by ∼15%, which is comparable with cells treated with 50 μm cystamine, a known inhibitor of Htt aggregation (Fig. 1, A and B) (10). The observed modest aggregate reductions were not due to potential SI-related toxicities because we did not detect loss of cell viability (data not shown).
To further define the role of SI on insoluble polyQ-Htt aggregates, we performed filter trap assays on SI-treated cells. Using an anti-polyQ antibody, we observed a dose-dependent reduction of SDS-insoluble aggregates after SI treatment (Fig. 1, C and D). The reductions were statistically significant from 25 to 100 μm SI (Fig. 1, C and D). Interestingly, compared with visible aggregate counts, aggregate reduction observed by filter trap was markedly higher (∼50% at 100 μm SI), possibly due to the filter trap assay detecting smaller insoluble aggregates in addition to denser inclusions visible as fluorescent puncta.
SI Promotes Clearance of Htt
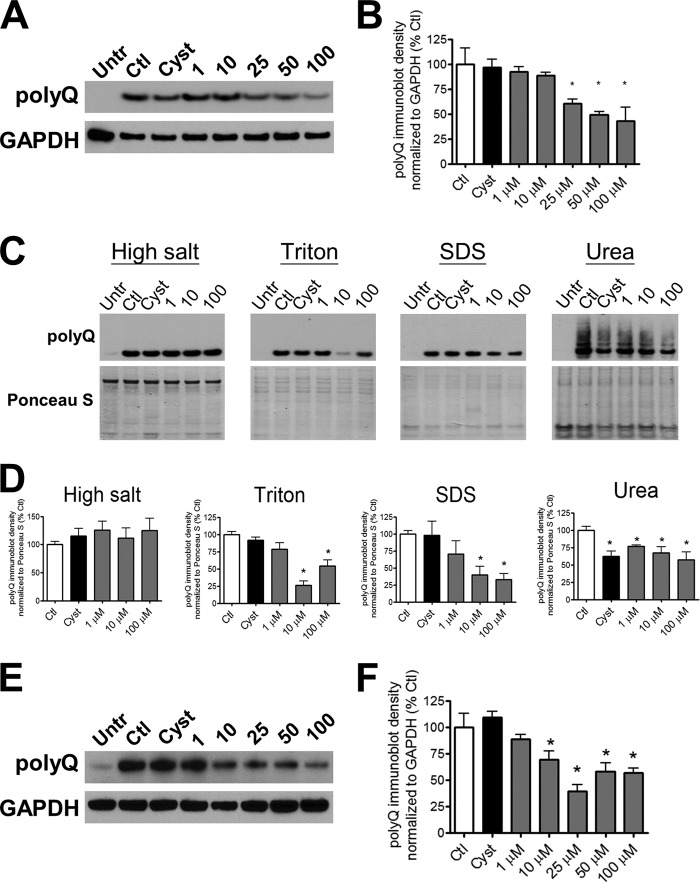
To investigate the possible mechanism by which SI reduced polyQ-Htt accumulation, we examined total protein levels of polyQ-Htt as a function of SI dose. We solubilized SI-treated and control cultures of Htt14A2.5 PC12 cells with a buffer containing Triton and SDS and probed with an anti-polyQ antibody to differentiate polyQ-expanded huntingtin protein from endogenous protein. We demonstrate that SI treatment dose-dependently decreased the abundance of polyQ-Htt protein with an effective dose of 25 μm (Fig. 2, A and B). Reduction in Htt protein abundance was confirmed by re-probing the same blots with a huntingtin-specific antibody (data not shown). As published previously (10), treatment with 50 μm cystamine decreased the number of visible polyQ-Htt aggregates but did not affect protein abundance of polyQ-Htt (Fig. 2, A and B). To examine the action of SI in greater detail, we sequentially fractionated the cells with four buffers of increasing solubilization strength. We found that SI dose-dependently lowered polyQ-Htt abundance in Triton- and SDS-soluble fractions; however, no treatment effect was detected in the high salt-soluble fractions (Fig. 2, C and D). Cystamine treatment in contrast had no effect on polyQ-Htt abundance in the high salt-, Triton-, or SDS-soluble fractions (Fig. 2, C and D). In the urea-soluble fractions containing visible aggregates, both SI and cystamine reduced polyQ-Htt abundance (Fig. 2, C and D). The differential activities of SI and cystamine on polyQ-Htt protein abundance suggest that the mechanism leading to decreased visible inclusions may differ for these two compounds. Notably, cystamine has been shown by others to inhibit tissue transglutaminase activity toward mutant Htt leading to decreased post-translational modification resulting in decreased aggregation (27, 28).
FIGURE 2.
SI dose-dependently promotes clearance of polyQ-Htt. PC12 cultures were induced with 5 μm ponasterone and were treated with either vehicle, 1–100 μm SI, or 50 μm cystamine. A, representative immunoblots; B, quantification of polyQ-Htt expression in untreated/uninduced (Untr) cells and induced cells treated with vehicle (Ctl), SI, or cystamine (Cyst) for 24 h (n = 3). The cells were solubilized in Lysis Buffer T/S. C, representative immunoblots; D, quantification of polyQ-Htt expression in sequentially extracted fractions of increasing solubilizing strength. The cells were either untreated/uninduced or induced with treatments of vehicle, SI, or cystamine (n = 4). E, representative immunoblots; F, quantification of polyQ-Htt expression in cultures first induced with ponasterone for 24 h, then removed of ponasterone, and treated with either SI or cystamine for another 24 h (n = 6). The cells were solubilized in Lysis Buffer T/S. * denotes p < 0.05 relative to control.
