Abstract
Global classification of the human proteins with regards to spatial expression patterns across organs and tissues is important for studies of human biology and disease. Here, we used a quantitative transcriptomics analysis (RNA-Seq) to classify the tissue-specific expression of genes across a representative set of all major human organs and tissues and combined this analysis with antibody-based profiling of the same tissues. To present the data, we launch a new version of the Human Protein Atlas that integrates RNA and protein expression data corresponding to ∼80% of the human protein-coding genes with access to the primary data for both the RNA and the protein analysis on an individual gene level. We present a classification of all human protein-coding genes with regards to tissue-specificity and spatial expression pattern. The integrative human expression map can be used as a starting point to explore the molecular constituents of the human body.
Central questions in human biology relate to how cells, tissues, and organs differ in the expression of genes and proteins and what consequences the global expression pattern has for the phenotype of various cells with different functions in the body. Therefore, the annotation of the human protein-coding genes with regards to the spatial, temporal, and functional space represents one of the greatest challenges in human biology (1). Important questions related to this are how many of the genes actually code for functional proteins, how many are expressed in a tissue-specific manner, and how many proteins have “housekeeping” functions and are therefore expressed in all cells? These questions have a major impact not only on efforts to try to understand human biology, but also for applied medical research, such as pharmaceutical drug development and biomarker discovery in the field of translational medicine.
Several efforts have been initiated in the aftermath of the genome project to systematically annotate the putative protein-coding part of the human genome. Genome annotation efforts, such as Ensembl (2) and RefSeq (3), have provided an increasingly accurate map with at present ∼20,000 protein-coding genes. Similarly, the ENCODE consortium has been launched to provide an integrated encyclopedia of DNA elements in the human genome (4). On the protein level, the UniProt consortium (5) has annotated 20,255 human genes as coding for proteins (release 2013_05), including a large number of isoforms. In addition, the protein distribution in human tissues have been explored using antibodies generating more than 13 million manually annotated immunohistochemistry images in the Human Protein Atlas (6). On the RNA level, the FANTOM consortium has been initiated to map the transcriptional space of the human genome and several gene expression atlases for RNA expression data have been launched, such as the original work to create a gene atlas by integrating mouse and human expression data from multiple tissues using micro arrays (7), the BioGPS portal with expression data from numerous sources (8), the repository ArrayExpress (9) and the RNA-Seq Atlas (10), with transcriptomics data based on deep sequencing from eleven normal human tissues. These resources are important tools for comparisons of gene expression patterns in different normal and disease tissues and applications span from in-depth analysis of specific genes to more global systems biology studies to understand human biology and disease.
Here, we describe an extension of these efforts by integrating protein expression data with a transcriptomics analysis based on deep sequencing (RNA-Seq)1 of tissue samples. Because the selection of samples cover a large portion of the major tissues in the human body, we have used the quantitative RNA expression analysis to allow a tissue-specific classification of all the human protein-coding genes. A new version of the Human Protein Atlas (www.proteinatlas.org) is presented with RNA and protein expression data corresponding to 91 and 80%, respectively, of the putative protein-coded genes.
MATERIALS AND METHODS
Transcript Profiling (RNA-seq)
The use of human tissue samples was approved by the Uppsala Ethical Review Board (Reference #2011/473). Tissues samples, collected within the infrastructure of an established biobank, were embedded in Optimal Cutting Temperature (O.C.T.) compound and stored at −80°C. A hematoxylin-eosin (HE) stained frozen section (4 μm) was prepared from each sample using a cryostat and the CryoJane® Tape-Transfer System (Instrumedics, St. Louis, MO, USA). Each slide was examined by a pathologist to ensure proper tissue morphology. Three sections (10 μm) were cut from each frozen tissue block and collected into a tube for subsequent RNA extraction. The tissue was homogenized mechanically using a 3 mm steel grinding ball (VWR). Total RNA was extracted from cell lines and tissue samples using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The extracted RNA samples were analyzed using either an Experion automated electrophoresis system (Bio-Rad Laboratories, Hercules, CA, USA) with the standard-sensitivity RNA chip or an Agilent 2100 Bioanalyzer system (Agilent Biotechnologies, Palo Alto, USA) with the RNA 6000 Nano Labchip Kit. Only samples of high-quality RNA (RNA Integrity Number ≥7.5) were used in the following mRNA sample preparation for sequencing. mRNA sequencing was performed on Illumina HiSeq2000 and 2500 machines (Illumina, San Diego, CA, USA) using the standard Illumina RNA-seq protocol with a read length of 2 × 100 bases. The samples were sequenced multiplexed 15 per lane, producing an average of 18 million mappable read pairs per sample.
The raw reads obtained from the sequencing system were trimmed for low quality ends with the software sickle (11), using a phred quality threshold of 20. All reads shorter than 54 bp after the trimming were discarded. The processed reads were mapped to the GRCh37 version of the human genome with Tophat v2.0.3 (12). Potential PCR duplicates were eliminated using the MarkDuplicates module of Picard 1.77 (13). To obtain quantification scores for all human genes and transcripts, FPKM (fragments per kilobase of exon model per million mapped reads) values were calculated using Cufflinks v2.0.2 (14), which corrects for transcript length and the total number of mapped reads from the library to compensate for different read depths for different samples. The gene models from Ensembl build 69 (2) were used in Cufflinks.
Sample Selection
At least three samples (from different individuals) of each tissue were selected for sequencing in order to ensure proper statistical power, in accordance to common best practice recommendations for RNA-seq. Five samples that did not group with other samples from the same tissue in the hierarchical clustering were discarded after closer investigation of the tissue cuts and, if possible, replaced with newly prepared samples. However, for three tissues (ovary, duodenum and pancreas), only two replicates are included in the final analyses.
