Abstract
Background
Most synovial sarcomas contain a chromosomal translocation t(X;18), which results in the formation of an oncoprotein SS18-SSX critical to the viability of synovial sarcoma.
Questions/purposes
We (1) established and characterized three novel synovial sarcoma cell lines and asked (2) whether inhibition of SS18-SSX1 decreases cell viability in these cell lines; and (3) whether reduction in viability after SS18-SSX1 knockdown is caused by apoptosis. After identifying a specific posttranscriptional splice variant in our cell lines, we asked (4) whether this provides a survival benefit in synovial sarcoma.
Methods
Cells lines were characterized. SS18-SSX1 knockdown was achieved using a shRNA system. Cell viability was assessed by WST-1 analysis and apoptosis examined by caspase-3 activity.
Results
We confirmed the SS18-SSX1 translocation in all cell lines and identified a consistent splicing variant. We achieved successful knockdown of SS18-SSX1 and with this saw a significant reduction in cell viability. Decreased viability was a result of increased apoptosis. Reintroduction of the exon 8 sequence into cells reduced cell viability in all cell lines.
Conclusions
We confirmed the presence of the SS18-SSX1 translocation in our cell lines and its importance in the survival of synovial sarcoma. We have also demonstrated that reduction in cell viability is related to an increase in apoptosis. In addition, we have identified a potential mediator of SS18-SSX function in exon 8.
Clinical Relevance
SS18-SSX represents a tumor-specific target in synovial sarcoma. Exploitation of SS18-SSX and its protein partners will allow us to develop potent tumor-specific therapeutic agents.
Introduction
Synovial sarcoma accounts for 5% to 10% of all soft tissue sarcoma cases and is most prevalent in young adults [22]. Unfortunately, synovial sarcoma is an aggressive disease and has a poor prognosis, especially when it presents with metastatic disease [24, 34]. Despite advancements in surgical techniques, radiation therapy, and chemotherapy, we have done little to change outcomes in patients with this disease. Metastasis will develop in approximately 50% of patients, most often to the lung, although metastasis to lymph nodes occurs in approximately 10% of cases [13, 26, 31]. Current chemotherapy regimens do not significantly alter oncologic outcome for the majority of patients with synovial sarcoma [16, 21, 25].
Current chemotherapy lacks benefit as a result of a very small therapeutic index of highly toxic options. Fortunately, most synovial sarcoma tumors contain a chromosomal rearrangement that is not present in normal patient cells [33]. This rearrangement results from a translocation between chromosomes X and 18 and is present in greater than 90% of patients. The translocation occurs between the SS18 gene on chromosome 18 and the SSX gene on the X chromosome. The resulting chimeric gene typically produces a transcript composed of exons 1 to 10 of the SS18 gene fused to exons 5 and 6 of an SSX gene [8, 9] and results in the formation of a fusion protein, SS18-SSX. To date, three different fusion protein subtypes have been identified, SS18-SSX1, SS18-SSX2, and SS18-SSX4 [1, 15, 28, 32]. The SS18-SSX1 subtype has been associated with more aggressive clinical behavior [23, 27]. Multiple splice variants of the SS18-SSX subtypes have been described in the literature [4, 14, 19].
Based on IUPred analysis, SS18-SSX1 is predicted to be an intrinsically disordered protein (IDP) (Fig. 1). IDPs are quickly becoming recognized as the most significant components of transcriptional complexes [37]. Disorder, in a protein, refers to the lack of a fixed folding pattern that occurs based on the primary amino acids present. Intrinsically disordered proteins have significant flexibility that facilitates their interaction with other proteins as well as small molecules. This flexibility not only contributes to the cell’s ability to interact with vital prooncogenic proteins, but may facilitate treatment strategies as well. The predicted disorder of SS18-SSX likely promotes its interaction with other transcription factors, but very little is known about how the SS18-SSX fusion protein contributes to the tumorigenesis of synovial sarcoma.
Fig. 1.
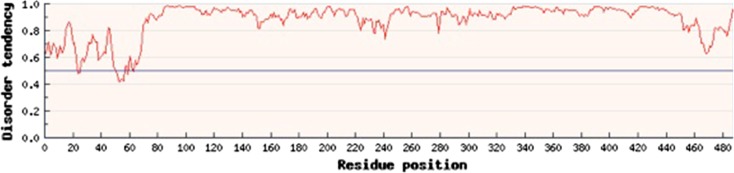
SS18-SSX1 is an intrinsically disordered protein. The IUPRed algorithm predicts disorder at 1.0 and an ordered protein at 0. By this algorithm, SS18-SSX1 is predicted to be almost entirely disordered.
In this study, we (1) established and characterized three novel human synovial sarcoma cell lines and asked (2) whether inhibition of SS18-SSX1 decreased cell viability in these cell lines; and (3) whether reduction in cell growth after SS18-SSX1 knockdown was caused by an increase in apoptosis. Finally, (4) after identifying a specific splice variant, we asked whether this provides a survival benefit in synovial sarcoma.
Materials and Methods
Institutional review board approval was obtained from Georgetown University, and informed consent was obtained from patients and/or their guardians before collection of patient specimens.
To test these hypotheses, we established three novel human synovial sarcoma cell lines. All tissue samples were obtained at the time of initial patient biopsy before any neoadjuvant treatment with the exception of GUSS-3b, which was obtained at the time of definitive surgery, after neoadjuvant chemotherapy and radiation. Patient demographics and tumor characteristics are presented (Table 1). Tissue specimens were emulsified and plated in tissue flasks pretreated with a 1:50 dilution of collagen. Initially cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 15% fetal bovine serum (FBS) and 5% fungizone and penicillin/streptomycin (Life Technologies, Grand Island, NY, USA). After 1 month in culture, cells were maintained in DMEM/15% FBS only. Cells underwent two to three passages per month and grew at relatively consistent rates.
Table 1.
Patient demographics and tumor characteristics
| Cell line | Tumor type | Patient age (years), sex | MSTS stage at presentation | Oncologic progression (months) | Disease status (months) |
|---|---|---|---|---|---|
| GUSS-1 | High grade/biphasic | 51, female | IIB | Date of initial metastasis unknown | DOD: 17 |
| GUSS-2 | High grade/monophasic | 36, male | IIB | LR: 14, LR: 27 | NED: 55 |
| GUSS-3 | High grade/biphasic | 12, male | IIB | LR: 11, metastasis: 15 | DOD: 22 |
MSTS = Musculoskeletal Tumor Society; LR = local recurrence; DOD = died of disease; NED = no evidence of disease.