The reduction in polyQ-Htt protein abundance after SI treatment suggests that the synthesis or degradation of polyQ-Htt may contribute to this reduction. To differentiate the effects of SI on synthesis versus degradation, we induced polyQ-Htt protein expression for 24 h, turned off protein expression by removing ponasterone, and subsequently treated with SI for 24 h. Under this paradigm in the absence of new polyQ-Htt synthesis, SI treatment dose-dependently decreased polyQ-Htt protein levels with a lower effective dose of 10 μm (Fig. 2, E and F). Furthermore, levels of ponasterone-induced mRNA of polyQ-Htt did not change with SI treatment (data not shown) demonstrating that SI is not a direct competitor at the level of mRNA. The combined data suggest that reduction in polyQ-Htt protein levels may involve enhanced protein degradation.
To determine the potential clearance mechanisms that contribute to SI reduction in polyQ-Htt abundance, we examined exocytosis and lysosomal-autophagic and proteasomal proteolysis pathways. Exocytosis of toxins and aggregated proteins as a mechanism of cell survival has been proposed to lead to enhanced disease exposure from cell to cell transfer and initiation of extracellular deposits (29). The potential clearance of polyQ-Htt by exocytosis was therefore examined for the presence of polyQ-Htt in the cell media. No detectable levels of polyQ-Htt were found in the cell media (data not shown), suggesting a mechanism directly involving intracellular degradation.
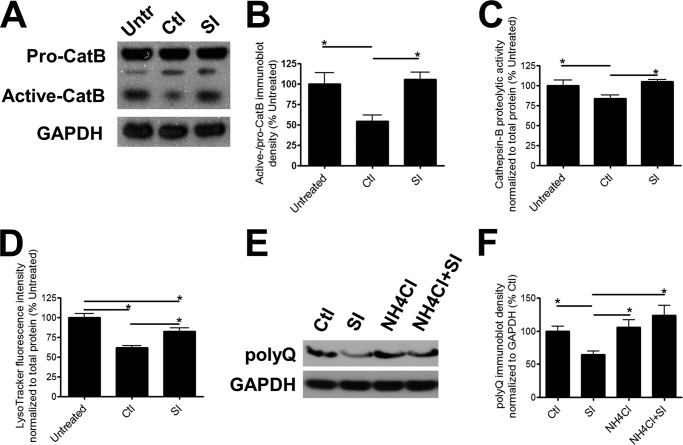
Lysosome-mediated proteolysis is a major degradation mechanism in mammalian cells, and lysosomal enzymes, such as cathepsins, have been previously implicated in polyQ-Htt degradation (30, 31). We found that induction of polyQ-Htt expression decreased the conversion of pro-cathepsin B to its active form indicating that, in the presence of accumulating polyQ-Htt protein levels, lysosomal activity is impaired compared with control cells not expressing polyQ-Htt (Fig. 3, A and B). Proteolytic activity of cathepsin B was also decreased upon ponasterone induction of polyQ-Htt protein expression (Fig. 3C), albeit to a lesser degree compared with the more potent reduction in pro-/active cathepsin B conversion. To reflect overall changes in lysosomal activity, we probed the cultures with LysoTracker, a fluorescent indicator specific for acidic organelles (32, 33). Fluorescence intensity from LysoTracker has been used as a measurement of lysosomal acidification and activity (32, 33). Similar to cathepsin B activity, lysosomal acidification decreased following induction of polyQ-Htt protein expression (Fig. 3D). After incubation with SI, the levels of pro- to active cathepsin B conversion, cathepsin B-mediated proteolysis, as well as lysosomal acidification were significantly higher than that of PC12 cells expressing polyQ-Htt demonstrating a rescue of lysosomal activity (Fig. 3, A–D). To further elucidate a role for lysosomal degradation, we inhibited lysosomal enzymatic activity by increasing intracellular pH using ammonium chloride. Treatment with ammonium chloride abrogated the SI-induced degradation of polyQ-Htt (Fig. 3, E and F), indicating that lysosomal proteolysis contributes to SI-mediated clearance of polyQ-Htt.
FIGURE 3.
Involvement of lysosomal proteolysis in SI-mediated clearance of polyQ-containing Htt. A, representative immunoblots; B, quantification of cathepsin B (CatB) conversion from pro-cathepsin B to active cathepsin B in untreated/uninduced (Untr) cultures and 5 μm ponasterone-induced cultures treated with either vehicle (Ctl) or 100 μm SI for 24 h (n = 6). The cells were lysed in Lysis Buffer T/S. C, cathepsin B activity in untreated/uninduced cultures and 5 μm ponasterone-induced cultures treated with either vehicle or 100 μm SI for 24 h (n = 6). D, fluorescence intensity of LysoTracker probes in untreated/uninduced cultures and 5 μm ponasterone-induced cultures treated with either vehicle or 100 μm SI for 24 h (n = 6). E, representative immunoblots; F, quantification of polyQ-Htt expression in induced cultures treated with either vehicle, 100 μm SI, 5 mm NH4Cl, or both SI and NH4Cl for 24 h (n = 6). The cells were solubilized in Lysis Buffer T/S. * denotes p < 0.05.