Barcode “Leakage”
As has been previously observed (15) multiplexing of samples on a single lane on the Illumina platform can sometimes lead to misidentification of barcodes, leading to what looks like a cross-contamination between samples. Here we observe a small quantity of misidentified reads (∼0.1%) for samples sequenced multiplexed on the same lane in the same run. Effectively this makes genes with very high expression in a certain tissue appear to be have low expression in the other samples run on the same lane.
This introduces a minor bias in the data, which could potentially mislead analyses. However, because most analyses here are looking at relative differences (fold-changes) compared the tissue with highest expression, a leakage of 0.1% hardly affects the analyses.
Specificity Classification
For each tissue, the average FPKM value of all individual samples was used to estimate the gene expression level. A cutoff value of 1 FPKM was used as the detection limit. Each of the 20,050 genes were classified into one of eight categories based on the FPKM levels in 27 tissues: 1. “Not detected” - < 1 FPKM in all 27 tissues; “Tissue specific” - 50-fold higher FPKM level in one tissue compared with all other tissues; “Tissue enriched” - fivefold higher FPKM level in one tissue compared with all other tissues; “Group enriched” - fivefold higher average FPKM level in a group of 2–7 tissues compared with all other tissues; “Mixed low” - detected in 1–26 tissues and at least one tissue < 10 FPKM; “Mixed high” - detected in 1–26 tissues and all detected tissues > 10 FPKM; “Expressed in all low” - detected in 27 tissues and at least one tissue < 10 FPKM; “Expressed in all high” - detected in 27 tissues and all tissues >10 FPKM′. For analyses performed in this study where a log2-scale of the data was used, pseudo-counts of +1 were added to the data set.
Hierarchical Clustering
FPKM values for all 20,050 genes were used to calculate a correlation matrix based on Spearman's rank correlation coefficient for each pairwise combination of individual samples (n = 95). The correlation matrix was used for unsupervised hierarchical clustering analyses of individual samples using the average linkage method to measure distances between clusters. The results were visualized in a heatmap of all pair-wise correlation coefficients.
Gene Ontology Analysis
A gene ontology (16) analysis was performed using the GOrilla tool (17) to determine overrepresented GO categories in the gene set of not detected genes. Genes were assigned to terms in order of overexpression of the terms. Meaning that genes placed in the first category (Sensory perception) were not assigned to less overexpressed terms even though they were associated with the term. A list of all genes analyzed in this study was used as the background list in GOrilla.
Network Analysis
A network analysis of all genes in the tissue enriched and group enriched categories was performed using Cytoscape 3.0 (19). The resulting network includes only group enriched nodes with at least three expressed genes and a maximum of five connections.
Tissue Profiling
Tissue microarrays (TMA) containing triplicate 1-mm cores of 44 different types of normal tissue and duplicate 1-mm cores of 216 different cancer tissues representing the 20 most common forms of human cancer were generated as previously described (18). TMA sections were immunostained as previously described (20). Briefly, slides were deparaffinized in xylene, hydrated in graded alcohols and blocked for endogenous peroxidase in 0.3% hydrogen peroxide diluted in 95% ethanol. For antigen retrieval, a Decloaking chamber® (Biocare Medical, Walnut Creek, CA) was used. Slides were immersed and boiled in Citrate buffer®, pH6 (Lab Vision, Freemont, CA) for 4 min at 125C and then allowed to cool to 90C. Automated IHC was performed essentially as previously described (21) in brief, using an Autostainer 480 instrument® (Lab Vision). Primary antibodies and a dextran polymer visualization system (UltraVision LP HRP polymer®, Lab Vision) were incubated for 30 min each at room temperature and slides were developed for 10 min using Diaminobenzidine (Lab Vision) as chromogen. All incubations were followed by rinse in wash buffer® (Lab Vision). Slides were counterstained in Mayers hematoxylin (Histolab) and cover slipped using Pertex® (Histolab) as mounting medium. Incubation with PBS instead of primary antibody served as negative control. TheAperioScanScope XT Slide Scanner (Aperio Technologies, Vista, CA) system was used to capture digital whole slide images with a 20× objective. Slides were de-arrayed to obtain individual cores. The outcome of IHC stainings in the screening phase, that included various normal and cancer tissues, was manually evaluated and scored by certified pathologists using a web-based annotation system (unpublished). In brief, the manual score of IHC-based protein expression was determined as the fraction of positive cells defined in different tissues: 0 = 0–1%, 1 = 2–25%, 2 = 26–75%, 3 > 75% and intensity of immunoreactivity: 0 = negative, 1 = weak, 2 = moderate, and 3 = strong staining. All of the tissues used as donor blocks were acquired from the archives at the Department of Pathology of Uppsala University Hospital in agreement with approval from the Research Ethics Committee at Uppsala University (Uppsala, Sweden)(Ups 02–577).
Antibodies
The IDs of all antibodies used in the immunohistochemical stainings shown in figure 5 are summarized here. All antibodies are from the Human Protein Atlas project, unless otherwise stated. For antibody validation profiles, see http://www.antibodypedia.com: a) NPHS2: P0372 (Sigma-Aldrich), SLC22A13: HPA035603, TMEM72: HPA039894. b) SLCO1B1: HPA50892, ADH4: HPA20525, CYP1A2: sc-53241 (Santa Cruz Biotechnology), CYP3A4: sc-53850 (Santa Cruz Biotechnology), CYP2E1: HPA29564, CYP2A13: HPA46713. c) SPATA3: HPA018254, ZPBP: HPA058673, HMGB4: HPA035699, RBMXL2: HPA051842, TSPYL6: HPA034700, SOCS7: HPA004475, C18ORF62: HPA041686.