Cells were grown to approximately 80% confluency in 15-cm dishes. Total RNA was isolated from cell lines by Trizol reagent (Invitrogen, Grand Island, NY, USA). RNA was reverse-transcribed with high-fidelity Taq polymerase (Invitrogen). The resulting complementary DNA was subjected to polymerase chain reaction (PCR) amplification with the following primers: SS18: 5′CAACAGCAAGATGCATACCA3′, SSX: 5′CACTTGCTATGCACCTGATG3′, SSX1: 5′GGTGCAGTTGTTTCCCATCG3′, SSX2: 5′GGCACAGCTCTTTCCCATCA3′. The amplification parameters used consisted of denaturation at 95° C for 1 minute, annealing at 60° C for 1 minute, and extension at 72° C for 1 minute for 35 cycles. The products were separated by electrophoresis in agarose gels and visualized with ethidium bromide.
Fresh PCR SS18-SSX1 product was obtained from each cell line. Bands were cut from agarose gels at the appropriate base pair length, and DNA was isolated with the GENECLEAN Turbo Kit (MP Biomedicals, Solon, OH, USA) per the manufacturer’s protocol. A TOPO cloning reaction was performed per the manufacturer’s protocol (Life Technologies). Bacterial transformation was performed using chemically competent DH5α Escherichia coli per the manufacturer’s protocol. Positive clones were selected and incubated overnight in LB medium containing 50 μg/mL kanamycin. Plasmid DNA was isolated using a RapidPURE Plasmid Mini kit (MP Biomedicals) per the manufacturer’s protocol. A restriction enzyme digest was performed using EcoRI. Resultant plasmid DNA corresponding to SS18-SSX1 band was sent for DNA sequencing analysis (GENEWIZ, South Plainfield, NJ, USA).
Protein disorder was evaluated by typing the amino acid sequence of the SS18-SSX1 protein present in our cell lines into the IUPred server (http://iupred.enzim.hu).
Whole cell lysates from cells were subject to SDS-PAGE and then transferred to an Immobilon-P membrane (Millipore Corp, Billerica, MA, USA). Membranes were subjected to blocking in 5% nonfat dry milk in 1× TTBS (20 mM Tris-HCL, pH 7.5; 150 mM NaCl; and 0.5% Tween 20) for 1 hour. Dilutions for primary antibodies were as follows: anti-SS18 (sc-365170) at 1:1000 (Santa Cruz Biotechnologies, Inc, Santa Cruz, CA, USA) antiactin-horseradish peroxidase (C-11) at 1:3000 (Santa Cruz Biotechnology, Inc), and anti-PARP (04-575) at 1:1000 (Millipore). Primary antibodies were added to the membrane in 5% nonfat dry milk in 1× TTBS for 1 hour. The membrane was then washed three times in 1× TTBS and horseradish peroxidase-linked antimouse secondary antibody (GE Healthcare, Waukesha, WI, USA) was added for 1 hour. Blots were washed three times in 1× TTBS and then developed using Millipore Immobilon Western Chemiluminescent HRP Substrate per the manufacturer’s instructions (Millipore Corp). Chemiluminescence was detected using a Fujifilm LAS-3000 (Life Science, Stamford, CT, USA) imaging system. Densitometry values were obtained using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Genomic DNA was isolated from cell lines with the Wizard Genomic DNA Purification Kit per the manufacturer’s protocol (Promega, Madison, WI, USA). Briefly, cells were lysed with nuclei lysis solution. Protein precipitation solution was added and the samples were chilled on ice for 5 minutes. Samples were then centrifuged for 4 minutes at 15,000 ×g. The supernatant was transferred to a clean tube and washed with 70% ethanol. The DNA pellet was allowed to air-dry for 15 minutes at room temperature. The pellet was rehydrated using DNA rehydration solution and incubated at 65° C for 1 hour. The following primers were used to amplify the DNA: SS18F: 5′-TTAAATATTTTTAGGGTTTTCACTCA-3′, SS18R: 5′-ATTTTCATTCCTGTAGAAGGGGAT-3′.
A nonsilencing shRNA construct control was purchased from Open Biosystems (Huntsville, AL, USA). Anti-SS18-SSX shRNA was purchased from Open Biosystems that contained six distinct shRNA species (Table 2) targeting different sequences of the SS18-SSX transcript (V2LHS_56392, V2LHS_56395, V2LHS_56396, V3LHS_320227, V3LHS-320228, V3LHS_320229). Cells were transfected with the shRNA using Fugene6 (Roche Applied Science, Indianapolis, IN, USA). After 48 hours, SS18-SSX protein expression was analyzed by Western blot analysis as described previously. Densitometry analysis was performed with ImageJ software. Untreated cells, empty vector, and scramble shRNA were used as negative controls.
Table 2.
SS18-SSX shRNA sequences sense:loop:antisense
| Construct | Sequence |
|---|---|
| shRNA1 | TGCTGTTGACAGTGAGCGCACAGTTAAACTGACATCTTAATAGTGAAGCCACAGATGTATTAAGATGTCAGTTTAACTGTATGCCTACTGCCTCGGA |
| shRNA2 | TGCTGTTGACAGTGAGCGATGAAGCAACCTCTTATGTATATAGTGAAGCCACAGATGTATATACATAAGAGGTTGCTTCAGTGCCTACTGCCTCGGA |
| shRNA3 | TGCTGTTGACAGTGAGCGAGGCCTTAATTGCAATAGCCATTAGTGAAGCCACAGATGTAATGGCTATTGCAATTAAGGCCCTGCCTACTGCCTCGGA |
| shRNA4 | TGCTGTTGACAGTGAGCGCCGCGACTGTTCTGCCAAGCAATAGTGAAGCCACAGATGTATTGCTTGGCAGAACAGTCGCGATGCCTACTGCCTCGGA |
| shRNA5 | TGCTGTTGACAGTGAGCGACTCAAGGATGATGATGCTGAATAGTGAAGCCACAGATGTATTCAGCATCATCATCCTTGAGGTGCCTACTGCCTCGGA |
| shRNA6 | TGCTGTTGACAGTGAGCGCCTGTAAGAAATGTTTGTTCAATAGTGAAGCCACAGATGTATTGAACAAACATTTCTTACAGATGCCTACTGCCTCGGA |
Forty-eight hours after transfection with shRNAs, cell lines were replated in 96-well plates and cell viability was evaluated using a WST-1 (Roche 11644807001) assay according to the manufacturer’s protocol. Apoptosis was analyzed using a caspase-3 assay, also according to the manufacturer’s protocol. Untreated cells, empty vector, and scramble shRNA were used as negative controls.