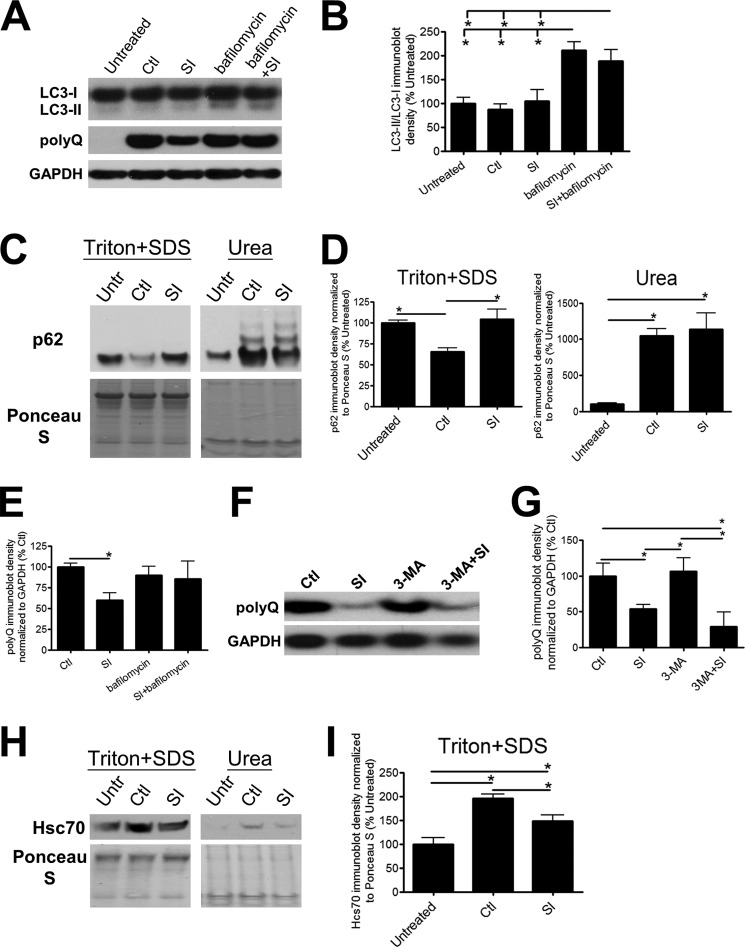
To define the degradation pathways upstream of the lysosome, we examined macroautophagy (hereafter referred to as autophagy). Autophagy is a process where a set of membranes form intracellularly to enclose cellular contents, forming autophagosomes that eventually fuse with the lysosome and “self-digest” (34). In vitro and in vivo stimulation and inhibition of autophagy has previously been shown to modulate polyQ-Htt degradation and decrease neuronal toxicity resulting from polyQ-Htt aggregates (4). Conversion of the autophagosomal protein light chain 3 (LC3), from LC3-I to LC3-II occurs during the maturation of autophagosomes, and changes in LC3 conversion have been used as a marker of autophagic modulation (34). We found that LC3 conversion did not vary as a function of polyQ-Htt expression or SI treatment suggesting that initiation of autophagocytosis is not impaired (Fig. 4, A and B). In contrast, expression of p62, often used as a marker of autophagic turnover (34), decreased after induction of polyQ-Htt accumulation compared with untreated and SI-treated cells (Fig. 4, C and D). Closer examination revealed a drastic increase in p62 protein trapped in the urea-soluble fraction upon induction of polyQ-Htt protein expression regardless of SI treatment (Fig. 4, C and D). This result suggests that decreased p62 in the Triton/SDS-soluble fraction is a consequence of sequestration in the insoluble aggregates rather than an indication of increased autophagic turnover. SI treatment (Fig. 4, C and D) did not modulate the majority of p62 accumulation sequestered within aggregates, suggesting that SI-mediated polyQ-Htt degradation does not require autophagic activity. To further characterize the autophagic pathway, we inhibited total autophagic activity with either 10 nm bafilomycin, an inhibitor of autophagosome-lysosome fusion (35), or 5 mm 3-MA, a suppressor of autophagic induction and turnover (34). We verified the inhibitory activities of bafilomycin and 3-MA by increased accumulation of LC3-II and p62 (Fig. 4, A and B, data not shown). Surprisingly, we found that although treatment with 3-MA did not inhibit the SI-mediated effects on polyQ-Htt protein clearance, treatment with bafilomycin did (Fig. 4, A and E–G). These results are consistent with bafilomycin inhibition of autophagosome-lysosomal fusion and the disruption of macroautophagy as well as bafilomycin-induced de-acidification of the lysosome leading to inhibition of lysosomal degradation pathways unrelated to macroautophagy (35). These combined data support an SI-mediated polyQ-Htt degradation pathway that is independent of macroautophagy but dependent on lysosomal activity.
FIGURE 4.
Autophagy is not the main mechanism involved in the clearance of polyQ-containing Htt mediated by SI. A, representative immunoblots of LC3 conversion from LC3-I to LC3-II and polyQ-Htt in untreated/uninduced cultures (Untr) and 5 μm ponasterone-induced cultures treated with either vehicle (Ctl), 100 μm SI, 10 nm bafilomycin, or both bafilomycin and SI for 24 h. The cells were solubilized in Lysis Buffer T/S. B, quantification of LC3-I to LC3-II conversion (n = 4). C, representative immunoblots; D, quantification of p62 in Lysis Buffer T/S-soluble and Urea Buffer-soluble fractions. The cells were either untreated/uninduced or ponasterone-induced with treatments of vehicle or 100 μm SI (n = 4). E, quantification of polyQ-Htt expression (n = 6). F, representative immunoblots; G, quantification of polyQ-Htt expression in induced cultures treated with either vehicle, 100 μm SI, 5 mm 3-MA, or both SI and 3-MA. The cells were solubilized in Lysis Buffer T/S (n = 5). H, representative immunoblots; I, quantification of Hsc70 expression in Lysis Buffer T/S-soluble and Urea Buffer-soluble fractions. The cells were either untreated/uninduced or ponasterone-induced with treatments of vehicle or 100 μm SI (n = 4). * denotes p < 0.05.
To further investigate the role of the lysosome in SI-mediated polyQ-Htt degradation, we examined markers of chaperone-mediated autophagy. Huntingtin protein has a number of KFERQ-like sequences that are recognized by the heat shock cognate protein, Hsc70, and necessary for targeting to lysosomal degradation (36, 37). In agreement with chaperone-mediated autophagic degradation of the N-terminal fragment of huntingtin (37), soluble Hsc70 protein levels are increased after induction of polyQ-Htt expression in our cell culture system (Fig. 4, H and I). This result suggests that chaperone-mediated autophagy is up-regulated upon induction of polyQ-Htt protein expression. SI-induced reduction of mt-Htt protein levels corresponds to a reduction in Hsc70 protein levels (Fig. 4, H and I). These results in combination with the lack of degradation through macroautophagic pathways suggest a role for chaperone-mediated autophagy in SI-induced degradation of polyQ-Htt.
