Background: Glyceraldehyde-3-phosphate dehydrogenase is a pivotal glycolytic enzyme and implicated in many human cancers.
Results: GAPDH acetylation promotes its activity in response to glucose via the action of PCAF and HDAC5.
Conclusion: GAPDH acetylation promotes cell proliferation and tumor growth.
Significance: This study demonstrates a critical role of GAPDH acetylation in tumor growth and provides potential therapy target for cancer.
Keywords: Cell Growth, Glycolysis, Histone Deacetylase, Lung Cancer, Metabolism, Acetylation, GAPDH, HDAC5, PCAF, Tumorigenesis
Abstract
The altered metabolism in most tumor cells consists of elevated glucose uptake and increased glycolysis even in the presence of high oxygen tension. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is an obligatory enzyme in glycolysis. Here, we report that acetylation at lysine 254 (K254) increases GAPDH activity in response to glucose. Furthermore, acetylation of GAPDH (K254) is reversibly regulated by the acetyltransferase PCAF and the deacetylase HDAC5. Substitution of K254 to glutamine compromises the ability of GAPDH to support cell proliferation and tumor growth. Our study reveals a mechanism of GAPDH enzyme activity regulation by acetylation and its critical role in cellular regulation.
Introduction
Most cancer cells rely on aerobic glycolysis, instead of mitochondrial oxidative phosphorylation, to generate the energy and metabolic intermediates needed for cellular processes, resulting in increased glucose uptake and consumption. This phenomenon, which is coined as “the Warburg effect” (1), provides the basis to image tumors in clinics using 2-(18F)-fluoro-2-deoxy-d-glucose (18F-FDG)2 by positron emission tomography (PET). 18F-FDG-PET combined with computer tomography (CT) is widely used for detecting most epithelial tumors (2, 3). Significantly elevated levels of glycolytic enzymes were observed in most human cancers (4).
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is an enzyme in glycolysis. GAPDH specifically catalyzes the simultaneous phosphorylation and oxidation of glyceraldehyde 3-phosphate using nicotinamide adenine dinucleotide (NAD+) as a cofactor to produce glycerate 1,3-biphosphate and NADH.
GAPDH is located mainly in the cytoplasm and primarily exists as a tetrameric structure composed of four identical 37 kDa subunits. GAPDH was once considered as a simple “housekeeping” protein and frequently used as a control in protein and gene studies, a number of studies however showed that the GAPDH gene and protein are actively regulated in cell proliferation (5, 6). In addition to its role in energy production, GAPDH has been implicated in many cellular processes including DNA repair (7), tRNA export (8), membrane fusion and transport (9), endocytosis and nuclear membrane assembly (10), and cell death (11, 12). Recently elegant studies also found GAPDH is involved in immunity (13) and senescence (14). The diverse functions of GAPDH are mainly regulated by its oligomerization, post-translational modification and sub-cellular localization.
Protein acetylation is a reversible post-translational modification, which has been recently identified as being broadly involved in the regulation of many cellular processes. In addition, several acetylation proteomics studies reported more than 2000 potential acetylation nonnuclear substrates (15–17). Most enzymes in glycolysis, including GAPDH, are acetylated in liver. In this study we investigated the GAPDH acetylation and its functional role in cellular proliferation and tumor growth.
EXPERIMENTAL PROCEDURES
Deacetylase Inhibitor Treatment
Deacetylase inhibitor treatments were carried out by adding TSA (0.5 mm) and NAM (5 mm) to the culture medium 18 and 6 h before harvesting, respectively; both concentrations are final concentrations in the culture medium.
Immunoprecipitation (IP) and Western Blotting Analysis
Protein lysate were prepared from HEK 293T, A549, and stable pool cells in a buffer containing 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.1% Nonidet P-40, and a mixture of protease inhibitors (Roche Applied Science). For IP experiments, 500 μg of cell lysate were incubated with anti-Flag M2-agarose for 3 h at 4 °C. Beads were washed three times with lysis buffer and centrifuged at 2,000 × g for 2 min between each wash. The Flag-tagged proteins were eluted by Flag peptides (Gilson Biochemical). Lysate was resolved on 8–10% SDS-PAGE and transferred onto nitrocellulose membrane (Bio-Rad) for Western blotting analysis. Antibodies to FLAG (catalogue no. A00170 from GenScript or catalogue no. A2220 from Sigma), GAPDH (used for IP, 5632-1, Epitomics; 1:100), GAPDH (6C5) (SC32233, Santa Cruz Biotechnology), HA (F7) (SC7392, Santa Cruz Biotechnology), and β-actin (13E5) (no. 4970, Cell Signaling) were purchased commercially.
Measurement of GAPDH Activity
The GAPDH assay was carried out as G9263-Enzyme Assay and followed the manufacturer's instruction (Sigma Aldrich).
In Vitro Deacetylation Assay
CobB was expressed in Escherichia coli, purified with nickel beads, and stored at −80 °C in 10% glycerol. For in vitro deacetylation assay, 10 μg CobB and GAPDH was added into the buffer (40 mm HEPES (pH 7.0), 6 mm MgCl2, 1 mm NAD+, and 1 mm DTT) at 37 °C for 1 h.
Cell Proliferation Analysis
4 × 104 cells were seeded in triplicate in 6-well plates, and cell numbers were counted every 24 h over a 5-day period using Cellometer Auto T4 and followed the manufacturer's instruction (Nexcelom Bioscience).
Cell Counting Kit-8 (CCK-8) Assay
2000 cells were seeded in triplicate in 96-well plates, and add 10 μl of CCK-8 solution to each well of the plate and incubate the cells for 2 h. Measure the absorbance at 450 nm using a microplate reader (Elx 800, Biotek instruments).
Xenograft Studies
BALB/C-Nude mice (nu/nu, male, 4-weeks old) were injected subcutaneously with 5 × 106 A549 cells. Around 7 weeks after injection, the tumors were dissected and weighed.