Results
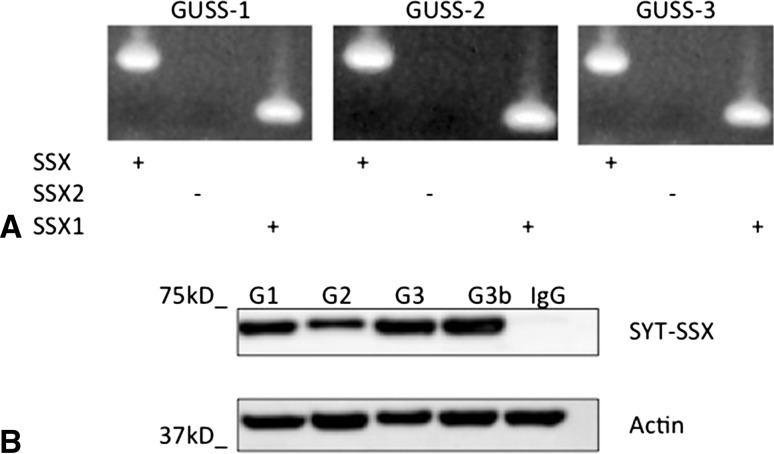
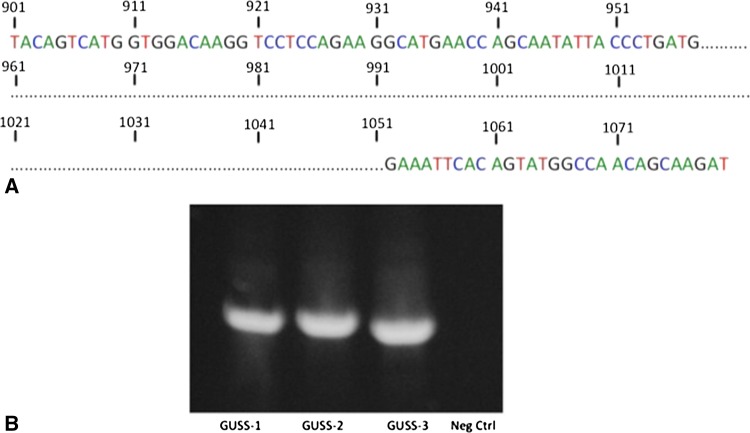
We have established three novel human synovial sarcoma cell lines: GUSS-1, GUSS-2, and GUSS-3. All three cell lines were established from separate patient tumor specimens at the time of initial biopsy. GUSS- 3b (as shown in Fig. 2B) was derived from the same patient as GUSS-3 but was obtained after the patient received neoadjuvant chemotherapy and radiation therapy. We determined by PCR analysis that all of our cell lines contained the SS18-SSX translocation and were all of the SS18-SSX1 subtype (Fig. 2A). To verify that all of our synovial sarcoma cell lines express the SS18-SSX oncoprotein, protein lysates were extracted analyzed by Western blot analysis. All cell lines express the SS18-SSX oncoprotein (Fig. 2B). In addition, we identified a specific deletion in the expected coding sequence (as published by the National Institutes of Health genebank) of SS18-SSX1 (Fig. 3A), which was present in all three cell lines. This resulted in the loss of a 93-base pair sequence (31 amino acids) in the expected coding sequence in the SS18 domain of the fusion protein corresponding to the loss of exon 8. The amino acid sequence (GHNDYGYQQPSYPEQGYDRPYEDSSQHYYEG) of this deleted region has no known functional significance but is large enough to function as an independent protein partner. After identifying this deletion, we examined the genomic DNA of all three cell lines to determine if this deletion was inherent to the cell line itself or was a result of a posttranscriptional splicing variant resulting in an alternatively expressed protein. We extracted genomic DNA and designed primers to identify this sequence in the genomic DNA. We illustrate that this deleted sequence is present in the genomic DNA of all cell lines (Fig. 3B), indicating that the cells are actively removing this sequence after transcription.
Fig. 2A–B.
Characterization of human synovial sarcoma cell lines GUSS-1, GUSS-2, and GUSS-3. We performed reverse transcription–PCR on total RNA from each of our synovial sarcoma cell lines using the SS18, SSX (consensus), SSX1, and SSX2 primers. The PCR products were then separated by electrophoresis on an agarose gel and visualized with ethidium bromide. All cell lines are of the SS18-SSX1 subtype (A). Protein lysates from each were analyzed by Western blot for SS18-SSX1 expression. All cell lines express SS18-SSX1 protein (B). G1 = GUSS-1; G2 = GUSS-2; G3 = GUSS-3 prechemotherapy; G3b = GUSS-3 postchemotherapy; IgG = negative IgG control. Actin was used as a protein concentration loading control.
Fig. 3A–B.
Identification of splicing variant with exon 8 deletion. Fresh PCR product corresponding to the SS18-SSX1 sequence from all of our cell lines was sent for sequencing analysis. The highlighted portion represents a 93 base pair deletion in the expected coding sequence for SS18-SSX1, which corresponds to the deletion of exon-8 in the SS18 domain of the fusion protein (A). Genomic DNA was isolated from synovial sarcoma cell lines and simplified by PCR with primers designed to flank the exon-8 sequence. Exon-8 was present in the genomic DNA of all three cell lines (B). cDNA from GUSS-3, which lacks the exon-8 sequence, was used as a negative control.