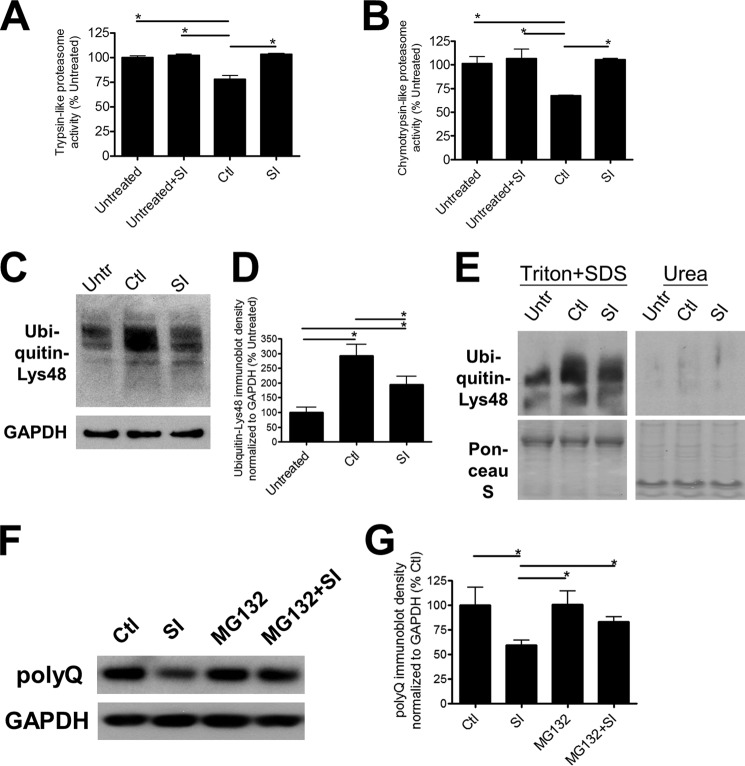
Although p62 is often used as a marker of autophagy, it has been shown to bind directly to both ubiquitin and LC3 for shuttling and targeting of proteins to the proteasome and the autophagic pathway, respectively (38, 39). In light of the observed changes in soluble p62 expression, we examined proteasomal activity utilizing the characteristic trypsin-like and chymotrypsin-like proteolytic activities of the proteasome (Fig. 5, A and B). We found that both activities decreased in polyQ-Htt-expressing Htt14A2.5 PC12 cells in comparison with untreated cells consistent with previous reports of proteasomal deficits in primary neuronal cultures expressing polyQ-Htt (Fig. 5, A and B) (40–42). To further characterize proteasomal function, we examined the abundance of Lys-48-polyubiquitinated chains by immunoblotting. Proteins polyubiquitinated at Lys-48 are targeted for degradation by the proteasome (5, 43). We found that cells expressing polyQ-Htt also accumulated higher levels of Lys-48-linked ubiquitin chains in the Triton/SDS-soluble fraction (Fig. 5, C and D), suggesting that degradation of Lys-48-ubiquitin-labeled proteins that are normally targeted for proteolysis by the proteasome are also inhibited (5, 43). The level of polyubiquitinated proteins in the insoluble urea fraction was negligible in comparison with the Triton/SDS-soluble fraction (Fig. 5E). These combined results suggest an overall dysfunctional proteasomal degradation system in the Htt14A2.5 PC12 cells after induction of polyQ-Htt.
FIGURE 5.
Degradation by proteasome is involved in the SI-mediated clearance of poly-Q-containing Htt. A, trypsin-like; B, chymotrypsin-like proteasome activities in untreated/uninduced (Untr) cultures and induced cultures treated with either vehicle (Ctl) or 100 μm SI (n = 6). C, representative immunoblots; D, quantification of Lys-48-linked ubiquitin expression in untreated/uninduced cultures and ponasterone-induced cultures treated with either vehicle or 100 μm SI (n = 4). Cells were solubilized with Lysis Buffer T/S. E, representative immunoblots of Lys-48-linked ubiquitin expression compared in Lysis Buffer T/S-soluble and Urea Buffer-soluble fractions. F, representative immunoblots; G, quantification of polyQ-Htt expression in induced cultures treated with either vehicle, 100 μm SI, 10 nm MG132, or both SI and MG132 (n = 5). The cells were solubilized with Lysis Buffer T/S. * denotes p < 0.05.
Proteasomal enzymatic activity and polyubiquitination, indicators of proteasomal impairment, were restored by SI treatment (Fig. 5, A–D). To determine whether the restoration of proteasomal activity was a direct effect of SI on the proteasome, we examined trypsin- and chymotrypsin activity of Htt14A2.5 PC12 cells in the absence of polyQ-Htt induction. SI treatment had no effect on proteasomal proteolytic activity in untreated cells (Fig. 5, A and B), suggesting that the observed restoration is not a direct effect on the proteasome. To further characterize the involvement of the proteasome, we treated polyQ-Htt-expressing Htt14A2.5 PC12 cells with an inhibitor of proteasomal activity, MG132 (Fig. 5, F and G). We found that proteasomal inhibition prevented the polyQ-Htt clearance mediated by SI, as polyQ-Htt levels in MG132-treated cultures were not significantly different from cells treated with both MG132 and SI (Fig. 5, F and G). These results implicate proteasome degradation as an important mechanism in SI-mediated clearance of polyQ-Htt. Further investigation revealed that in the absence of SI and continuous ponasterone induction, intrinsic degradation by Htt14A2.5 PC12 cells also involved the proteasome (Fig. 6, A–D, see under “Discussion” for details).