Data Availability
All the data (FPKM values for all the samples) are available for downloads without any restrictions (www.proteinatlas.org/about/download). The primary data (reads) are available through the Array Express Archive (www.ebi.ac.uk/arrayexpress/) under the accession number: E-MTAB-1733. The transcript profiling data (FPKM values) for each gene in each cell and tissue will also be available in the next version of the Human Protein Atlas (www.proteinatlas.org). The classification according to Fig. 2C will be included on the Protein Atlas gene summary page for each of the genes. FPKM values for all 20,050 genes included in the study are provided as downloads to facilitate comparative studies with other “omics” data.
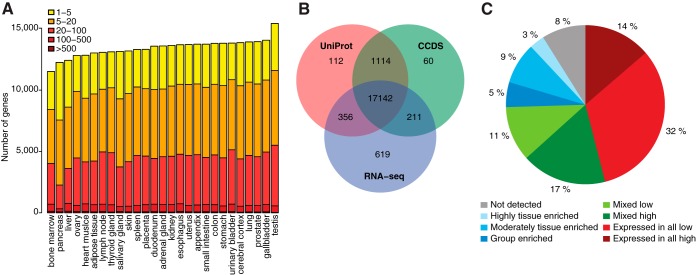
Fig. 2.
The classification of all human protein-coding genes with regards to transcriptional levels in 27 tissues. A, The total number of genes with detected transcripts in each cell type using five different abundance levels for FPKM values; 1–5 FPKM (yellow), 5–20 FPKM (orange), 20–100 FPKM (light red), 100–500 FPKM (red), >500 FPKM (dark red). B, Venn-diagram showing the overlap between a total of 20,083 human Ensembl genes based on three different datasets: (1) genes detected in one or more tissues used in this study (RNA-seq), (2) the core set of genes defined by the Consensus CDS project (CCDS), (3) genes with transcript or protein evidence in UniProt. C, Piechart showing the distribution of all 20,050 genes into eight different categories based on the transcript abundance as well as the number of detected tissues.
RESULTS
The transcriptomes of 27 different human organs and tissues were analyzed using next generation sequencing based on specimens from altogether 95 individuals. These tissues were selected to represent tissue types with specialized body functions and to include all major organs and tissues (Fig. 1A). All tissues were microscopically examined to ensure the representation of normal tissue and to estimate the fraction of normal cell types in each sample. The transcriptome of each sample was quantified using RNA-Seq to determine the normalized mRNA abundance, calculated as FPKM-values (22). In these analyses we have used a cutoff of FPKM 1, roughly representing one mRNA molecule per average cell in the sample (23). The distribution of average FPKM values in the various organ and tissues samples range from 1 to more than 10,000, as exemplified by 60,000 for some of the transcripts encoding digestive enzymes in the pancreas (chymotrypsinogen, amylase and protease), to yield a dynamic range of more than 104 within most tissues. Analysis of biological replicates across all tissues showed high reproducibility with a Spearman correlation between samples from the same tissue, but different individuals, of 0.94 to 0.98 (supplemental Table S1 and supplemental Fig. S1). The high correlation coefficients suggest good technical reproducibility between samples and low biological inter-individual variability between different individuals.
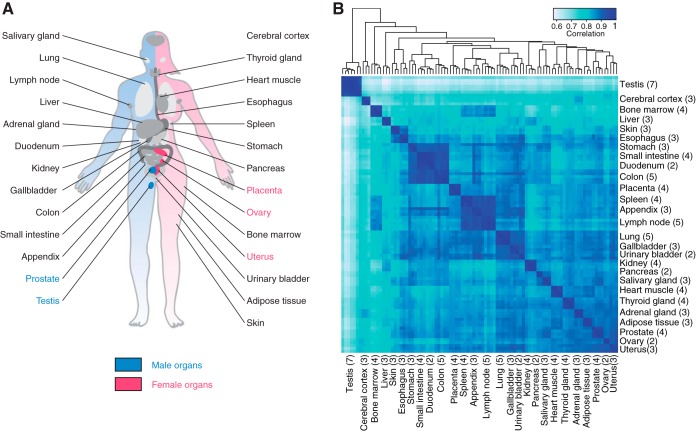
Fig. 1.
The human tissues and organs analyzed by the transcriptomics analysis. A, The location of all 27 analyzed tissues and organs in the human body. B, The relationship between the tissues. Hierarchical clustering results showing the relationships between the 27 different tissues and organs based and a heat map showing the pairwise Spearman correlation. The numbers in parentheses show the number of replicate samples for each tissue.
The global expression profiles were investigated using hierarchical clustering and a correlation heat map including all the 95 biological replicates for the 27 organs and tissues (Fig. 1B). The results reveal the relationship between the samples with distinct clusters for each tissue type and larger clusters representing the similar tissue from the gastro-intestinal tract (colon, duodenum, stomach, and small intestine) and tissues dominated by cells from the blood and immune system (lymph node, appendix, spleen, and bone marrow).