RESULTS
GAPDH Is Acetylated at Lysine 254
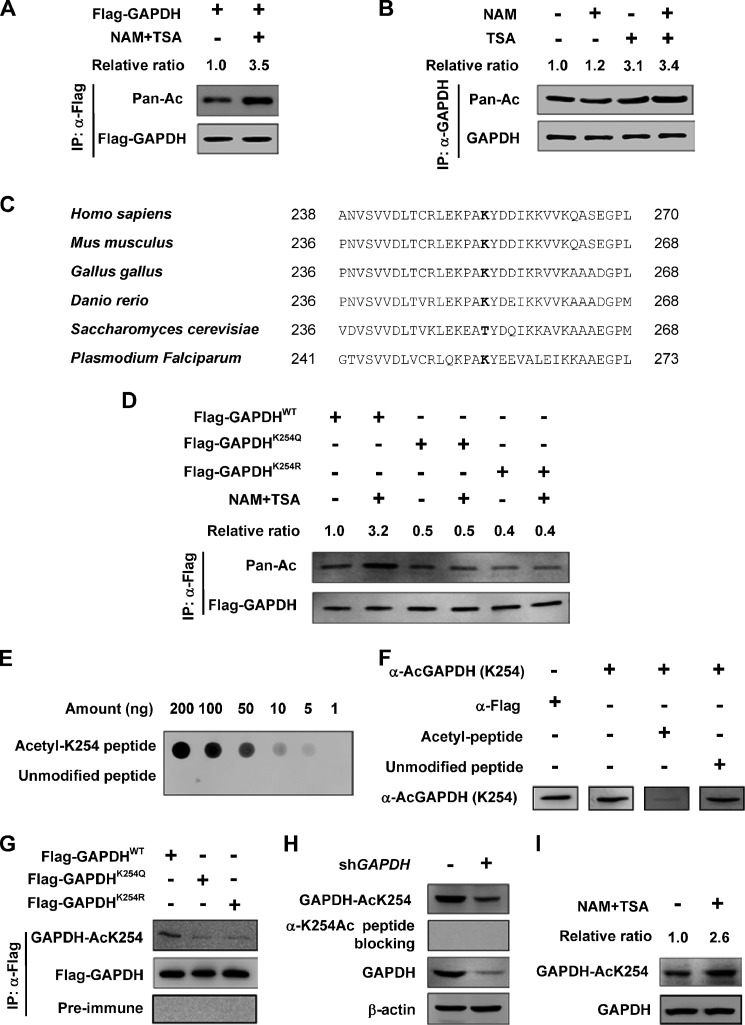
Several acetylation proteomic studies identified GAPDH as a putative substrate of acetylation. To confirm this observation, we transfected Flag-GAPDH into HEK 293T cells and analyzed GAPDH acetylation by the pan anti-acetyl lysine antibody and found that GAPDH was indeed acetylated. Furthermore, Flag-GAPDH acetylation level was enhanced by over 3-fold after treatment with trichostatin A (TSA), an inhibitor of histone deacetylase (HDAC) class I and class II, and Nicotinamide (NAM), an inhibitor of the SIRT family deacetylase (Fig. 1A). Moreover, endogenous GAPDH was purified, and GAPDH acetylation was analyzed. We found that the acetylation level increased more dramatically by TSA (3.1-fold) than by NAM (1.2-fold) (Fig. 1B). These results indicate that both exogenously and endogenously expressed GAPDH are acetylated.
FIGURE 1.
GAPDH is acetylated at lysine 254. A, exogenous GAPDH is acetylated. Flag-GAPDH was expressed in HEK 293T cells followed by treatment with or without deacetylase inhibitors NAM and TSA. The acetylation and protein levels were analyzed by Western blot. Relative ratios of GAPDH acetylation were calculated from normalizing against Flag-GAPDH. B, endogenous GAPDH is acetylated. Endogenous GAPDH protein was purified from HEK 293T cells after NAM and TSA treatment as indicated. The acetylation and protein levels were analyzed by Western blot. Relative ratios of GAPDH acetylation were calculated from normalizing against GAPDH. C, K254 site in GAPDH is conserved. The sequences around K254 from different species were aligned. Conserved lysine residues corresponding to human GAPDH K254 are bolded. D, inhibition of deacetylase increases the acetylation level of wild-type (WT), but not the mutants of GAPDH. GAPDH was expressed in HEK 293T cells and affinity purified, followed by Western blot. Relative ratios of GAPDH acetylation were calculated from normalizing against Flag-GAPDH. E, specificity of the antibody was determined by dot blot. Nitrocellulose membrane was spotted with different amounts of either acetyl-K254 peptide or unmodified peptide and probed with anti-AcGAPDH (K254) antibody. F, K254-acetylated GAPDH peptide can absorb the acetyl-K254 antibody. The anti-AcGAPDH (K254) antibody was incubated with acetylated peptide or unmodified peptide for 1 h and used for Western blotting as indicated. G, GAPDH was expressed in HEK 293T cells and affinity-purified protein was measured by Western blot using anti-AcGAPDH (K254) antibody or pre-immune serum. H, cell lysate from scramble or GAPDH shRNA knockdown stable cells were probed. I, endogenous GAPDH is acetylated at K254. HEK 293T cells were treated with NAM and TSA. The protein levels and acetylation of K254 were determined by Western blot. Relative ratios of GAPDH K254 acetylation were calculated from normalizing against GAPDH.
Acetylation of K254 in peptide RLEKPAKAcYDDIKK was hit five times with a high score and matched the human GAPDH sequence with acetylation at K254 by two different proteomic studies (15, 17). Notably, K254 in GAPDH is conserved from Danio rerio to Homo sapiens (Fig. 1C). To identify whether K254 is indeed acetylated, we mutated lysine 254 individually to glutamine (Q) or arginine (R) and examined their acetylation by using the pan anti-acetyl lysine antibody and found that mutations of K254 resulted in a significant reduction in GAPDH acetylation (Fig. 1D). To further confirm the acetylation of K254, we transfected GAPDH wild-type (WT) and mutants into HEK 293T cells and treated the cells with NAM and TSA or not. Then we measured GAPDH acetylation and found the inhibition of deacetylases increased the acetylation level of GAPDH WT, but not the mutants of K254Q and K254R (Fig. 1D), indicating that under this condition, K254 is the major acetylation site of GAPDH.
To determine if K254 is acetylated in vivo, we generated a site-specific antibody against an acetylated K254 GAPDH peptide (VVDLTCRLEKPAKAcYDDIKKVVKQAS). The specificity of the anti-AcGAPDH (K254) antibody was first confirmed by its ability to recognize the K254 acetylated, but not the unmodified peptide (Fig. 1E). The peptide blocking experiment showed K254-acetylated GAPDH absorbed anti-AcGAPDH (K254) antibody (Fig. 1F). This antibody strongly recognized the ectopically expressed WT GAPDH, but only weakly recognized the K254 mutated GAPDH (Fig. 1G), confirming the specificity of the antibody. Next, we used the anti-AcGAPDH (K254) antibody to detect endogenous GAPDH acetylation. We identified a band of expected molecular weight for GAPDH in Chang's liver cell lysate, which was diminished when the antibody was pre-incubated with competing antigen peptide or when the endogenous GAPDH was knocked down (Fig. 1H). Lysate of HEK 293T cells treated with or without NAM and TSA were analyzed by Western blotting using the anti-AcGAPDH (K254) antibody. The acetylation level of K254 was increased by nearly 3-fold after the inhibition of deacetylase (Fig. 1I). Together, these characterizations demonstrated GAPDH is acetylated at K254.