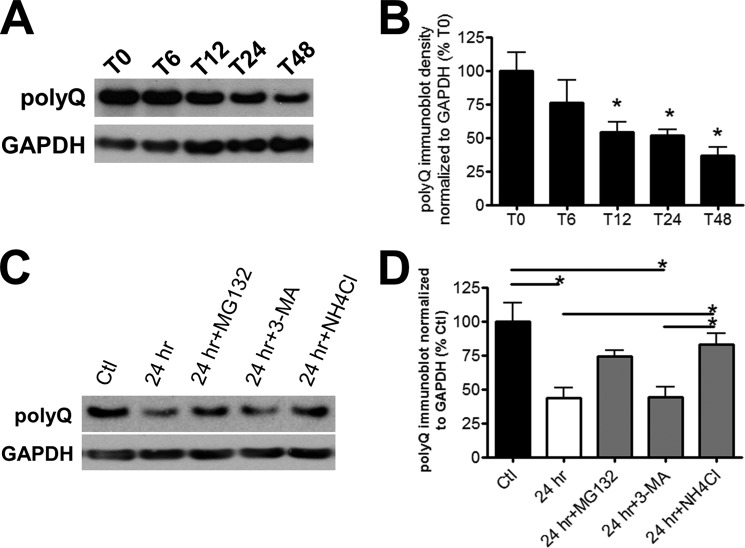
FIGURE 6.
Intrinsic degradation pathways activated in the absence of SI. A, representative immunoblots and B, quantification of polyQ-Htt expression 0 h (T0) to 48 h (T48) after removal of ponasterone induction. The cells were first induced with ponasterone for 24 h prior to removal of ponasterone. They were solubilized in Lysis Buffer T/S (n = 4). * denotes p < 0.05 relative to T0. C, representative immunoblots and D, quantification of polyQ-Htt expression 24 h after removal of ponasterone induction. The cells were treated with vehicle (24 h), 10 nm MG132 (24 h+MG132), 5 mm 3-MA (24 h+3-MA), or 5 mm NH4Cl (24 h+NH4Cl). A set of cultures did not have ponasterone removed and served as the control (Ctl). The cells were solubilized in Lysis Buffer T/S (n = 4). * denotes p < 0.05.
DISCUSSION
The objective of this study was to determine whether SI modulates polyQ-Htt accumulation and the downstream cellular degradation pathways. Collectively, our results show that SI promotes clearance of polyQ-Htt protein via pathways dependent on lysosomal and proteasomal degradation, but not autophagic induction.
The ameliorating effects of SI are mediated via either a primary effect on reduction of polyQ-Htt levels through enhanced clearance or an indirect mechanism by limiting the polyQ-Htt-induced detrimental effects on degradation pathways. In this study, SI in the absence polyQ-Htt expression did not affect proteasomal activity (Fig. 5, A and B). In our previous study, SI was incubated with isolated lysosomes, and conversion to active cathepsin B protein levels and enzymatic activity levels were unaffected (20). Thus, we propose a direct interaction between SI and polyQ-Htt resulting in enhanced clearance. Molecular modeling experiments demonstrated that SI binding to aggregation-prone peptides depends on both the morphology and the surface physiochemical properties of the protein/peptide (44, 45). Although significantly smaller, the planar structure of SI, rendering two hydrophobic faces and hydroxyl groups for hydrogen bonding, is similar to that of polyphenols previously shown to bind to multiple aggregation-prone peptides/proteins, including mutant huntingtin (46, 47). These combined results suggest that the specific binding mechanisms and the degradation pathways may be protein-specific. Further investigation is needed to better understand the binding mechanisms between SI and polyQ-Htt.
The brain has a need for constitutive selective autophagy and ubiquitin-proteasome system due to the intricate control of neuronal activity. In light of this, both pathways have been shown to contribute to decreased accumulation of aggregated proteins (39). Although disruption of autophagosome-lysosome fusion by bafilomycin abrogated the clearance effects of SI (Fig. 4, A and E), bafilomycin also elevates lysosomal pH and inhibits overall lysosomal proteolysis (35). In the absence of SI, the intrinsic clearance mechanisms of polyQ-Htt in the current cellular model involve a pathway independent of autophagosome formation. We examined polyQ-Htt degradation 6–48 h after removal of ponasterone induction and found that ∼50% of Triton/SDS-soluble polyQ-Htt was degraded 24 h post-removal (Fig. 6, A and B). This degradation was not inhibited by 3-MA but was inhibited by both ammonium chloride and MG132 (Fig. 6, C and D). Taken together, involvement of a lysosomal degradation pathway independent of autophagosomes, such as chaperone-mediated autophagy (37), is consistent with our results. A previous study has shown that as the length of the polyQ expansion increases, the ability of Htt to cross the lysosomal membrane is decreased suggesting that degradation through chaperone-mediated autophagy is impaired (37). This is consistent with our findings of decreased lysosomal activity in the presence of normal macroautophagy.
The proteasomal deficit detected upon the induction of polyQ-Htt that we report is consistent with previous cellular models that overexpress the N-terminal fragment of mutant Htt and is shown in an acute overexpression in vivo model before inclusion body formation (48–51). Recently, a genetic screen identified an endogenous protein, NUB1, as a regulator of mutant huntingtin protein abundance by promotion of degradation via the proteasomal pathway (18). Furthermore, this study demonstrated that targeting mutant huntingtin protein degradation is sufficient to inhibit in vitro toxicity and in vivo neurodegeneration. This is consistent with previous studies that have inhibited mutant huntingtin protein expression and rescued motor and pathological disease readouts (52, 53). In light of these results, small molecules that promote degradation of mutant huntingtin protein abundance may lead to similar benefit for disease progression.