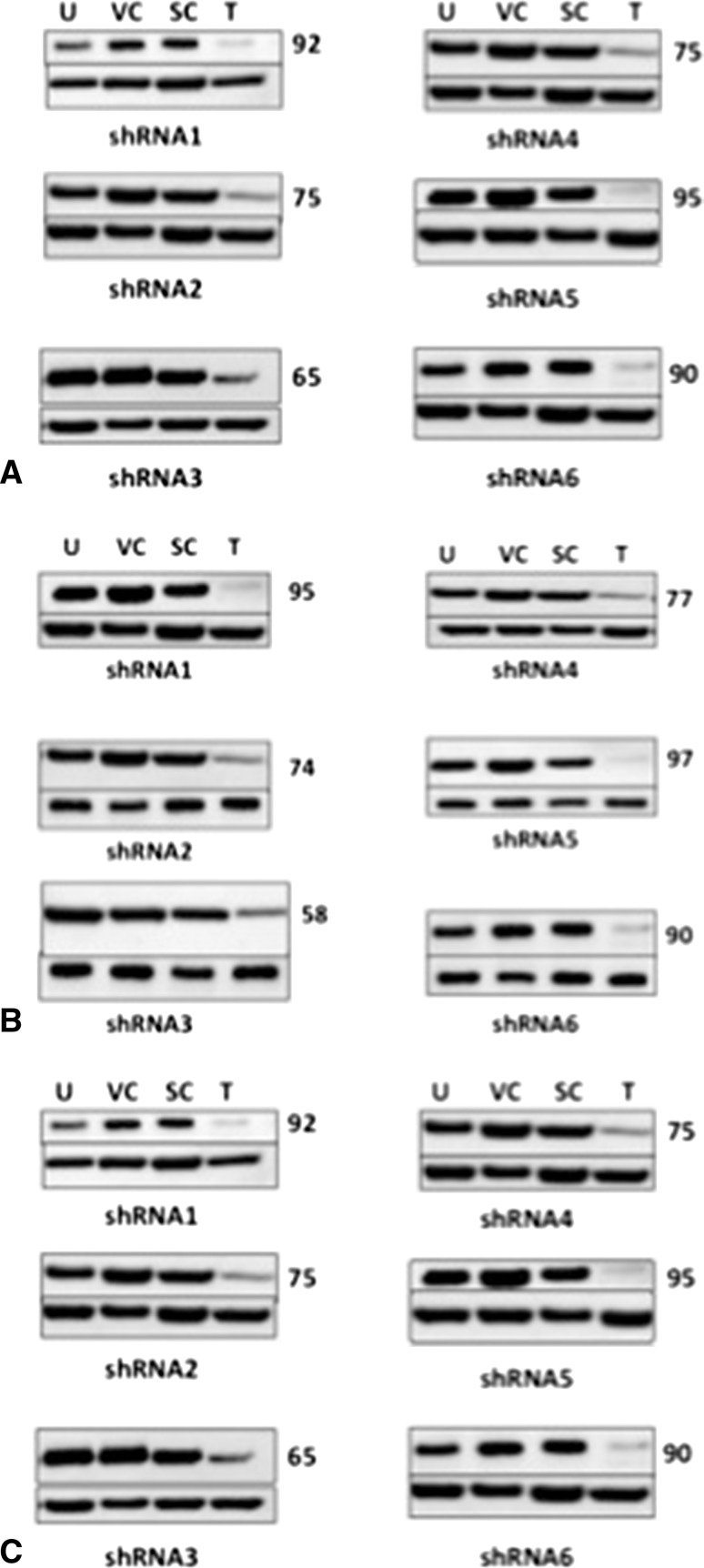
To determine whether inhibition of SS18-SSX1 inhibits cell viability in our cell lines, we first investigated our ability to knockdown SS18-SSX1 after treatment with SS18-SSX1 shRNAs corresponding to six independent regions of the SS18-SSX1 fusion message (Table 2). We achieved successful knockdown in all cell lines. Average reduction with shRNA-1 was 95% (± 3) when compared with untreated, empty vector, and scramble shRNA controls. Average reduction with shRNA-5 was 94% (± 3.6) and shRNA-6 illustrated an average reduction of 93% (± 4.6) (Fig. 4A–C). Our cell viability assays support our hypotheses that SS18-SSX1 is critical to the survival of synovial sarcoma. We saw a significant reduction in cell viability in all of our cell lines when treated with SS18-SSX1 shRNA constructs (Fig. 5). Cell viability was reduced to 5% (± 1.2) in our cell lines after treatment with shRNA-1 when compared with untreated, empty vector, and scramble shRNA negative controls. Treatment with shRNA-5 illustrated a reduction in cell viability to 5% (± 1.1) and shRNA-6 illustrated a reduction in cell viability to 7% (± 1.7).
Fig. 4A–C.
Successful reduction of SS18-SSX1 protein by shRNA. Protein lysates were extracted from synovial sarcoma cell lines transfected with anti-SS18-SSX shRNAs. (A) GUSS-1; (B) GUSS-2; (C) GUSS-3. U = untreated cells; VC = empty vector control; SC = shRNA scramble control; T = treated cells. Numbers to the right of the panel quantify percent reduction in protein expression by densitometry analysis. Variable knockdown of SYT-SSX was seen with various shRNA constructs, but > 90% reduction was seen in all three cell lines with shRNAs 1, 5, and 6.
Fig. 5.
Reduction of SS18-SSX reduces cell viability. Cell lines were treated with shRNAs against SS18-SSX and cell viability was examined by WST-1 assay. We saw a significant reduction in cell viability in all three cell lines with each of the shRNA constructs.