Inhibition of Deacetylase Increases GAPDH Activity
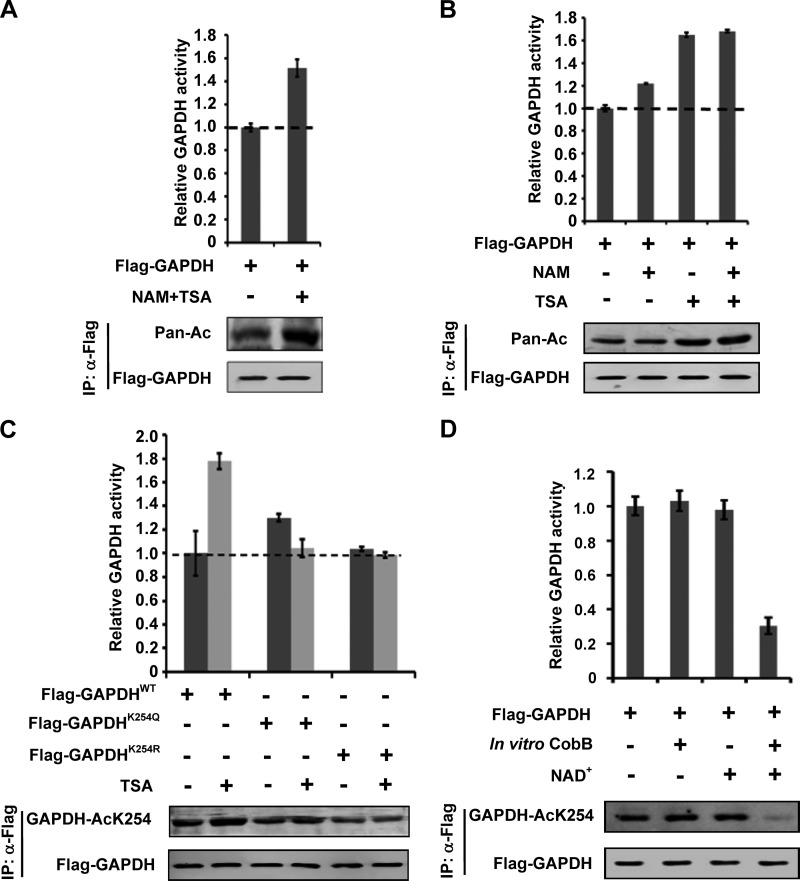
Acetylation has several different mechanisms in regulating metabolic enzymes (18). To determine the effect of K254 acetylation on GAPDH, we measured GAPDH enzyme activity. Flag-GAPDH was transfected into HEK 293T cells and the transfected cells were treated with or without TSA and NAM. Flag-GAPDH was immunoprecipitated, and enzyme activity was measured in vitro. We observed that the GAPDH activity was increased over 40% in the TSA and NAM treated cells (Fig. 2A). As expected, NAM and TSA treatment also increased the acetylation level of Flag-GAPDH. Compared with TSA, NAM had a much weaker effect (Fig. 2B). The above results suggest that a TSA-sensitive member of class I and/or II HDAC regulates GAPDH acetylation and activity.
FIGURE 2.
Acetylation at K254 increases GAPDH enzyme activity. A, acetylation increases GAPDH enzyme activity. Flag-GAPDH was expressed in HEK 293T cells followed by treatment with NAM and TSA, and GAPDH acetylation and protein levels were analyzed by Western blot. The enzyme activity was measured and normalized against protein level. Mean values of relative enzyme activity of triplicate experiments ±S.D. are presented. B, TSA increases GAPDH enzyme activity more dramatically than NAM. Flag-GAPDH was expressed in HEK 293T cells followed by treatment with NAM or TSA, acetylation levels and enzyme activity of GAPDH were determined as A. C, inhibition of deacetylase increases the enzyme activity of WT, but not mutants of GAPDH. The indicated plasmids were expressed in HEK 293T cells with or without TSA treatment and purified by IP, acetylation levels and enzyme activity of GAPDH were determined as A. D, CobB decreases GAPDH acetylation level and enzyme activity. GAPDH was expressed in HEK 293T cells, affinity purified and incubated with or without recombinant CobB at the presence of NAD+ or not. Different samples were analyzed for acetylation levels and enzyme activities as A.
Acetylation Increases GAPDH Enzyme Activity
To determine the effect of K254 acetylation, we measured the activity of WT and K254 mutants GAPDH in the presence or absence of TSA. The K254Q mutant displayed activity slightly higher than the WT GAPDH whereas the K254R mutant has activity indistinguishable from that of the WT enzyme (Fig. 2C). In contrast to the WT GAPDH that was strongly activated by TSA, a similar TSA treatment had little effect on the activity of either GAPDHK254Q or GAPDHK254R mutants (Fig. 2C). This result suggests that acetylation of K254 is responsible for GAPDH activation, although K254Q mutant did not mimic the effect of acetylation. To further determine the function of acetylation in GAPDH activity regulation, we incubated the purified Flag-GAPDH with the bacterial deacetylase CobB. NAD+ is a co-factor required for CobB-catalyzed deacetylation. As expected, CobB could not deacetylate nor inhibit GAPDH activity in the absence of NAD+. GAPDH activity decreased by as much as 65% by CobB, this reduction is associated with almost complete loss of K254 acetylation (Fig. 2D). Collectively, these results provide convincing in vitro biochemical evidence that demonstrating K254 acetylation increases GAPDH catalytic activity.
PCAF Acetylates GAPDH at K254
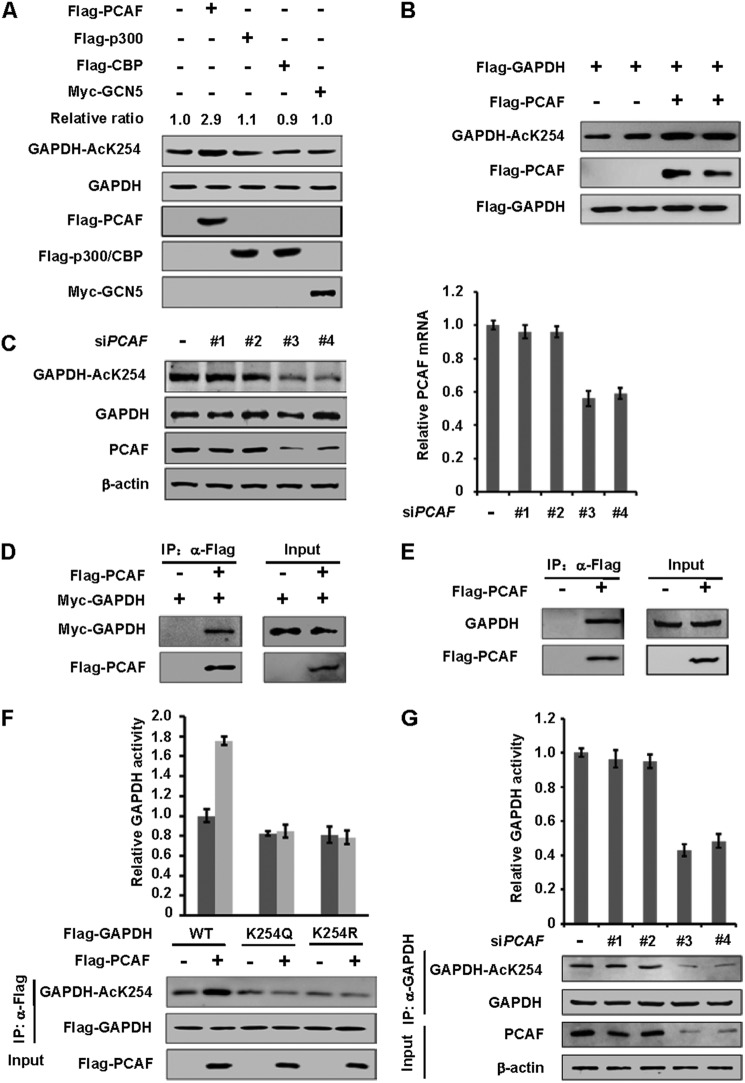
To determine the acetyltransferase responsible for GAPDH K254 acetylation, we transfected four acetyltransferases, p300 (E1A-binding protein 300kD), CBP (cAMP response element-binding protein (CREB)-binding protein), PCAF (p300/(CREB-binding protein)-associated factor), and GCN5 (KAT2A), individually into HEK 293T cells and observed GAPDH K254 acetylation. We found that only PCAF transfection increased endogenous GAPDH acetylation at K254 by nearly 3-fold, while the other three acetyltransferases had little effect (Fig. 3A). Co-expression experiments also showed the enhancement of K254 acetylation of exogenously expressed GAPDH by PCAF (Fig. 3B). To complement the transfection data, PCAF was knocked-down by small interfering RNA (siRNA). The knock down efficiency was measured by qPCR and Western blotting analysis. Knocking down PCAF significantly reduced K254 acetylation level (Fig. 3C). These data indicate that PCAF is a potential K254 acetyltransferase in vivo. To determine if PCAF can interact with GAPDH, we co-transfected Flag-PCAF and Myc-GAPDH into HEK 293T cells, and proteins were purified by anti-Flag beads. We found that PCAF interacted with GAPDH (Fig. 3D). Furthermore, the endogenous GAPDH could be readily detected in a PCAF immune-complex derived from HEK 293T cells (Fig. 3E), demonstrating a physical association between GAPDH and PCAF in vivo. We also found that PCAF co-expression increased the enzyme activity of GAPDH WT, but had no significant effect on the activity of GAPDH mutants (Fig. 3F), indicating that PCAF stimulates GAPDH activity mostly via acetylation of K254. Furthermore, knocking down PCAF significantly reduced GAPDH activity by more than 50% (Fig. 3G). Taken together, the above data support a model that PCAF may be one of the acetyltransferases of GAPDH and PCAF acetylates GAPDH at K254.