There have been a number of small molecules identified in vitro as mutant Htt aggregation inhibitors, as evidenced by a decrease in the number of intracellular aggregates (10–14). None of these studies investigated the degradation of mutant Htt, and many of the compounds did not alter intracellular mutant Htt levels suggesting that degradation was not promoted with these compounds (10–14). The mechanism of polyQ-Htt modulation may vary between compounds and be independent of direct interaction of the compound with mutant Htt. The decrease in polyQ-Htt aggregation after SI treatment is consistent with previous studies on Aβ peptide aggregation that demonstrated a decrease in Aβ oligomer and fibril formation in vitro (19, 54), stabilization of preformed oligomers in vitro (54), and decreased oligomers and prevention of amyloid plaques in vivo (55–57). The lack of toxicity detected in this study is also in agreement with the SI-induced decrease in α-synuclein toxicity in an inducible neuronal cell model (21) and in amyloid-β peptide toxicity in the 7PA2 cell line (54). Furthermore, SI promotes degradation of Aβ by autophagic digestion (20). These combined results suggest that SI may lead to cellular homeostasis by efficiently targeting aggregation-prone proteins/peptides to the most efficient substrate-specific degradation pathway.
Acknowledgment
We are grateful of the generosity of Dr. Leslie M. Thompson.
This work was supported by Canadian Institutes of Health Research Grants PRG37857 (to J. M.) and 120894 (to A. L.) and an Ontario postdoctoral scholarship (to A. L.). J. M. declares a conflict of interest as a named inventor on patents and patent applications relating to scyllo-inositol and derivatives.
- mt-Htt
- mutant Htt
- SI
- scyllo-inositol
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- EGFP
- enhanced green fluorescent protein
- T/S
- Trition X-100/sodium dodecyl sulfate
- 3-MA
- 3-methyladenine
- AMC
- aminomethylcoumarin.
REFERENCES
- 1. Ross C. A., Tabrizi S. J. (2011) Huntington's disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 10, 83–98 [DOI] [PubMed] [Google Scholar]
- 2. Vonsattel J. P. (2008) Huntington disease models and human neuropathology: similarities and differences. Acta Neuropathol. 115, 55–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arrasate M., Finkbeiner S. (2012) Protein aggregates in Huntington's disease. Exp. Neurol. 238, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sarkar S., Rubinsztein D. C. (2008) Huntington's disease: degradation of mutant huntingtin by autophagy. FEBS J. 275, 4263–4270 [DOI] [PubMed] [Google Scholar]
- 5. Bennett E. J., Shaler T. A., Woodman B., Ryu K. Y., Zaitseva T. S., Becker C. H., Bates G. P., Schulman H., Kopito R. R. (2007) Global changes to the ubiquitin system in Huntington's disease. Nature. 448, 704–708 [DOI] [PubMed] [Google Scholar]
- 6. Miller J., Arrasate M., Shaby B. A., Mitra S., Masliah E., Finkbeiner S. (2010) Quantitative relationships between huntingtin levels, polyglutamine length, inclusion body formation, and neuronal death provide novel insight into huntington's disease molecular pathogenesis. J. Neurosci. 30, 10541–10550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Miller J., Arrasate M., Brooks E., Libeu C. P., Legleiter J., Hatters D., Curtis J., Cheung K., Krishnan P., Mitra S., Widjaja K., Shaby B. A., Lotz G. P., Newhouse Y., Mitchell E. J., Osmand A., Gray M., Thulasiramin V., Saudou F., Segal M., Yang X. W., Masliah E., Thompson L. M., Muchowski P. J., Weisgraber K. H., Finkbeiner S. (2011) Identifying polyglutamine protein species in situ that best predict neurodegeneration. Nat. Chem. Biol. 7, 925–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Southwell A. L., Ko J., Patterson P. H. (2009) Intrabody gene therapy ameliorates motor, cognitive, and neuropathological symptoms in multiple mouse models of Huntington's disease. J. Neurosci. 29, 13589–13602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Butler D. C., Messer A. (2011) Bifunctional anti-huntingtin proteasome-directed intrabodies mediate efficient degradation of mutant huntingtin exon 1 protein fragments. PLoS One 6, e29199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Apostol B. L., Kazantsev A., Raffioni S., Illes K., Pallos J., Bodai L., Slepko N., Bear J. E., Gertler F. B., Hersch S., Housman D. E., Marsh J. L., Thompson L. M. (2003) A cell-based assay for aggregation inhibitors as therapeutics of polyglutamine-repeat disease and validation in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 100, 5950–5955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chopra V., Fox J. H., Lieberman G., Dorsey K., Matson W., Waldmeier P., Housman D. E., Kazantsev A., Young A. B., Hersch S. (2007) A small-molecule therapeutic lead for Huntington's disease: preclinical pharmacology and efficacy of C2–8 in the R6/2 transgenic mouse. Proc. Natl. Acad. Sci. U.S.A. 104, 16685–16689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fuentealba R. A., Marasa J., Diamond M. I., Piwnica-Worms D., Weihl C. C. (2012) An aggregation sensing reporter identifies leflunomide and teriflunomide as polyglutamine aggregate inhibitors. Hum. Mol. Genet. 21, 664–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Bebber F., Paquet D., Hruscha A., Schmid B., Haass C. (2010) Methylene blue fails to inhibit Tau and polyglutamine protein dependent toxicity in zebrafish. Neurobiol. Dis. 39, 265–271 [DOI] [PubMed] [Google Scholar]