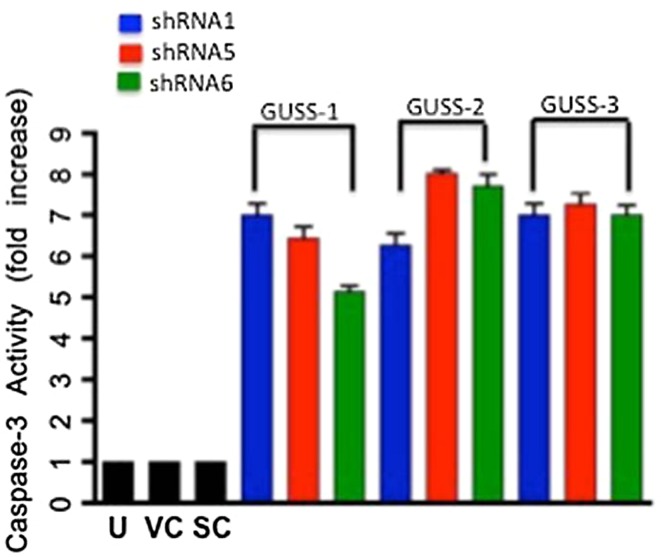
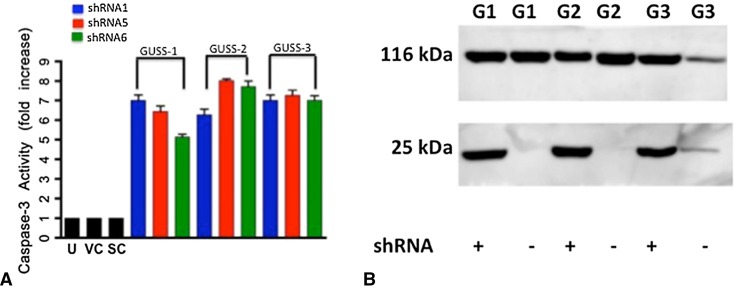
We validated our hypothesis that reduction in synovial sarcoma cell growth after inhibition of SS18-SSX1 is caused by an increase in apoptosis (Fig. 6A). We saw a 6.7 (± 1.7)-fold increase in caspase-3 activity after treatment with shRNA-1 when compared with negative controls. Treatment with shRNA-5 resulted in a 7.8 (± 1.6)-fold increase in caspase-3 activity in the respective cell lines, whereas we saw a 7.2 (± 1.4)-fold increase in caspase-3 activity in our respective cell lines after reduction with shRNA-6. To further validate that this reduction in cell viability is a result of apoptosis, we performed a PARP cleavage assay (Fig. 6B). Cells treated with anti-SS18-SSX shRNA expressed both full-length PARP and cleaved PARP compared with untreated cells that did not demonstrate any PARP cleavage.
Fig. 6A–B.
SS18-SSX knockdown induces apoptosis. Caspase-3 activity was measured after disruption of SS18-SSX (A). We saw a significant fold increase in caspase-3 activity in all three cell lines with each of the shRNA constructs. Untreated cells (U), empty vector (VC), and scramble shRNA (SC) were used as negative controls. These results depict the average of three separate experiments, each run in triplicate (n = 9). Total cell lysates were analyzed by Western blot analysis for full-length PARP and cleaved PARP. Cells treated with anti-SS18-SSX shRNA expressed cleaved PARP, whereas untreated cells did not. G1 = GUSS-1; G2 = GUSS-2; G3 = GUSS-3.
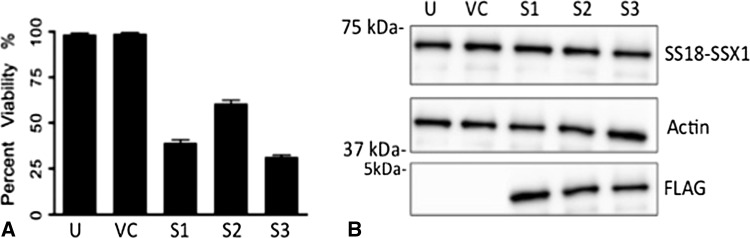
Finally we examined the effect of the splicing variant present in our cells on cell survival by reexpressing exon 8 in our cell lines. After restoration of exon 8, we demonstrate a significant reduction in cell viability by WST-1 assay (Fig. 7A). After reinsertion, we saw a 60% reduction in cell viability in GUSS-1 cells, a 40% reduction in GUSS-2 cells, and a 67% reduction in GUSS-3 cells. Reinsertion of exon 8 into our synovial sarcoma cell lines did not affect SS18-SSX1 protein expression (Fig. 7B) indicating that exon 8’s ability to reduce the tumorigenicity of our synovial sarcoma cell lines is not by inhibiting SS18-SSX1 directly, but perhaps is preventing the SS18-SSX1 oncoprotein from interacting with a critical protein partner.
Fig. 7A–B.
Reinsertion of exon-8 into synovial sarcoma cell lines reduces cell viability without affecting SS18-SSX expression. Synovial sarcoma cell lines were transfected with a pcDNA4TO vector containing the exon-8 sequence. Cell viability was examined by WST-1 analysis. We saw a significant reduction in cell viability after the exon-8 peptide was reintroduced into the cell (A). SS18-SSX expression was examined by Western blot analysis (B). There was no change in SS18-SSX expression after reinsertion of exon-8 into the cell lines. Actin was used as a protein loading control. S1 = GUSS-1; S2 = GUSS-2; S3 = GUSS-3. Positive FLAG expression confirms the presence of exon-8. U = untreated cells; VC = empty vector control. A depicts the results of three separate experiments each run in triplicate (n = 9).
Discussion
Synovial sarcoma contains a chromosomal rearrangement resulting from a translocation of chromosomes X and 18 [5, 17]. This chromosomal rearrangement results in a tumor = specific oncoprotein, SS18-SSX. Although previous studies have identified this oncoprotein as being important to survival in synovial sarcoma, the exact mechanism of how it contributes to the tumorigenicity of synovial sarcoma is unknown [3, 11, 20, 30]. We have established and characterized three novel human synovial sarcoma cell lines and identified an important splicing variant. We have demonstrated that knockdown of the SS18-SSX1 protein decreases cell viability in synovial sarcoma and that this is likely the result of an increase in apoptosis. Finally, we have demonstrated that reintroduction of the deleted exon 8 sequence into our synovial sarcoma cell lines causes a decrease in cell viability, indicating that this splicing variant is important for the survival of synovial sarcoma.
One of the weaknesses of the study is that previous in vitro and in vivo studies have already verified that knockdown of the SS18-SSX protein inhibits the growth of synovial sarcoma. However, in this study, we have established three novel human synovial sarcoma cell lines. To use these cell lines in further experiments to explore the oncogenesis of synovial sarcoma, we need to validate that they behave in a similar manner to those that have already been established in the literature. The fact that they do behave in a similar fashion validates our sarcoma model. A second weakness of the study is, although we have identified the importance of SS18-SSX1 in the viability of synovial sarcoma, we have been unable to provide any mechanistic evidence of why this occurs. We have, however, identified a potential mediator of its function by demonstrating that the specific splice variant of SS18-SSX1 in our cell lines does offer a survival advantage. Further work will focus on establishing the mechanisms of how this occurs.