The Human Transcriptome
The total number of genes with detected transcripts was calculated for each sample and the results are shown in Fig. 2A. The genes were categorized based on mRNA abundance to reveal the number of genes in each category. With a cut-off of 1 FPKM, the number of detected genes ranged from 11,520 in the bone marrow to 15,224 in the testis. The overlap between genes identified in this study with putative human protein-coding genes annotated by UniProt (5) and the consensus gene consortium (CCDS) are shown in Fig. 2B. The Venn-diagram shows that more than 17,000 genes of the genes identified as transcribed here have previously been proposed as protein-coding by both the UniProt and CCDS consortia, whereas transcripts corresponding to 619 genes that were detected in the tissues analyzed in the present study have not yet been annotated as protein-coding genes by either UniProt or CCDS. These are important genes for further in-depth studies to establish if they indeed code for proteins.
Classification of all Protein-coding Genes Based on Transcriptomics Analysis
The transcriptomics analysis across the 27 organs and tissues allowed us to classify all the protein-coding genes (n = 20,050) according to pattern of tissue-specific expression (Fig. 2C). The definition of the various classes used in this study is shown in supplemental Table S2 with four major classes further subdivided into subclasses. The largest group of genes (46%) is the “housekeeping genes” that are expressed in all tissues, supporting earlier results (24) that many genes are ubiquitously expressed in human cells. The second class (28%) consists of the mixed categories with gene expression detected in 2–26 of the tissues, but not identified as tissue-specific. The third class is the tissue-specific genes that consist of two major subclasses; the tissue-enriched genes with at least fivefold higher expression in a specified tissue as compared with all other tissues and the group-enriched genes with at least fivefold higher expression in a small group of tissues as compared with all other tissues. The tissue-enriched genes constitute ∼12% of all genes (n = 2,473), with ∼3% highly tissue enriched with at least 50-fold higher mRNA level than any other tissue, and 9% moderately tissue enriched with at least a fivefold higher expression than any other tissue. Another 5% (n = 1026) of all genes are classified as group enriched in 2–7 tissues. Eight percent of all genes were not detected in any of the tissues analyzed here and a functional analysis (supplemental Fig. S2) shows that many of the undetected genes belong to olfactory receptors, keratin-associated proteins and genes involved in development. Thus, these genes will most likely be found when more specialized tissues and different developmental stages are included and they will most likely be added to the tissue-specific class of proteins. Almost half of the genes in the undetected group were unknown according to the Gene Ontology (GO) analysis and the question remains if these genes are true protein-coding genes or if they should be re-classified as pseudogenes or noncoding genes.
The Tissue-specific Human Proteome
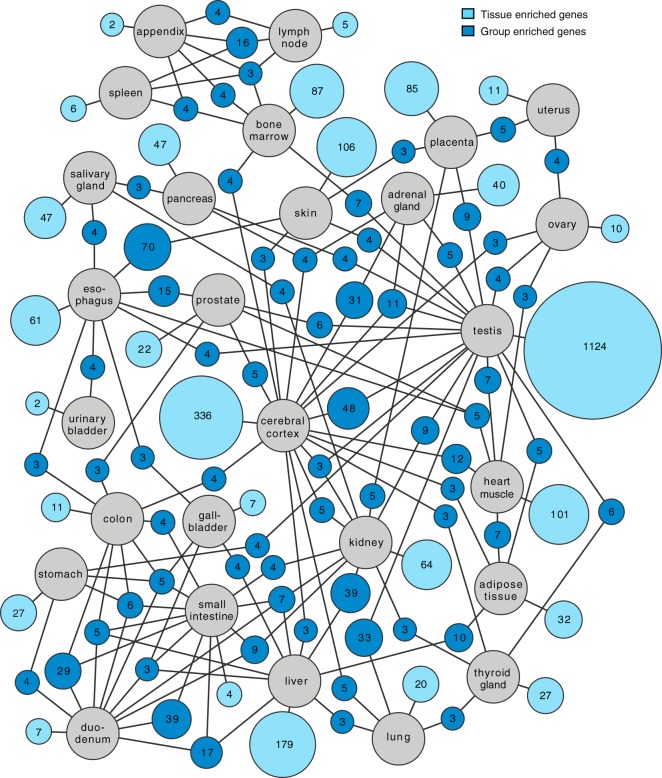
Altogether 3499 genes were defined as tissue-specific across the 27 tissues. These include genes identified as tissue-enriched and group-enriched (supplemental Table S3). Many well-known genes previously suggested to be tissue-specific were identified as tissue-enriched (supplemental Table S4) in our study, including insulin (pancreas), troponin (heart muscle), protamine (testis), haptoglobin (liver) and prostate specific antigen (prostate). A network analysis was performed for the tissue enriched and group enriched genes and the results are shown in Fig. 3, with the number of tissue enriched genes for each tissue displayed as well as group-enriched genes shared with another tissue. Most tissue-enriched genes are found in the testis followed by cerebral cortex, liver, and skin. Note that the tissues from the GI-tract and immune system have relatively few tissue-enriched genes, but instead have a large fraction of group-enriched genes. This is expected, because these tissues are similar and the cell type specific genes will thus be detected in more than one tissue.
Fig. 3.
Network plot showing the relationship of tissue enriched and group enriched genes in the various tissues and organs analyzed here. Blue circle nodes represent a group of expressed genes and are connected to tissues represented by gray circles. Dark blue nodes show the number of genes that are group enriched in up to five different tissue types (gray circles), with a minimum of three genes. The light blue nodes show the total number of highly and moderately tissue enriched genes in each tissue. The size of each blue node is related to the square root of the number of genes enriched in a particular combination of tissues.