- 14. Sontag E. M., Lotz G. P., Yang G., Sontag C. J., Cummings B. J., Glabe C. G., Muchowski P. J., Thompson L. M. (2012) Detection of mutant huntingtin aggregation conformers and modulation of SDS-soluble fibrillar oligomers by small molecules. J. Huntingtons Dis. 1, 127–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sontag E. M., Joachimiak L. A., Tan Z., Tomlinson A., Housman D. E., Glabe C. G., Potkin S. G., Frydman J., Thompson L. M. (2013) Exogenous delivery of chaperonin subunit fragment ApiCCT1 modulates mutant Huntingtin cellular phenotypes. Proc. Natl. Acad. Sci. U.S.A. 110, 3077–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gillis J., Schipper-Krom S., Juenemann K., Gruber A., Coolen S., van den Nieuwendijk R., van Veen H., Overkleeft H., Goedhart J., Kampinga H. H., Reits E. A. (2013) The DNAJB6 and DNAJB8 protein chaperones prevent intracellular aggregation of polyglutamine peptides. J. Biol. Chem. 288, 17225–17237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tashiro E., Zako T., Muto H., Itoo Y., Sörgjerd K., Terada N., Abe A., Miyazawa M., Kitamura A., Kitaura H., Kubota H., Maeda M., Momoi T., Iguchi-Ariga S. M., Kinjo M., Ariga H. (2013) Prefoldin protects neuronal cells from polyglutamine toxicity by preventing aggregation formation. J. Biol. Chem. 288, 19958–19972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lu B., Al-Ramahi I., Valencia A., Wang Q., Berenshteyn F., Yang H., Gallego-Flores T., Ichcho S., Lacoste A., Hild M., Difiglia M., Botas J., Palacino J. (2013) Identification of NUB1 as a suppressor of mutant Huntington toxicity via enhanced protein clearance. Nat. Neurosci. 16, 562–570 [DOI] [PubMed] [Google Scholar]
- 19. McLaurin J., Golomb R., Jurewicz A., Antel J. P., Fraser P. E. (2000) Inositol stereoisomers stabilize an oligomeric aggregate of Alzheimer amyloid β peptide and inhibit Aβ-induced toxicity. J. Biol. Chem. 275, 18495–18502 [DOI] [PubMed] [Google Scholar]
- 20. Lai A. Y., McLaurin J. (2012) Inhibition of amyloid-β peptide aggregation rescues the autophagic deficits in the TgCRND8 mouse model of Alzheimer disease. Biochim. Biophys. Acta 1822, 1629–1637 [DOI] [PubMed] [Google Scholar]
- 21. Vekrellis K., Xilouri M., Emmanouilidou E., Stefanis L. (2009) Inducible overexpression of wild type α-synuclein in human neuronal cells leads to caspase-dependent non-apoptotic death. J. Neurochem. 109, 1348–1362 [DOI] [PubMed] [Google Scholar]
- 22. Fenili D., Weng Y. Q., Aubert I., Nitz M., McLaurin J. (2011) Sodium/myo-inositol transporters: substrate transport requirements and regional brain expression in the TgCRND8 mouse model of amyloid pathology. PLoS One 6, e24032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Larson M. E., Lesné S. E. (2012) Soluble Aβ oligomer production and toxicity. J. Neurochem. 120, 125–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S. (2004) Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 [DOI] [PubMed] [Google Scholar]
- 25. Lichtenberg M., Mansilla A., Zecchini V. R., Fleming A., Rubinsztein D. C. (2011) The Parkinson's disease protein LRRK2 impairs proteasome substrate clearance without affecting proteasome catalytic activity. Cell Death Dis. 2, e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khandros E., Thom C. S., D'Souza J., Weiss M. J. (2012) Integrated protein quality-control pathways regulate free α-globin in murine β-thalassemia. Blood 119, 5265–5275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Karpuj M. V., Garren H., Slunt H., Price D. L., Gusella J., Becher M. W., Steinman L. (1999) Transglutaminase aggregates huntingtin into nonamyloidogenic polymers, and its enzymatic activity increases in Huntington's disease brain nuclei. Proc. Natl. Acad. Sci. U.S.A. 96, 7388–7393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karpuj M. V., Becher M. W., Springer J. E., Chabas D., Youssef S., Pedotti R., Mitchell D., Steinman L. (2002) Prolonged survival and decreased abnormal movements in transgenic model of Huntington disease, with administration of the transglutaminase inhibitor cystamine. Nat. Med. 8, 143–149 [DOI] [PubMed] [Google Scholar]
- 29. Brundin P., Melki R., Kopito R. (2010) Prion-like transmission of protein aggregates in neurodegenerative diseases. Nat. Rev. Mol. Cell Biol. 11, 301–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liang Q., Ouyang X., Schneider L., Zhang J. (2011) Reduction of mutant huntingtin accumulation and toxicity by lysosomal cathepsins D and B in neurons. Mol. Neurodegener 6, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bhutani N., Piccirillo R., Hourez R., Venkatraman P., Goldberg A. L. (2012) Cathepsins L and Z are critical in degrading polyglutamine-containing proteins within lysosomes. J. Biol. Chem. 287, 17471–17482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee J. H., Yu W. H., Kumar A., Lee S., Mohan P. S., Peterhoff C. M., Wolfe D. M., Martinez-Vicente M., Massey A. C., Sovak G., Uchiyama Y., Westaway D., Cuervo A. M., Nixon R. A. (2010) Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 141, 1146–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kazmi F., Hensley T., Pope C., Funk R. S., Loewen G. J., Buckley D. B., Parkinson A. (2013) Lysosomal sequestration (trapping) of lipophilic amine (cationic amphiphilic) drugs in immortalized human hepatocytes (Fa2N-4 cells). Drug Metab. Dispos. 41, 897–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheung Z. H., Ip N. Y. (2011) Autophagy deregulation in neurodegenerative diseases-recent advances and future perspectives. J. Neurochem. 118, 317–325 [DOI] [PubMed] [Google Scholar]