FIGURE 3.
PCAF is one of the acetyltransferases of GAPDH. A, overexpression of PCAF, but not other acetyltransferases, increases endogenous GAPDH K254 acetylation level. Indicated plasmids were expressed in HEK 293T cells. Endogenous acetylation levels of GAPDH were determined by anti-AcGAPDH (K254) antibody. Relative ratios of GAPDH K254 acetylation were calculated from normalizing against GAPDH. B, overexpression of PCAF increases GAPDH K254 acetylation levels. The indicated plasmids were expressed in HEK 293T cells, and the acetylation and protein levels were analyzed by Western blot. C, PCAF knockdown decreases endogenous GAPDH acetylation at K254. A549 cells were transfected with siPCAF (four different oligos) or negative control (siNC) as previously described, and the acetylation and protein levels were analyzed by Western blot (left panel). PCAF transcription level was measured by qPCR (right panel). D, exogenous GAPDH interacts with PCAF. The indicated plasmids were expressed in HEK 293T cells and proteins were affinity purified. GAPDH and PCAF protein levels were determined against indicated antibodies, respectively. E, endogenous GAPDH interacts with PCAF. Flag-PCAF was expressed in HEK 293T cells and proteins were affinity purified. GAPDH and PCAF protein levels were determined against indicated antibodies, respectively. F, PCAF increases the enzyme activity of WT, but not mutants of GAPDH. The indicated plasmids were expressed in HEK 293T cells and affinity purified. Different samples were analyzed by Western blot and enzyme activities were measured as described in Fig. 2A. Mean values of relative enzyme activity of triplicate experiments ±S.D. are presented. G, PCAF knockdown decreases endogenous GAPDH activity. HEK 293T cells were transfected with siPCAF or negative control (siNC) as previously described and endogenous GAPDH were purified. Different samples were analyzed by Western blot and enzyme activities were measured as described in Fig. 2A. Mean values of relative enzyme activity of triplicate experiments ±S.D. are presented.
HDAC5 Deacetylates GAPDH at K254
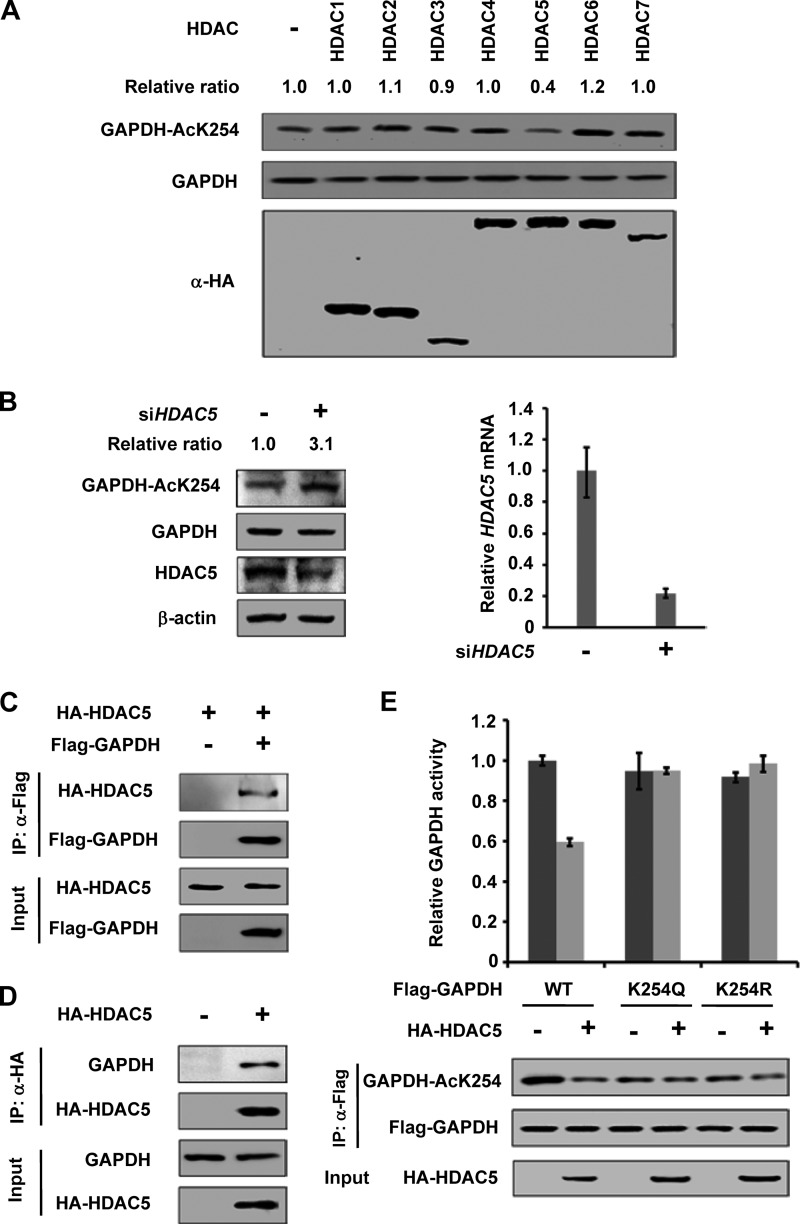
Because TSA, but not NAM, significantly increased GAPDH acetylation, we speculated that a HDAC deacetylase is involved in GAPDH deacetylation. To identify the specific HDAC, we co-expressed GAPDH with seven HDAC deacetylases, HDAC 1–7 individually. We found that HDAC5, but not the other six HDACs, decreased endogenous GAPDH K254 acetylation by 60% (Fig. 4A). Knocking down of HDAC5 by siRNA significantly increased K254 acetylation level (by over 3-fold) (Fig. 4B). This data indicates that HDAC5 is a potential GAPDH K254 deacetylase in vivo. We then performed Co-IP experiments and found that HDAC5 readily not only co-precipitated with the ectopically expressed GAPDH (Fig. 4C) but also with the endogenous GAPDH (Fig. 4D), further supporting a role of HDAC5 in GAPDH deacetylation.