Integration of the Transcriptomics Data on the Human Protein Atlas
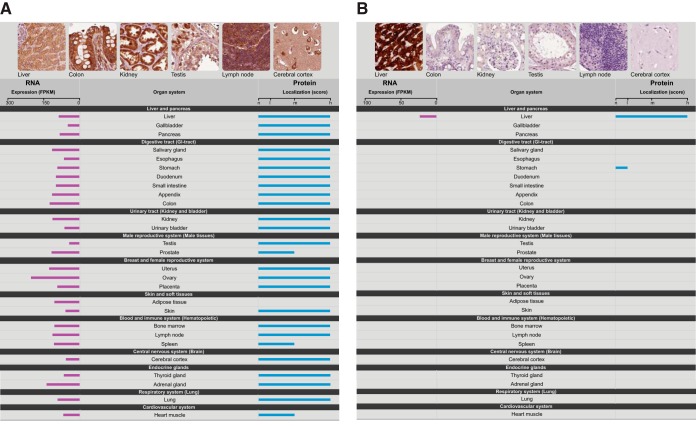
The transcriptomics data has been imported into the Human Protein Atlas to allow for visualization of the RNA expression patterns integrated with the results from more than 13 million individual antibody-based immunostainings (6) corresponding to more 80% of the protein coding genes. A new summary page for all genes has been designed to provide for comparative analysis of RNA/protein expression on an individual gene basis. In Fig. 4, two examples of summary pages are shown for a “house-keeping” gene (RPL24) and a tissue-specific gene (CYP2A13) with the RNA and protein level as assessed from the RNA-Seq data and antibody-based protein profiling data across the same 27 tissues. Note that the antibody for CYP2A13, in addition to the strong staining in liver, also stains one of the tissues in the GI-tract (stomach) weakly, whereas the RNA-Seq data show no transcripts for this gene in this tissue suggesting that the protein staining might be because of technical issues related to off-target binding. The comparative analysis of both RNA and protein levels can therefore be used as a validation tool to generate a protein atlas with higher reliability in the future.
Fig. 4.
Visualization of the RNA and protein expression levels across the 27 tissues. Examples of the presentation of expression data from the Protein Atlas, showing (A) a ubiquitously expressed gene (ribosomal protein L24, RPL24) and (B) a protein expressed specifically in one tissue (cytochrome P450, 2A13, CYP2A13).
In the new version of the Human Protein Atlas portal, the comparative analysis is shown for more than 80% of the putative protein-coding genes, including a visualization of the underlying primary data for the RNA-Seq with the number of reads corresponding to each base in the exons and introns of the analyzed gene. The lists of tissue-specific genes (supplemental Table S3) are available in the new version of the Human Protein Atlas (www.proteinatlas.org) portal to allow convenient exploration of the tissue-specific genes in various parts of the human body. The raw sequencing reads as well as the calculated mRNA levels (FPKM values) in each tissue, are also available at ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) under the accession number E-MTAB-1733. All the data including the classification of human protein-coding genes can also be downloaded from the Human Protein Atlas portal.
Antibody-based Profiling of the Tissue-specific Proteins
The analysis has given us lists of potential tissue-specific proteins, defined as highly or moderately tissue enriched, or group enriched. In order to validate the results, we have used the results from more than 13 million individual antibody-based protein profiles available in the Human Protein Atlas (6) to confirm the tissue-specificity also on the protein level. Immunohistochemistry allows a high resolution spatial mapping on a single cell level in the composite tissues and organs, to yield a more precise map of human protein expression. The various tissue-specific proteomes, such as cerebral cortex, testis, liver, kidney, fat, and pancreas, with accompanied protein data, will be published separately, but here we give a few examples illustrating the usefulness of combining the transcriptomics analysis with the antibody-based protein profiling.
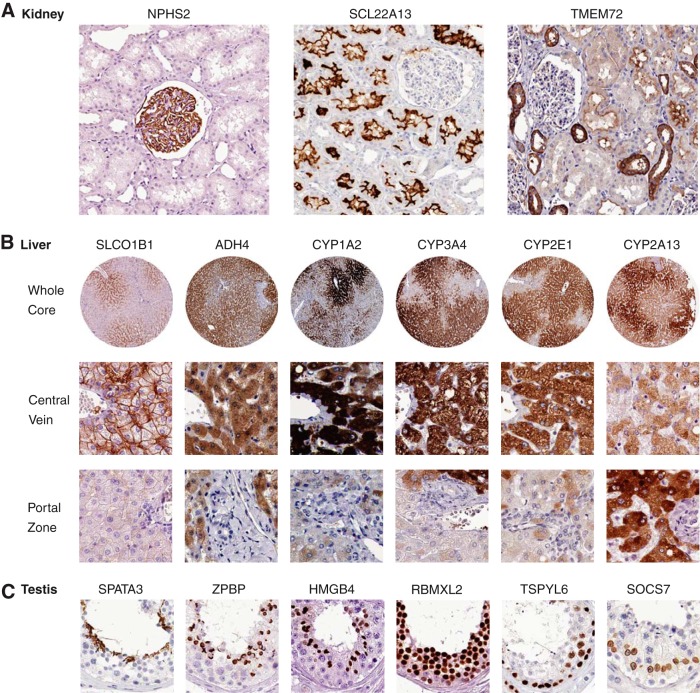
The first example is from the kidney-specific proteome for which 65 tissue-enriched genes were defined in our study. Immunohistochemistry was available for 62 of the corresponding proteins and the results show a strong bias for membrane-bound localization. In Fig. 5A, three examples of such proteins are shown, localized to distinct compartments in the human kidney, such as glomeruli (NPHS2), proximal tubuli (SLC22A13), and distal tubuli (TMEM72). Of the highly tissue enriched proteins, 18 out of 20 are transmembrane proteins as predicted by bioinformatics analysis and 16 of these are shown to localize to various subcompartments of the renal nephron by the immunohistochemistry analysis. Twelve of these proteins have previously been annotated as members of different solute carrier proteins (SLC), which mediate trans-membrane movement of electrolytes, endogenous metabolites, nutrients, micronutrients, and vitamins (25). Among the 65 kidney-enriched genes, we identified 15 genes previously not described as expressed in the kidney and we were able to confirm the localization of the corresponding proteins in the kidney by immunohistochemistry for a majority of proteins, as exemplified by TMEM72.