- 35. Klionsky D. J., Elazar Z., Seglen P. O., Rubinsztein D. C. (2008) Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy. 4, 849–950 [DOI] [PubMed] [Google Scholar]
- 36. Dice J. F. (1990) Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem. Sci. 15, 305–309 [DOI] [PubMed] [Google Scholar]
- 37. Qi L., Zhang X. D., Wu J. C., Lin F., Wang J., DiFiglia M., Qin Z. H. (2012) The role of chaperone-mediated autophagy in huntingtin degradation. PLoS One 7, e46834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wooten M. W., Hu X., Babu J. R., Seibenhener M. L., Geetha T., Paine M. G., Wooten M. C. (2006) Signaling, polyubiquitination, trafficking, and inclusions: sequestosome 1/p62's role in neurodegenerative disease. J. Biomed. Biotechnol. 2006, 62079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jana N. R. (2010) Role of the ubiquitin-proteasome system and autophagy in polyglutamine neurodegenerative diseases. Future Neurol. 5, 105–112 [Google Scholar]
- 40. Wang J., Wang C. E., Orr A., Tydlacka S., Li S. H., Li X. J. (2008) Impaired ubiquitin-proteasome system activity in the synapses of Huntington's disease mice. J. Cell Biol. 180, 1177–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tydlacka S., Wang C. E., Wang X., Li S., Li X. J. (2008) Differential activities of the ubiquitin-proteasome system in neurons versus glia may account for the preferential accumulation of misfolded proteins in neurons. J. Neurosci. 28, 13285–13295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hunter J. M., Lesort M., Johnson G. V. (2007) Ubiquitin-proteasome system alterations in a striatal cell model of Huntington's disease. J. Neurosci. Res. 85, 1774–1788 [DOI] [PubMed] [Google Scholar]
- 43. Newton K., Matsumoto M. L., Wertz I. E., Kirkpatrick D. S., Lill J. R., Tan J., Dugger D., Gordon N., Sidhu S. S., Fellouse F. A., Komuves L., French D. M., Ferrando R. E., Lam C., Compaan D., Yu C., Bosanac I., Hymowitz S. G., Kelley R. F., Dixit V. M. (2008) Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134, 668–678 [DOI] [PubMed] [Google Scholar]
- 44. Li G., Pomès R. (2013) Binding mechanism of inositol stereoisomers to monomers and aggregates of Aβ(16–22). J. Phys. Chem. B. 117, 6603–6613 [DOI] [PubMed] [Google Scholar]
- 45. Li G., Rauscher S., Baud S., Pomès R. (2012) Binding of inositol stereoisomers to model amyloidogenic peptides. J. Phys. Chem. B. 116, 1111–1119 [DOI] [PubMed] [Google Scholar]
- 46. Török B., Bag S., Sarkar M., Dasgupta S., Török M. (2013) Structural features of small molecule amyloid-β self-assembly inhibitors. Curr. Bioact. Compd. 9, 37–63 [Google Scholar]
- 47. Ehrnhoefer D. E., Duennwald M., Markovic P., Wacker J. L., Engemann S., Roark M., Legleiter J., Marsh J. L., Thompson L. M., Lindquist S., Muchowski P. J., Wanker E. E. (2006) Green tea (−)-epigallocatechin-gallate modulates early events in huntingtin misfolding and reduces toxicity in Huntington's disease models. Hum. Mol. Genet. 15, 2743–2751 [DOI] [PubMed] [Google Scholar]
- 48. Jana N. R., Zemskov E. A., Wang Gh, Nukina N. (2001) Altered proteasomal function due to the expression of polyglutamine-expanded truncated N-terminal huntingtin induces apoptosis by caspase activation through mitochondrial cytochrome c release. Hum. Mol. Genet. 10, 1049–1059 [DOI] [PubMed] [Google Scholar]
- 49. Bennett E. J., Bence N. F., Jayakumar R., Kopito R. R. (2005) Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol. Cell 17, 351–365 [DOI] [PubMed] [Google Scholar]
- 50. Mitra S., Tsvetkov A. S., Finkbeiner S. (2009) Single neuron ubiquitin-proteasome dynamics accompanying inclusion body formation in huntington disease. J. Biol. Chem. 284, 4398–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ortega Z., Díaz-Hernández M., Maynard C. J., Hernández F., Dantuma N. P., Lucas J. J. (2010) Acute polyglutamine expression in inducible mouse model unravels ubiquitin/proteasome system impairment and permanent recovery attributable to aggregate formation. J. Neurosci. 30, 3675–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamamoto A., Lucas J. J., Hen R. (2000) Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell 101, 57–66 [DOI] [PubMed] [Google Scholar]
- 53. Kordasiewicz H. B., Stanek L. M., Wancewicz E. V., Mazur C., McAlonis M. M., Pytel K. A., Artates J. W., Weiss A., Cheng S. H., Shihabuddin L. S., Hung G., Bennett C. F., Cleveland D. W. (2012) Sustained therapeutic reversal of Huntington's disease by transient repression of huntingtin synthesis. Neuron. 74, 1031–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Townsend M., Cleary J. P., Mehta T., Hofmeister J., Lesne S., O'Hare E., Walsh D. M., Selkoe D. J. (2006) Orally available compound prevents deficits in memory caused by the Alzheimer amyloid-β oligomers. Ann. Neurol. 60, 668–676 [DOI] [PubMed] [Google Scholar]
- 55. McLaurin J., Kierstead M. E., Brown M. E., Hawkes C. A., Lambermon M. H., Phinney A. L., Darabie A. A., Cousins J. E., French J. E., Lan M. F., Chen F., Wong S. S., Mount H. T., Fraser P. E., Westaway D., St George-Hyslop P. (2006) Cyclohexanehexol inhibitors of Aβ aggregation prevent and reverse Alzheimer phenotype in a mouse model. Nat. Med. 12, 801–808 [DOI] [PubMed] [Google Scholar]
- 56. Fenili D., Brown M., Rappaport R., McLaurin J. (2007) Properties of scyllo-inositol as a therapeutic treatment of AD-like pathology. J. Mol. Med. 85, 603–611 [DOI] [PubMed] [Google Scholar]
- 57. Aytan N., Choi J. K., Carreras I., Kowall N. W., Jenkins B. G., Dedeoglu A. (2013) Combination therapy in a transgenic model of Alzheimer's disease. Exp. Neurol. 250, 228–238 [DOI] [PMC free article] [PubMed] [Google Scholar]