FIGURE 4.
HDAC5 is the deacetylase of GAPDH. A, HDAC5 deacetylates K254 of GAPDH. HA-tagged HDACs were expressed in HEK 293T cells, and endogenous GAPDH proteins were purified. The acetylation and protein levels were analyzed by Western blot. Relative ratios of GAPDH K254 acetylation were calculated from normalizing against GAPDH. B, knockdown of HDAC5 increases K254 acetylation level. HEK 293T cells were transfected with siHDAC5 or siNC as previously described, and the acetylation and protein levels were analyzed by Western blot (left panel). HDAC5 transcription level was measured by qPCR (right panel). C, exogenous GAPDH interacts with HDAC5. The indicated plasmids were expressed in HEK 293T cells, and proteins were affinity purified. GAPDH and HDAC5 protein levels were determined against indicated antibodies, respectively. D, endogenous GAPDH interacts with HDAC5. HA-tagged HDAC5 was expressed in HEK 293T cells and proteins were affinity purified. GAPDH and HDAC5 protein levels were determined against indicated antibodies, respectively. E, HDAC5 decreases the enzyme activity of WT, but not mutants of GAPDH. The indicated plasmids were expressed in HEK 293T cells and purified by IP. Different samples were analyzed by Western blot with indicated antibody, respectively. Enzyme activities were measured as described in Fig. 2A. Mean values of relative enzyme activity of triplicate experiments ±S.D. are presented.
To determine the functional significance of HDAC5 in GAPDH regulation, we measured the enzyme activity of GAPDH WT and mutants co-expressed with HDAC5 or not. We observed that co-expression of HDAC5 decreased the activity of GAPDHWT but had no significant effect on the activity of GAPDHK254Q or GAPDHK254R mutants (Fig. 4E). Collectively, these data establish a specific and prominent role of HDAC5 in GAPDH K254 deacetylation and enzymatic inactivation.
Glucose Activates GAPDH by Stimulating K254 Acetylation
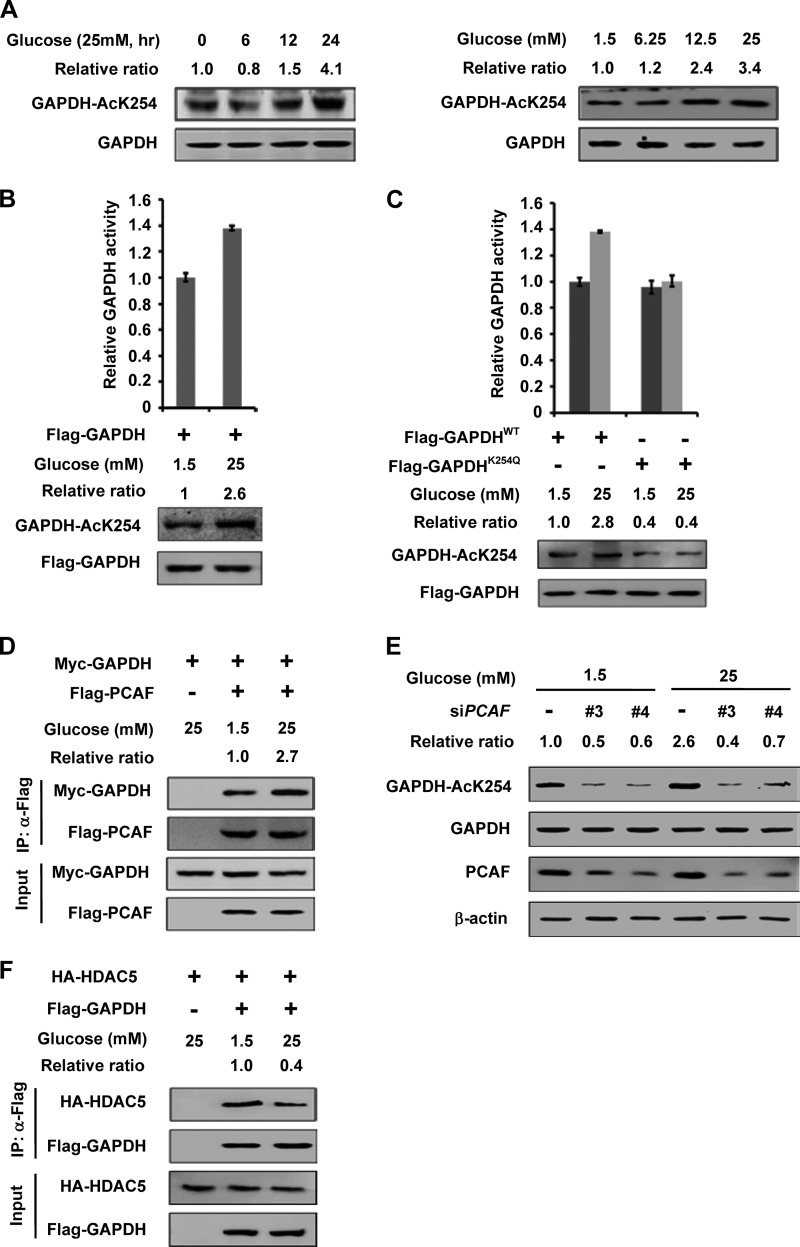
GAPDH catalyzes an obligatory step in glycolysis. To determine if K254 acetylation of GAPDH is dynamically regulated in vivo, we tested whether glucose affects GAPDH acetylation. HEK 293T cells were cultured in medium with different glucose conditions. We found that high glucose significantly increased the acetylation levels of GAPDH K254 by more than 3-fold (Fig. 5A, right panel). The acetylation of GAPDH K254 not only increased with the increasing glucose concentration but also with the extension of exposure time (Fig. 5A, left panel). To determine the effect of glucose on GAPDH activity, HEK 293T cells were transfected with Flag-GAPDH and maintained in 1.5 mm or 25 mm glucose. We found that the cells maintained in high (25 mm) glucose media displayed GAPDH activity 40% higher than cells maintained in low (1.5 mm) glucose media (Fig. 5B). Notably, high glucose had no significant effect on the acetylation level and enzyme activity of GAPDHK254Q (Fig. 5C). We conclude that glucose stimulates GAPDH activity mainly via increasing K254 acetylation.
FIGURE 5.