Fig. 5.
Examples of genes with selective expression pattern in kidney, liver and testis. All examples show immunohistochemistry images from the Human Protein Atlas (www.proteinatlas.org). A, Examples of kidney-specific proteins localized to different parts of the human kidney (glomerus, proximal and distal tubuli). B, Examples of liver-specific enzymes with a gradient-like expression in hepatocytes from the central vein to the portal zone. C, Examples of testis specific proteins localized to the various cell types representing the different phases of the spermatogenesis.
The second example is from the liver-specific proteome for which 179 tissue-enriched proteins were defined in our study. Immunohistochemistry-based protein profiling data was available for 141 of these proteins. Of the highly tissue enriched proteins, half (27 out of 54) are plasma proteins as predicted by bioinformatics analysis and 25 of these show plasma positivity by the immunohistochemistry analysis, including serpin peptidase inhibitors, haptoglobulin, and fibrinogens. Many of the other liver specific proteins are associated with known liver specific functions, such as drug, retinol and xenobiotics metabolism as well as primary bile acid biosynthesis. The antibody-based profiling allows a more in depth analysis of the protein expression pattern in the liver and in Fig. 5B, examples of detoxification enzymes with a heterogeneous protein expression pattern in the hepatocytes are displayed. A majority of the enzymes show a gradient-like expression pattern with high expression in hepatocytes surrounding the central vein and low expression adjacent to the portal zone, whereas CYP2A13 shows the opposite expression pattern.
The third example is from the testis. This male organ has by far the largest number of tissue-specific proteins (n = 1,378), accounting for more than one third of all identified tissue-specific proteins. Immunohistochemistry was available for 1061 of the testis tissue-specific proteins and a preference toward cytoplasmic and nuclear localization was observed, which is not surprising because many of the genes are involved in intracellular events, such as meiosis. Among the highest expressed genes are PRM1, PRM2, and TNP1, which are nuclear proteins known to be involved in the condensation of sperm chromatin during the haploid phase of spermatogenesis (26). We identified several previously uncharacterized proteins shown to be involved in various stages of spermatogenesis, such as SPATA3, ZPBP, HMGB4, RBMXL2, TSPYL6, and SOCS7 (Fig. 5C). The majority of the tissue-enriched genes belong to cell types representing various stages of spermatogenesis, whereas only a few proteins were localized in Sertoli and Leydig cells.
Splice Variants
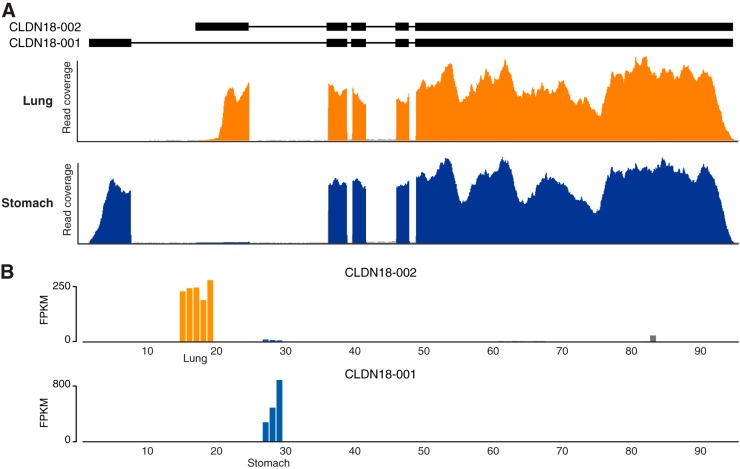
The RNA-Seq data allow us to investigate the presence of various transcripts in the different tissues. The question arises if analysis of splice variants coding for different proteins might yield evidence of additional tissue specificity. An example of this is displayed in Fig. 6, showing the transcript data for Claudin 18 (CLDN18), which is a gene encoding an integral membrane protein in tight junction strands. Tight junction strands serve as a physical barrier to prevent solutes and water from passing freely through the paracellular space between epithelial or endothelial cell sheets, and also play critical roles in maintaining cell polarity and signal transductions. The RNA-Seq data (Fig. 6) show that alternative initial exons are used in lung and stomach, respectively. A detailed analysis of the corresponding coding regions reveal that, although similar, the N-terminal region of the proteins (coded by the initial exons) differ in almost 1/3 of the residues between lung and stomach. Further studies are needed to elucidate how wide-spread this phenomenon is and to gain a more in-depth understanding of the molecular consequences in these two tissues and other similar cases.
Fig. 6.
Example of differential splicing in lung and stomach for the gene CLDN18. A, The exon structure (with introns scaled down 20-fold) of two different splice variants of CLDN18 are shown on top. Read coverage plots for lung (yellow) and stomach (blue) highlight the use of different initial exons in the two tissues. B, Transcript abundance (FPKM) plotted for all 95 samples, showing that each splice variant is specific for either of the two tissues. The yellow bars show expression levels for the five individual lung samples and the blue bars for the three stomach samples. Also some slight expression for one of the transcripts is detected in a single sample of heart muscle, shown by the gray bar.