We have demonstrated that our cell lines express the oncoprotein SS18-SSX1. Most importantly, we have identified a consistent deletion in the expected coding sequence of the SS18 domain of the SS18-SSX1 oncoprotein, which corresponds to the loss of exon 8 in the SS18 domain of the fusion protein. Although this deletion has been described by Tamborini et al. [36], the nature of its contribution to the oncogenic potential of synovial sarcoma has not been elucidated. In addition, we demonstrate that this deleted sequence is present in the genomic DNA of the cell lines, indicating that this deletion is a posttranscriptional event. This alternative splice variant could be instrumental in promoting the malignant potential of synovial sarcoma. Alternative processing of pre-mRNA allows an organism to increase the diversity of its proteome. Although the exact mechanisms are unknown, splice variants of RNA isoforms have been implicated in a variety of cancers including Kaposi’s sarcoma, soft tissue sarcoma, and bone sarcomas [2, 6, 7, 18]. The deletion of the exon 8 portion of the SS18-SSX1 protein potentially confirms a structural change that enhances its ability to interact with important protein partners, thereby increasing its malignant potential. These findings validate a highly specific cancer target, SS18-SSX, and a potential key regulator of its function in exon 8.
We have confirmed that SS18-SSX is vital to synovial sarcoma cell survival in our cell lines by demonstrating reduction in cell viability after knockdown of the protein. We have shown > 90% reduction in SS18-SSX1 protein expression with at least three independent shRNA constructs. Takenaka et al. [35] have shown previously that knockdown of SS18-SSX inhibits cell viability in vitro as well as inhibits tumor growth in an animal model. In addition, Peng et al. [29] has shown that disruption of SS18-SSX with siRNA inhibits cell growth and induces apoptosis in human synovial sarcoma cell lines. Knockdown of SS18-SSX1 correlated with significant reduction in cell viability in all of our cell lines, confirming SS18-SSX1 is vital to the survival of our cell lines. Despite our successful in vitro studies with antisense shRNA, this is not practical as a human therapy based on inadequate delivery and stability.
We have confirmed that the reduction in cell viability after SS18-SSX knockdown is a result of increased apoptosis. We saw a significant increase in caspase-3 activity in all of our cell lines after knockdown of SS18-SSX and have demonstrated the cleavage of PARP after SS18-SSX knockdown. Previous studies by Peng et al. [29] demonstrated a reduction in antiapoptotic gene B-cell lymphoma-2 (bcl-2) and a parallel increase in proapoptotic caspase-3 after knockdown of SS18-SSX by siRNA. In addition, D’Arcy et al. demonstrated expression of SS18-SSX1 in U2OS osteosarcoma cells inhibited apoptosis induced by cytotoxic agents such as doxorubicin [10]. We have validated that in our cell models SS18-SSX1 reduction is also linked to apoptosis activation.
After identifying the exon 8 deletion, we aimed to identify its contribution to the malignant potential of synovial sarcoma. We demonstrate that when exon 8 is reintroduced into our cell lines, we see a significant reduction in cell viability, indicating that its presence inhibits the growth of synovial sarcoma. This specific splice variant of SS18-SSX1 offers the synovial sarcoma cell lines a survival advantage. Protein levels of SS18-SSX1 are unaffected after exon 8 is reintroduced into the cell, indicating that exon 8 may be responsible for preventing a key interaction between SS18-SSX and a critical protein partner as opposed to preventing translation of the protein itself. This may result from an altered structure of SS18-SSX, which affords the oncoprotein the opportunity to bind with protein partners, which in its nonmutant form it cannot. We know based on IUPred analysis that SS18-SSX1 is predicted to be an IDP. It is known that these proteins have considerably favored thermodynamics for binding to other proteins and even small molecules. The targeting of disordered proteins with small molecules is an emerging concept yet is favored thermodynamically. Ewing’s sarcoma, which also has a chromosomal translocation t(11;22) resulting in a fusion oncoprotein EWS-FLI1, has been of particular interest in the development of small molecule inhibitors of key protein interactions. Erkizan et al. demonstrated reduced Ewing’s sarcoma growth in vitro and in vivo after disruption of the interaction of EWS-FLI1 and its protein partner RHA with a small molecule inhibitor [12]. Because IDPs bind more readily to small molecules, disruption of protein-protein complexes are a clear opportunity to create potent and specific anticancer reagents against synovial sarcoma.
Establishing therapeutic molecular targets in oncology presents a difficult challenge. Fortunately, synovial sarcoma contains a tumor-specific target in the oncoprotein SS18-SSX. We have confirmed SS18-SSX as an essential player in the viability of synovial sarcoma and have identified a potential key mediator of its function in exon 8. SS18-SSX is an IDP, which favors its interaction with protein partners as well as small molecule inhibitors of these partners. Exploitation of this flexibility and disruption of key protein interactions with SS18-SSX1 will provide the opportunity for the development of tumor-specific agents to fight this devastating disease.
Acknowledgments
We acknowledge Felasfa M. Wodajo MD, for providing tumor samples from which two of the cell lines were started.
Footnotes
Each author certifies that he or she, or a member of his or her immediate family, has no funding or commercial associations (eg, consultancies, stock ownership, equity interest, patent/licensing arrangements, etc) that might pose a conflict of interest in connection with the submitted article.
All ICMJE Conflict of Interest Forms for authors and Clinical Orthopaedics and Related Research editors and board members are on file with the publication and can be viewed on request.
Clinical Orthopaedics and Related Research neither advocates nor endorses the use of any treatment, drug, or device. Readers are encouraged to always seek additional information, including FDA-approval status, of any drug or device prior to clinical use.
Each author certifies that his or her institution approved the human protocol for this investigation, that all investigations were conducted in conformity with ethical principles of research, and that informed consent for participation in the study was obtained.
This work was performed at the Lombardi Comprehensive Cancer Institute, Washington, DC, USA.