Glucose increases the GAPDH K254 acetylation and enzyme activity. A, high glucose increases GAPDH acetylation at K254 in time and dose-dependent manner. HEK 293T cells were cultured with 25 mm glucose medium for various time lengths as indicated (left panel), or with different concentrations of glucose for 6 h (right panel) and endogenous GAPDH protein and acetylation levels were determined against indicated antibodies, respectively. Relative ratios of GAPDH K254 acetylation were calculated from normalizing against GAPDH. B, high glucose increases the activity of GAPDH. Flag-GAPDH was expressed in HEK 293T cells and maintained under indicated glucose concentrations. Different samples were analyzed for acetylation levels and enzyme activities were measured as described in Fig. 2A. Relative ratios of acetylation were calculated from normalizing against Flag-GAPDH. Mean values of relative enzyme activity of triplicate experiments ±S.D. are presented. C, high glucose increases the activity of GAPDHWT not the GAPDHK254Q. The indicated plasmids were expressed in HEK 293T cells and the cells were maintained under indicated glucose concentrations. Different samples were analyzed for acetylation levels and enzyme activities as described in Fig. 2A. Relative ratios of acetylation were calculated from normalizing against Flag-GAPDH. Mean values of relative enzyme activity of triplicate experiments ±S.D. are presented. D, GAPDH-PCAF interaction is enhanced under high glucose. Myc-GAPDH and Flag-PCAF were co-expressed in HEK 293T cells. The interaction between GAPDH and PCAF was determined by IP-Western. The Co-IP efficiency of Myc-GAPDH was calculated from normalizing against Flag-PCAF. E, PCAF knockdown blocks the increase of GAPDH acetylation level induced by high glucose. HEK 293T cells were maintained in 1.5 mm or 25 mm glucose for 6 h after transfected with siPCAF (two different oligos) or siNC as previously described. The proteins and acetylation levels were analyzed by Western blot. Relative ratios of acetylation were calculated from normalizing against GAPDH. F, GAPDH-HDAC5 interaction is decreased under high glucose. Flag-GAPDH and HA-HDAC5 were co-expressed in HEK 293T cells. The interaction between GAPDH and HDAC5 was determined by IP-Western. The Co-IP efficiency of HA-HDAC5 was calculated from normalizing against Flag-GAPDH.
We next examined the effect of glucose on the PCAF-GAPDH interaction. Co-IP experiment showed that the interaction between PCAF and GAPDH was enhanced by nearly 3-fold under high glucose than under low glucose condition (Fig. 5D). Notably, knocking down of PCAF blocked the increase of GAPDH acetylation by glucose (Fig. 5E), suggesting an important role of PCAF in mediating high glucose-stimulated K254 acetylation in GAPDH. Furthermore, high glucose decreased the interaction between HDAC5 and GAPDH by 60% (Fig. 5F). These data indicate that glucose stimulates GAPDH K254 acetylation and activity mainly by increasing the GAPDH-PCAF interaction and decreasing the GAPDH-HDAC5 interaction.
K254Q Inhibits the Function of GAPDH in Supporting Cell Proliferation
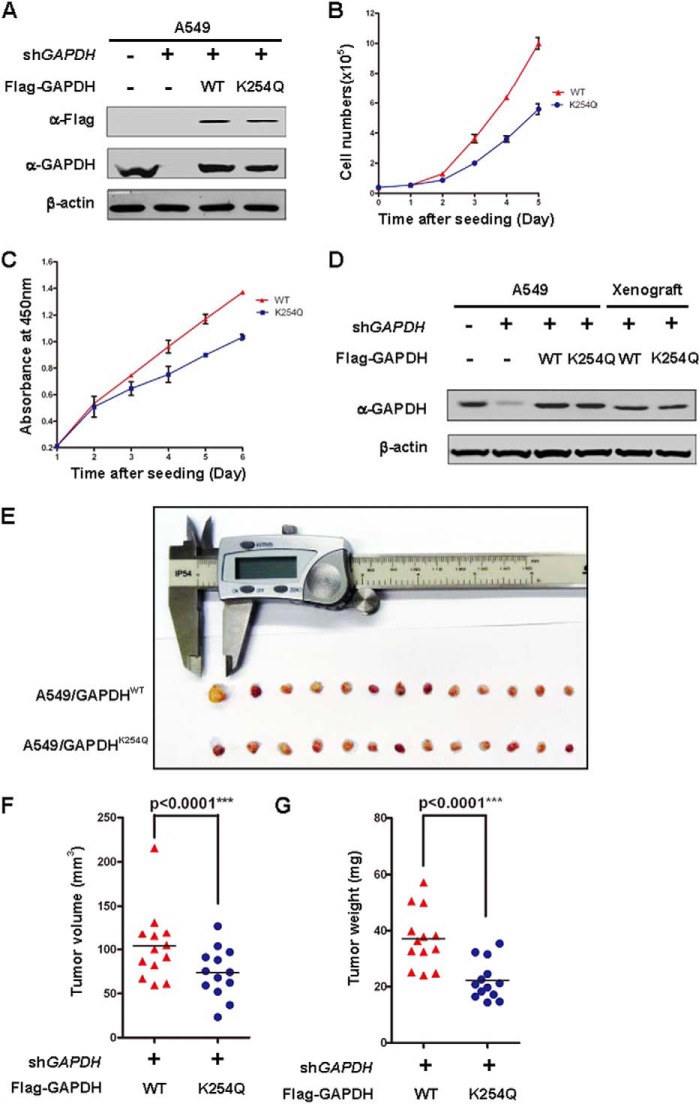
Elevated GAPDH protein levels are reported in different types of cancer cells, especially in lung cancer (4, 19). As a required enzyme in glycolysis, GAPDH is essential for cell growth in vitro and in vivo. We investigated the effect of GAPDH acetylation on cell proliferation and tumor growth. We generated GAPDH knock down and put back stable cell lines in the lung cancer cell line A549. First, the endogenous GAPDH was knocked down by shRNA and then the shRNA-resistant GAPDHWT and GAPDHK254Q were stably expressed in the knock down cells. Western blotting analysis demonstrated that the endogenous GAPDH was effectively knocked down. GAPDHWT and GAPDHK254Q were expressed at a level similar to the endogenous GAPDH (Fig. 6A). We observed that A549/GAPDHWT cells proliferated faster than A549/GAPDHK254Q cells (Fig. 6B), indicating a growth reduction conferred by the mutant at K254. The CCK8 assay also showed that A549/GAPDHWT cells had a prominent growth advantage over the A549/GAPDHK254Q cells (Fig. 6C).
FIGURE 6.