DISCUSSION
The comprehensive analysis presented here has identified ∼3500 human genes that display a tissue- or group-enriched expression pattern across the human body. Functional analysis of these tissue-specific proteins identified in our analysis is well in line with the function of the respective tissue or organ. Thus, the kidney specific proteome consists of many membrane-bound transport proteins, whereas the most abundant tissue-specific proteins in liver are secreted plasma proteins, e.g. albumin and haptoglobin, and detoxification proteins, such as alcohol dehydrogenase and a large number of proteins belonging to the cytochrome P450 superfamily of enzymes. The most abundant pancreas-specific proteins are digestive enzymes, whereas many of the fat-specific proteins are involved in lipid metabolism. The analysis identified a multitude of tissue-specific genes with no or little previous evidence on the protein level and the combined RNA- and antibody-based profiling can thus be used to confirm the functional existence of these protein-coding genes lacking previous annotation. These proteins are of course interesting starting points for further in-depth studies to gain better molecular understanding of the cellular phenotypes that define the function of each respective tissue and organ.
The data presented here has allowed us to build a new resource with integration of RNA and protein expression data. As expected, the RNA expression levels as measured by RNA-Seq and the protein levels detected by staining with immunohistochemistry display low correlation for many genes. This is not surprising, because immunohistochemistry based on enzymatic amplification technology is not quantitative (27) and in addition often yields off-target binding to unrelated proteins. An important task for the future will therefore be to investigate the genes with low correlation between the RNA and protein levels. In many cases, this lack of correlation could also be because of biology, such as secretion of the protein out of the expressed cell type or post-translational events because of protein turn-over or other proteolytic processing, but the integrative RNA and protein data can also be used to indicate genes with potential technical issues, such as a context-depending off-target binding of the antibody or erroneous mapping of the RNA-Seq reads to the genome. The antibody-based protein profiling can be used for more precise localization patterns within the mixture of cell populations in a tissue, as shown here for the subcompartments of the renal nephron in the kidney, well-defined stages of spermatogenesis in the testis or gradient-like expression patterns in the liver.
The analysis presented here can be expanded in many different directions to provide an even more refined atlas of protein expression in the human body. Integration of this data with similar global efforts, such as the Cancer Genome Atlas program (28), the ENCODE (4) and the UniProt (5) efforts, will allow for a knowledge-base for molecular studies of the individual components of human proteome. Analysis of protein isoforms, in particular splice variant analysis of mRNA molecules, is an attractive extension. The results also facilitate comparative studies where the involvement of tissue-specific proteins are explored to identify targets for drug development and diagnostic assays for personalized medicine, including stratification of patients.
The integration of protein and RNA expression data is aimed toward the generation of a global, high-resolution expression map covering most of the tissues and cell types in the human body and this will be facilitated by alternative efforts based on transcriptomics analysis of specialized tissues, cells of different development origin and inclusion of cells and tissues originating from patients with different diseases. The classification of genes described here can thus be seen as step toward integration of RNA and protein data from various sources, including proteomics (29), to generate a high-resolution knowledge-base resource to allow for in-depth studies on individual genes and their protein counterparts, as well as more global studies using systems biology approaches.
Supplementary Material
Acknowledgments
We thank the entire staff of the Human Protein Atlas program and the Science for Life Laboratory for valuable contributions. We thank the Uppsala Biobank and the Department of Pathology at the Uppsala Akademiska hospital, Uppsala, Sweden and Uppsala Biobank for kindly providing clinical diagnostics and specimens used in this study.
Footnotes
Author Contributions. MU and FP conceived and designed the study. MU, LF and BMH wrote the paper. LF and BMH performed the bioinformatics analysis. FP performed the antibody-based profiling comparison. AD, KE, ST, MS, LM, FD and AA prepared the RNA samples. LF, MH, FP and BMH and PO performed the RNA-seq data analysis. FP, ES, CK and AA performed the tissue profiling. MS, PO, EL, FD, FP, ÅS, MZ, KJ, MH and JN contributed analysis, text and comments to the paper.
Author Information. The authors declare that they have no conflict of interest. Correspondence and requests for materials should be addressed to MU (mathias.uhlen@scilifellab.se).
* This work was supported by the Knut and Alice Wallenberg Foundation and PROSPECTS, a 7th Framework grant by the European Directorate (grant agreement HEALTH-F4-2008-201648/PROSPECTS).
 This article contains supplemental Figs. S1 and S2 and Tables S1 to S4 and Data Set S1.
This article contains supplemental Figs. S1 and S2 and Tables S1 to S4 and Data Set S1.
1 The abbreviations used are:
- RNA-seq
- transcriptomic analysis
- SLC
- solute carrier proteins
- FPKM
- fragments per kilobase of exon model per million mapped reads.