References
- 1.Amary MF, Berisha F, Bernardi FC, Herbert A, James M, Reis-Filho JS, Fisher C, Nicholson AG, Tirabosco R, Diss TC, Flanagan AM. Detection of SS18-SSX fusion transcripts in formalin-fixed paraffin-embedded neoplasms: analysis of conventional RT-PCR, qRT-PCR, and dual color FISH as diagnostic tools for synovial sarcoma. Mod Pathol. 2004;20:482–496. doi: 10.1038/modpathol.3800761. [DOI] [PubMed] [Google Scholar]
- 2.Bartel F, Taylor AC, Taubert H, Harris LC. Novel mdm2 splice variants identified in pediatric rhabdomyosarcoma tumors and cell lines. Oncol Res. 2001;12:451–457. doi: 10.3727/096504001108747459. [DOI] [PubMed] [Google Scholar]
- 3.Brett D, Whitehouse S, Antonson P, Shipley J, Cooper C, Goodwin G. The SYT protein involved in the t(X;18) synovial sarcoma translocation is a transcriptional activator localized in nuclear bodies. Hum Mol Genet. 1997;6:1559–1564. doi: 10.1093/hmg/6.9.1559. [DOI] [PubMed] [Google Scholar]
- 4.Brodin B, Haslam K, Yang K, Bartolazzi A, Xie Y, Starborg M, Lundeberg J, Larsson O. Cloning and characterization of spliced fusion transcript variants of synovial sarcoma: SYT/SSX4, SYT/SSX4v, SYT/SSX2v. Possible regulatory role of the fusion gene product in wild type SYT expression. Gene. 2001;268:173–182. doi: 10.1016/S0378-1119(01)00412-7. [DOI] [PubMed] [Google Scholar]
- 5.de Bruijn DR, Baats E, Zechner U, de Leeuw B, Balemans M, Olde Weghuis D, Hirning-Folz U, Geurts van Kessel AG. Isolation and characterization of the mouse homolog of SYT, a gene implicated in the development of human synovial sarcomas. Oncogene. 1996;13:643–648. [PubMed] [Google Scholar]
- 6.Busto R, Schally AV, Braczkowski R, Plonowski A, Krupa M, Groot K, Armatis P, Varga JL. Expression of mRNA for growth hormone-releasing hormone and splice variants of GHRH receptors in human malignant bone tumors. Regul Pept. 2002;108:47–53. doi: 10.1016/S0167-0115(02)00109-X. [DOI] [PubMed] [Google Scholar]
- 7.Chang TY, Wu YH, Cheng CC, Wang HW. Differentially regulated splice variants and systems biology analysis of Kaposi’s sarcoma-associated herpesvirus-infected lymphatic endothelial cells. Nucleic Acids Res. 2011;39:6970–6985. doi: 10.1093/nar/gkr405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark J, Rocques PJ, Crew AJ, Gill S, Grimer R, Chand A, Shipley J, Gusterson BA, Cooper CS. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet. 1994;7:502–508. doi: 10.1038/ng0894-502. [DOI] [PubMed] [Google Scholar]
- 9.Crew AJ, Clark J, Fisher C, Gill S, Grimer R, Chand A, Shipley J, Gusterson BA, Cooper CS. Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. EMBO J. 1995;14:2333–2340. doi: 10.1002/j.1460-2075.1995.tb07228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Arcy P, Maruwge W, Ryan BA, Brodin B. The oncoprotein SS18-SSX1 promotes p53 ubiquination and degradation by enhancing HDM2 stability. Mol Cancer Res. 2008;6:127–138. doi: 10.1158/1541-7786.MCR-07-0176. [DOI] [PubMed] [Google Scholar]
- 11.Dos Santos NR, de Bruijn DRH, Balemans M, Janssen B, Gartner F, Lopes JM, de Leeuw B, van Kessel AG. Nuclear localization of SYT, SSX, and the synovial sarcoma-associated SYT-SSX fusion proteins. Hum Mol Genet. 1997;6:1549–1558. doi: 10.1093/hmg/6.9.1549. [DOI] [PubMed] [Google Scholar]
- 12.Erkizan HV, Kong Y, Merchant M, Schlottmann S, Barber-Rotenberg JS, Yuan L, Abaan OD, Chou TH, Dakshanamurthy S, Brown ML, Uren A, Toretsky JA. A small molecule blocking oncogenic protein EWS-FLI1 interaction with RNA helicase A inhibits growth of Ewing’s sarcoma. Nat Med. 2009;5:750–756. doi: 10.1038/nm.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrari A, Gronchi A, Casanova M, Meazza C, Gandola L, Collini P, Lozza L, Bertulli R, Olmi P, Casali PG. Synovial sarcoma: a retrospective analysis of 271 patients of all ages treated at a single institution. Cancer. 2004;101:627–634. doi: 10.1002/cncr.20386. [DOI] [PubMed] [Google Scholar]
- 14.Fligman I, Lonardo F, Jhanwar SC, Gerald W, Woodruff J, Ladanyi M. Molecular diagnosis of synovial sarcoma and characterization of a variant SYT-SSX2 fusion transcript. Am J Pathol. 1995;147:1592–1599. [PMC free article] [PubMed] [Google Scholar]
- 15.Gaffney R, Chakerian A, O’Connell JX, Mathers J, Garner K, Joste N, Viswanatha DS. Novel fluorescent ligase detection reaction and flow cytometric analysis of SYT-SSX fusions in synovial sarcoma. Mol Diagn. 2003;5:127–135. doi: 10.1016/S1525-1578(10)60462-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gortzak E, Azzarelli A, Buesa J, Bramwell VH, van Coevorden F, van Geel AN, Ezzat A, Santoro A, Oosterhuis JW, van Glabbeke M, Kirkpatrick A, Verweij J. A randomized phase II study on neoadjuvant chemotherapy for ‘high-risk’ adult soft tissue sarcoma. Eur J Cancer. 2001;37:1096–1103. doi: 10.1016/S0959-8049(01)00083-1. [DOI] [PubMed] [Google Scholar]