Acetylation of GAPDH at K254 promotes tumor cell proliferation. A, verification of GAPDH expressing A549 stable cell lines. A549 cells stably expressed GAPDHWT and GAPDHK254Q were transfected with shRNA-resistant WT or K254Q of GAPDH thus the knockdown and put back stable cells were generated. GAPDH knockdown efficiency and re-expression level were determined by Western blot. B, GAPDHK254Q is compromised to support cell proliferation. GAPDHWT and GAPDHK254Q cells were seeded as the same number in each well. Cell numbers were counted every 24 h. Error bars represent cell numbers ±S.D. for experiments performed in triplicate. C, absorbance at 450 nm was measured after adding CCK-8 solution to each well of the plate and incubating for 2 h. Error bars represent cell numbers ±S.D. for experiments performed in triplicate. D, expression of GAPDHWT and GAPDHK254Q in xenograft. Whole cell lysate were prepared from either original stable A549/GAPDHWT and A549/GAPDHK254Q pools or xenograft tumors, followed by Western blot. E—G, GAPDHK254Q is defective in supporting tumor growth in vivo. Xenograft was performed using the A549 cell lines characterized in A. 7 weeks after injection, mice were sacrificed and tumors were extracted and photographed (E). Tumor diameters were measured, and the volumes were calculated (F). Tumor weight was measured (G). The p value was calculated by paired t test.
To determine whether the K254Q expressing cells have a growth disadvantage in vivo, we performed a xenograft study using the A549 cell lines generated above. Five million A549/GAPDHWT and A549/GAPDHK254Q cells were injected into nude mice and tumor cell growth was monitored over a period of 7 weeks. Sustained expression of Flag-tagged GAPDH WT and K254Q mutant in xenograft tumors was verified by Western blot (Fig. 6D). We found that tumors developed in mice injected with A549/GAPDHWT cells were significantly (p < 0.0001) larger than those injected with A549/GAPDHK254Q cells (Fig. 6, E and F). The difference in tumor volume of cells expressing wild-type or K254Q mutant GAPDH remained statistically significant (p < 0.0001) even when the largest tumor sample of the wild type expressing cells was excluded. A549/GAPDHK254Q cells also displayed tumor weight significantly (p < 0.0001) lighter than the A549/GAPDHWT cells (Fig. 6G). Taken together, our results suggest a positive role of GAPDH K254 acetylation in modulating cell growth.
DISCUSSION
The GAPDH gene encodes a single mRNA leading to the production of a single protein without any alternative splicing. However, accumulating evidence reveals its diverse functions in various cellular processes (7–14). In this report, we identified a novel acetylation site (K254) in GAPDH. More interestingly, K254 acetylation is stimulated by high glucose, leading to the increase of GAPDH enzyme activity, eventually promote the cell proliferation and tumor formation. This reveals a potential mechanism of cellular metabolic adaptation to environmental conditions.
The protein acetylation modification is a reversible process via the control of acetyltransferase and deacetylase. The proteins that can add the acetyl group to the ϵ-amino group of lysine are called histone acetyltransferases (HATs) (20) and many HATs have more than one substrates and one substrate may have more than one HATs. Here we reported PCAF is one of the acetyltransferases of GAPDH. Other HATs might be involved in acetylating GAPDH. Similarly to the HATs, histone deacetylases (HDACs) also deacetylate many non-histone proteins. The classical HDAC family includes three phylogenetic classes, namely class I, II and class IV. Class I HDACs have homology to the yeast transcriptional regulator Rpd3 and comprises HDAC1, -2, -3, and -8, all of which are located in the nucleus. Class II HDACs (HDAC4, -5, -6, -7, -9, and -10) show homology to yeast Hda1 and all of them can be localized in both the nucleus and the cytoplasm (21). Class IV has only one member-HDAC11, which was newly found. Here we tested the overexpression of HDAC 1–7 and found that HDAC5 deacetylates GAPDH at K254 and reduces its enzyme activity. For many substrates can be deacetylated by more than one HDAC (21); it is possible that other HDACs could also deacetylate GAPDH at different sites.
Rapidly proliferating cancer cells showed a high glucose uptake and glycolysis was dramatically elevated (22). Here we found upon high glucose, the interaction between PCAF and GAPDH is increased while the binding between HDAC5 and GAPDH is decreased, promoting K254 acetylation and its enzyme activity. In contrast, upon low glucose, the binding between HDAC5 and GAPDH is increased while the interaction between PCAF and GAPDH is decreased, repressing K254 acetylation and its enzyme activity. Mutants at K254 to glutamine or arginine are defective of acetylation and cannot response to the glucose status change or the effect of PCAF and HDAC5. Consistently, compared with A549/GAPDHWT, A549/GAPDHK254Q showed a reduction in cell proliferation and tumor growth in vivo. Our study reveals a molecular mechanism of GAPDH regulation by K254 acetylation via the action of PCAF and HDAC5 in response to extracellular environment, indicating K254 acetylation of GAPDH might be readout of cellular glucose status.
Modification of GAPDH by S-nitrosylation, acetylation, and phosphorylation has been linked to nuclear translocation of GAPDH (23–25). During apoptosis, the activated inducible nitric oxide synthase (iNOS) or neuronal NOS (nNOS) could generate nitric oxide (NO) S-nitrosylating GAPDH to promote its binding with Siah1, an E3-ubiquitin-ligase with a nuclear localization signal (NLS) (23). Nuclear GAPDH is acetylated at lysine 160 by the acetyltransferase p300/CBP, which in turn stimulates the acetylation and catalytic activity of p300/CBP (26). In addition, it is also reported that Sirt1 restrains GAPDH entering nuclear via direct interaction with GAPDH in a deacetylase activity-independent manner (27). It is an open question whether K254 acetylation affects the nuclear translocation of GAPDH.
GAPDH is dramatically elevated in various human cancers and often associated with reduced survival (4, 28–30). Our study demonstrated that the critical role of acetylation regulation of GAPDH in the coordination of glucose utilization to promote cell growth and tumorigenesis. Regarding the promotive effect of K254 acetylation on GAPDH, drugs inhibiting GAPDH acetylation by targeting the acetyltransferase or deacetylase should inhibit GAPDH, therefore may have potential therapeutic value for cancers with high GAPDH activity. Collectively, our study not only identifies a novel site of GAPDH acetylation regulation, but also provides a potential therapeutic target for GAPDH overexpressed cancers.
Acknowledgments
We thank the members of the Fudan Molecular and Cell Biology laboratory for discussions throughout this study.
This work was supported by 973 (Grant 2011CB910600, 2009CB918401), NSFC (Grant 31071192, 81225016, 31271454), Shanghai Key Basic Research Program (12JC1401100), “100 Talents” Program of Shanghai Health (Grant XBR2011041), Scholar of “Dawn” Program of Shanghai Education Commission, and Shanghai Outstanding Academic Leader (Grant No.13XD1400600) (to Q. Y. L.). This work was also supported by the 985 Program, the Shanghai Leading Academic Discipline Project (Project Number B110).
- 18F-FDG
- 2-(18F)-fluoro-2-deoxy-d-glucose
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- TSA
- trichostatin A
- NAM
- nicotinamide
- IP
- immunoprecipitation
- PET
- positron emission tomography.