REFERENCES
- 1. Paik Y. K., Jeong S. K., Omenn G. S., Uhlen M., Hanash S., Cho S. Y., Lee H. J., Na K., Choi E. Y., Yan F., Zhang F., Zhang Y., Snyder M., Cheng Y., Chen R., Marko-Varga G., Deutsch E. W., Kim H., Kwon J. Y., Aebersold R., Bairoch A., Taylor A. D., Kim K. Y., Lee E. Y., Hochstrasser D., Legrain P., Hancock W. S. (2012) The Chromosome-Centric Human Proteome Project for cataloging proteins encoded in the genome. Nat. Biotechnol. 30, 221–223 [DOI] [PubMed] [Google Scholar]
- 2. Flicek P., et al. (2013) Ensembl 2013. Nucleic Acids Res. 41, D48–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pruitt K. D., Tatusova T., Brown G. R., Maglott D. R. (2012) NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 40, D130–D135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. The Encode Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Magrane M., and UniProt Consortium (2011) UniProt Knowledgebase: a hub of integrated protein data. Database 2011, bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S., Wernerus H., Björling L., Ponten F. (2010) Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 28, 1248–1250 [DOI] [PubMed] [Google Scholar]
- 7. Su A. I., Wiltshire T., Batalov S., Lapp H., Ching K. A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Cooke M. P., Walker J. R., Hogenesch J. B. (2004) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 101, 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S., Hodge C. L., Haase J., Janes J., Huss J. W., Su AI. (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 10, R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brazma A., Parkinson H., Sarkans U., Shojatalab M., Vilo J., Abeygunawardena N., Holloway E., Kapushesky M., Kemmeren P., Lara G. G., Oezcimen A., Rocca-Serra A., Sansone S-A. (2012) ArrayExpress–a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 31, 68–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krupp M., Marquardt J. U., Sahin U., Galle P. R., Castle J., Teufel A. (2012) RNA-Seq Atlas–a reference database for gene expression profiling in normal tissue by next-generation sequencing. Bioinformatics 28, 1184–1185 [DOI] [PubMed] [Google Scholar]
- 11. https://github.com/najoshi/sickle. Sickle - A windowed adaptive trimming tool for FASTQ files using quality.
- 12. Trapnell C., Pachter L., Salzberg S. L. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. http://picard.sourceforge.net/. Picard.
- 14. Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L., Wold B. J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kircher M., Sawyer S., Meyer M. (2012) Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40, e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. (2009) GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ponten F., Jirstrom K., Uhlen M. (2008) The Human Protein Atlas–a tool for pathology. J. Pathol. 216, 387–393 [DOI] [PubMed] [Google Scholar]
- 19. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kampf C., Andersson A-C., Wester K., Björling E., Uhlen M., Ponten F. (2004) Antibody-based tissue profiling as a tool in clinical proteomics. Clin. Proteomics 1, 285–300 [Google Scholar]
- 21. Paavilainen L., Edvinsson A., Asplund A., Hober S., Kampf C., Pontén F., Wester K. (2010) The impact of tissue fixatives on morphology and antibody-based protein profiling in tissues and cells. J. Histochem. Cytochem 58, 237–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2009) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 [DOI] [PubMed] [Google Scholar]
- 23. Hebenstreit D., Fang M., Gu M., Charoensawan V., van Oudenaarden A., Teichmann S. A. (2011) RNA sequencing reveals two major classes of gene expression levels in metazoan cells. Mol. Syst. Biol 7, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lundberg E., Fagerberg L., Klevebring D., Matic I., Geiger T., Cox J., Algenäs C., Lundeberg J., Mann M., Uhlen M. (2010) Defining the transcriptome and proteome in three functionally different human cell lines. Mol. Syst. Biol 6, 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schlessinger A., Matsson P., Shima J. E., Pieper U., Yee S. W., Kelly L., Apeltsin L., Stroud R. M., Ferrin T. E., Giacomini K. M., Sali A. (2010) Comparison of human solute carriers. Protein Sci. 19, 412–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sassone-Corsi P. (2002) Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science 296, 2176–2178 [DOI] [PubMed] [Google Scholar]
- 27. Rimm D. L. (2006) What brown cannot do for you. Nat. Biotechnol. 24, 914–916 [DOI] [PubMed] [Google Scholar]
- 28. The Cancer Genome Atlas Research Network, Weinstein J. N., Collisson E. A., Mills G. B., Mills Shaw K. R., Ozenberger B. A., Ellrott K., Shmulevich I., Sander C., Stuart J. M. (2013) The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lane L., Argoud-Puy G., Britan A., Cusin I., Duek P. D., Evalet O., Gateau A., Gaudet P., Gleizes A., Masselot A., Zwahlen C., Bairoch A. (2012) neXtProt: a knowledge platform for human proteins. Nucleic Acids Res. 40, D76–D83 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data (FPKM values for all the samples) are available for downloads without any restrictions (www.proteinatlas.org/about/download). The primary data (reads) are available through the Array Express Archive (www.ebi.ac.uk/arrayexpress/) under the accession number: E-MTAB-1733. The transcript profiling data (FPKM values) for each gene in each cell and tissue will also be available in the next version of the Human Protein Atlas (www.proteinatlas.org). The classification according to Fig. 2C will be included on the Protein Atlas gene summary page for each of the genes. FPKM values for all 20,050 genes included in the study are provided as downloads to facilitate comparative studies with other “omics” data.
Fig. 2.
The classification of all human protein-coding genes with regards to transcriptional levels in 27 tissues. A, The total number of genes with detected transcripts in each cell type using five different abundance levels for FPKM values; 1–5 FPKM (yellow), 5–20 FPKM (orange), 20–100 FPKM (light red), 100–500 FPKM (red), >500 FPKM (dark red). B, Venn-diagram showing the overlap between a total of 20,083 human Ensembl genes based on three different datasets: (1) genes detected in one or more tissues used in this study (RNA-seq), (2) the core set of genes defined by the Consensus CDS project (CCDS), (3) genes with transcript or protein evidence in UniProt. C, Piechart showing the distribution of all 20,050 genes into eight different categories based on the transcript abundance as well as the number of detected tissues.