- 17.Gure AO, Türeci O, Sahin U, Tsang S, Scanlan MJ, Jäger E, Knuth A, Pfreundschuh M, Old LJ, Chen YT. SSX: a multigene family with several members transcribed in normal testis and human cancer. Int J Cancer. 1997;72:965–971. doi: 10.1002/(SICI)1097-0215(19970917)72:6<965::AID-IJC8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 18.Hahnel A, Wichmann H, Greither T, Kappler M, Würl P, Kotzsch M, Taubert H, Vordermark D, Bache M. Prognostic impact of mRNA levels of osteopontin splice variants in soft tissue sarcoma patients. BMC Cancer. 2012;12:131. doi: 10.1186/1471-2407-12-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiraga H, Nojima T, Abe S, Sawa H, Yamashiro K, Yamawaki S, Kaneda K, Nagashima K. Diagnosis of synovial sarcoma with the reverse transcriptase-polymerase chain reaction: analyses of 84 soft tissue and bone tumors. Diagn Mol Pathol. 1998;7:102–110. doi: 10.1097/00019606-199804000-00007. [DOI] [PubMed] [Google Scholar]
- 20.Ishida M, Miyamoto M, Naitoh S, Tatsuda D, Hasegawa T, Nemoto T, Yokozeki H, Nishioka K, Matsukage A, Ohki M, Ohta T. The SYT-SSX fusion protein down-regulates the cell proliferation regulator COM1 in t(x;18) synovial sarcoma. Mol Cell Biol. 2007;27:1348–1355. doi: 10.1128/MCB.00658-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Italiano A, Penel N, Robin YM, Bui B, Le Cesne A, Piperno-Neumann S, Tubiana-Hulin M, Bompas E, Chevreau C, Isambert N, Leyvraz S, du Chaterlard PP, Thyss A, Coindre JM, Blay JY. Neo/adjuvant chemotherapy does not improve outcome in resected primary synovial sarcoma: a study of the French sarcoma group. Ann Oncol. 2009;20:425–430. doi: 10.1093/annonc/mdn678. [DOI] [PubMed] [Google Scholar]
- 22.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics. Cancer J Clin. 2004;54:8–29. doi: 10.3322/canjclin.54.1.8. [DOI] [PubMed] [Google Scholar]
- 23.Kawai A, Woodruff J, Healey JH, Brennan MF, Antonescu CR, Ladanyi M. SYT-SSX gene fusion as a determinant of morphology and prognosis in synovial sarcoma. N Engl J Med. 1998;338:153–160. doi: 10.1056/NEJM199801153380303. [DOI] [PubMed] [Google Scholar]
- 24.Lewis JJ, Antonescu CR, Leung DH, Blumberg D, Healey JH, Woodruff JM, Brennan MF. Synovial sarcoma: a multivariate analysis of prognostic factors in 112 patients with primary localized tumors of the extremity. J Clin Oncol. 2000;18:2087–2094. doi: 10.1200/JCO.2000.18.10.2087. [DOI] [PubMed] [Google Scholar]
- 25.McCarter MD, Jaques DP, Brennan MF. Randomized clinical trials in soft tissue sarcoma. Surg Oncol Clin N Am. 2002;11:11–22. doi: 10.1016/S1055-3207(03)00073-5. [DOI] [PubMed] [Google Scholar]
- 26.Mullen JR, Zagars GK. Synovial sarcoma outcome following conservation surgery and radiotherapy. Radiother Oncol. 1994;33:23–30. doi: 10.1016/0167-8140(94)90082-5. [DOI] [PubMed] [Google Scholar]
- 27.Nilsson G, Skytting B, Xie Y, Brodin B, Perfekt R, Mandahl N, Lundeberg J, Uhlén M, Larsson O. The SYT-SSX1 variant of synovial sarcoma is associated with a high rate of tumor cell proliferation and poor clinical outcome. Cancer Res. 1999;59:3180–3184. [PubMed] [Google Scholar]
- 28.Otsuka S, Nishijo K, Nakayama T, Aoyama T, Ishibe T, Shibata KR, Shima Y, Nakamura T, Otsuka T, Toguchida J. A variant of the SYT-SSX2 fusion gene in a case of synovial sarcoma. Cancer Genet Cytogenet. 2006;167:82–88. doi: 10.1016/j.cancergencyto.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 29.Peng C, Guo W, Yang Y, Zhao H. Downregulation of SS18-SSX1 expression by small interfering RNA inhibits growth and induces apoptosis in human synovial sarcoma cell line HS-SY-II in vitro. Eur J Cancer Prev. 2008;17:392–398. doi: 10.1097/CEJ.0b013e328305a11b. [DOI] [PubMed] [Google Scholar]
- 30.Perani M, Ingram CJE, Cooper CS, Garrett MD, Goodwin GH. Conserved SNH domain of the proto-oncoprotein SYT interacts with components of the human chromatin remodeling complexes, while the QPGY repeat domain forms homo-oligomers. Oncogene. 2003;22:8156–8167. doi: 10.1038/sj.onc.1207031. [DOI] [PubMed] [Google Scholar]
- 31.Ryan JR, Baker LH, Benjamin RS. The natural history of metastatic synovial sarcoma: experience of the southwest oncology group. Clin Orthop Relat Res. 1982;164:257–260. [PubMed] [Google Scholar]
- 32.Skytting B, Nilsson G, Brodin B, Xie Y, Lundeberg J, Uhlén M, Larsson O. A novel fusion gene, SYT-SSX4, in synovial sarcoma. J Natl Cancer Inst. 1999;91:974–975. doi: 10.1093/jnci/91.11.974. [DOI] [PubMed] [Google Scholar]
- 33.Smith S, Reeves BR, Wong L, Fisher C. A consistent chromosome translocation in synovial sarcoma. Cancer Genet Cytogenet. 1987;26:179–180. doi: 10.1016/0165-4608(87)90147-6. [DOI] [PubMed] [Google Scholar]
- 34.Spillane AJ, A’Hern R, Judson IR, Fisher C, Thomas JM. Synovial sarcoma: a clinicopathologic, staging and prognostic assessment. J Clin Oncol. 2000;18:3794–3803. doi: 10.1200/JCO.2000.18.22.3794. [DOI] [PubMed] [Google Scholar]
- 35.Takenaka S, Naka N, Araki N, Hashimoto N, Ueda T, Yoshioka K, Yoshikawa H, Itoh K. Downregulation of SS18-SSX1 expression in synovial sarcoma by small interfering RNA enhances the focal adhesion pathway and inhibits anchorage-independent growth in vitro and tumor growth in vivo. Int J Oncol. 2010;36:823–831. doi: 10.3892/ijo_00000559. [DOI] [PubMed] [Google Scholar]
- 36.Tamborini E, Agus V, Mezzelani A, Riva C, Sozzi G, Azzarelli A, Pierotti MA, Pilotti S. Identification of a novel spliced variant of the SYT gene expressed in normal tissues and in synovial sarcoma. Br J Cancer. 2001;84:1087–1094. doi: 10.1054/bjoc.2000.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uversky VN. Intrinsically disordered proteins from A to Z. Int J Biochem Cell Biol. 2011;43:1090–1103. doi: 10.1016/j.biocel.2011.04.001. [DOI] [PubMed] [Google Scholar]