REFERENCES
- 1. Warburg O. (1956) On the origin of cancer cells. Science 123, 309–314 [DOI] [PubMed] [Google Scholar]
- 2. Strauss L. G., Conti P. S. (1991) The applications of PET in clinical oncology. J. Nucl. Med. 32, 623–648; discussion 649–650 [PubMed] [Google Scholar]
- 3. Mankoff D. A., Eary J. F., Link J. M., Muzi M., Rajendran J. G., Spence A. M., Krohn K. A. (2007) Tumor-specific positron emission tomography imaging in patients: [18F] fluorodeoxyglucose and beyond. Clin. Cancer Res. 13, 3460–3469 [DOI] [PubMed] [Google Scholar]
- 4. Altenberg B., Greulich K. O. (2004) Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics 84, 1014–1020 [DOI] [PubMed] [Google Scholar]
- 5. Alexander-Bridges M., Dugast I., Ercolani L., Kong X. F., Giere L., Nasrin N. (1992) Multiple insulin-responsive elements regulate transcription of the GAPDH gene. Adv. Enzyme Regul. 32, 149–159 [DOI] [PubMed] [Google Scholar]
- 6. Meyer-Siegler K., Rahman-Mansur N., Wurzer J. C., Sirover M. A. (1992) Proliferative dependent regulation of the glyceraldehyde-3-phosphate dehydrogenase/uracil DNA glycosylase gene in human cells. Carcinogenesis 13, 2127–2132 [DOI] [PubMed] [Google Scholar]
- 7. Meyer-Siegler K., Mauro D. J., Seal G., Wurzer J., deRiel J. K., Sirover M. A. (1991) A human nuclear uracil DNA glycosylase is the 37-kDa subunit of glyceraldehyde-3-phosphate dehydrogenase. Proc. Natl. Acad. Sci. U.S.A. 88, 8460–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Singh R., Green M. R. (1993) Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science 259, 365–368 [DOI] [PubMed] [Google Scholar]
- 9. Tisdale E. J. (2001) Glyceraldehyde-3-phosphate dehydrogenase is required for vesicular transport in the early secretory pathway. J. Biol. Chem. 276, 2480–2486 [DOI] [PubMed] [Google Scholar]
- 10. Nakagawa T., Hirano Y., Inomata A., Yokota S., Miyachi K., Kaneda M., Umeda M., Furukawa K., Omata S., Horigome T. (2003) Participation of a fusogenic protein, glyceraldehyde-3-phosphate dehydrogenase, in nuclear membrane assembly. J. Biol. Chem. 278, 20395–20404 [DOI] [PubMed] [Google Scholar]
- 11. Ishitani R., Chuang D. M. (1996) Glyceraldehyde-3-phosphate dehydrogenase antisense oligodeoxynucleotides protect against cytosine arabinonucleoside-induced apoptosis in cultured cerebellar neurons. Proc. Natl. Acad. Sci. U.S.A. 93, 9937–9941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakajima H., Amano W., Fujita A., Fukuhara A., Azuma Y. T., Hata F., Inui T., Takeuchi T. (2007) The active site cysteine of the proapoptotic protein glyceraldehyde-3-phosphate dehydrogenase is essential in oxidative stress-induced aggregation and cell death. J. Biol. Chem. 282, 26562–26574 [DOI] [PubMed] [Google Scholar]
- 13. Chang C. H., Curtis J. D., Maggi L. B., Jr., Faubert B., Villarino A. V., O'Sullivan D., Huang S. C., van der Windt G. J., Blagih J., Qiu J., Weber J. D., Pearce E. J., Jones R. G., Pearce E. L. (2013) Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nicholls C., Pinto A. R., Li H., Li L., Wang L., Simpson R., Liu J. P. (2012) Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) induces cancer cell senescence by interacting with telomerase RNA component. Proc. Natl. Acad. Sci. U.S.A. 109, 13308–13313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 16. Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., Zhao Y. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 17. Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Li Y., Shi J., An W., Hancock S. M., He F., Qin L., Chin J., Yang P., Chen X., Lei Q., Xiong Y., Guan K. L. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guan K. L., Xiong Y. (2011) Regulation of intermediary metabolism by protein acetylation. Trends Biochem. Sci. 36, 108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Puzone R., Savarino G., Salvi S., Dal Bello M. G., Barletta G., Genova C., Rijavec E., Sini C., Esposito A. I., Ratto G. B., Truini M., Grossi F., Pfeffer U. (2013) Glyceraldehyde-3-phosphate dehydrogenase gene over expression correlates with poor prognosis in non small cell lung cancer patients. Mol. Cancer 12, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brown C. E., Lechner T., Howe L., Workman J. L. (2000) The many HATs of transcription coactivators. Trends Biochem. Sci. 25, 15–19 [DOI] [PubMed] [Google Scholar]
- 21. de Ruijter A. J., van Gennip A. H., Caron H. N., Kemp S., van Kuilenburg A. B. (2003) Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370, 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hanahan D., Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 23. Hara M. R., Agrawal N., Kim S. F., Cascio M. B., Fujimuro M., Ozeki Y., Takahashi M., Cheah J. H., Tankou S. K., Hester L. D., Ferris C. D., Hayward S. D., Snyder S. H., Sawa A. (2005) S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 7, 665–674 [DOI] [PubMed] [Google Scholar]
- 24. Huang Q., Lan F., Zheng Z., Xie F., Han J., Dong L., Xie Y., Zheng F. (2011) Akt2 kinase suppresses glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-mediated apoptosis in ovarian cancer cells via phosphorylating GAPDH at threonine 237 and decreasing its nuclear translocation. J. Biol. Chem. 286, 42211–42220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ventura M., Mateo F., Serratosa J., Salaet I., Carujo S., Bachs O., Pujol M. J. (2010) Nuclear translocation of glyceraldehyde-3-phosphate dehydrogenase is regulated by acetylation. Int. J. Biochem. Cell Biol. 42, 1672–1680 [DOI] [PubMed] [Google Scholar]
- 26. Sen N., Hara M. R., Kornberg M. D., Cascio M. B., Bae B. I., Shahani N., Thomas B., Dawson T. M., Dawson V. L., Snyder S. H., Sawa A. (2008) Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat. Cell Biol. 10, 866–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Joo H. Y., Woo S. R., Shen Y. N., Yun M. Y., Shin H. J., Park E. R., Kim S. H., Park J. E., Ju Y. J., Hong S. H., Hwang S. G., Cho M. H., Kim J., Lee K. H. (2012) SIRT1 interacts with and protects glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from nuclear translocation: implications for cell survival after irradiation. Biochem. Biophys. Res. Commun. 424, 681–686 [DOI] [PubMed] [Google Scholar]
- 28. Lu H., Zhang Y., Roberts D. D., Osborne C. K., Templeton N. S. (2002) Enhanced gene expression in breast cancer cells in vitro and tumors in vivo. Mol. Ther. 6, 783–792 [DOI] [PubMed] [Google Scholar]
- 29. Révillion F., Pawlowski V., Hornez L., Peyrat J. P. (2000) Glyceraldehyde-3-phosphate dehydrogenase gene expression in human breast cancer. Eur. J. Cancer 36, 1038–1042 [DOI] [PubMed] [Google Scholar]
- 30. Detterbeck F. C., Vansteenkiste J. F., Morris D. E., Dooms C. A., Khandani A. H., Socinski M. A. (2004) Seeking a home for a PET, part 3: Emerging applications of positron emission tomography imaging in the management of patients with lung cancer. Chest 126, 1656–1666 [DOI] [PubMed] [Google Scholar]