Abstract
Tetrahydrolipstatin (THL) is bactericidal but its precise target spectrum is poorly characterized. Here, we used a THL analog and activity-based protein profiling to identify target proteins after enrichment from whole cell lysates of Mycobacterium bovis Bacillus Calmette-Guérin cultured under replicating and non-replicating conditions. THL targets α/β-hydrolases, including many lipid esterases (LipD, G, H, I, M, N, O, V, W, and TesA). Target protein concentrations and total esterase activity correlated inversely with cellular triacylglycerol upon entry into and exit from non-replicating conditions. Cellular overexpression of lipH and tesA led to decreased THL susceptibility thus providing functional validation. Our results define the target spectrum of THL in a biological species with particularly diverse lipid metabolic pathways. We furthermore derive a conceptual approach that demonstrates the use of such THL probes for the characterization of substrate recognition by lipases and related enzymes.
Mycobacterium tuberculosis, the causative agent of tuberculosis (TB), is responsible for nearly 2 million deaths each year. The host immune response toward aerosol infection is to quarantine tubercle bacilli in a granulomatous structure (1, 2). However, granuloma-associated mycobacteria can switch to a non-replicative, “dormant” state and successfully evade immune response for decades after infection (3, 4). The metabolic events that permit tubercle bacilli to enter host cells and revive from states of persistence suggest that lipids are utilized as a carbon source (5–7). During times of oxygen deprivation and in the absence of host cells, cultivated mycobacteria store fatty acids (FAs) in the form of triacylglycerol (TAG)1-enriched lipid droplets (8–10). Upon resuscitation (by the re-introduction of oxygen), these lipid droplets vanish and TAGs are hydrolyzed (11). Unfortunately, the molecular mechanisms for TAG build-up and breakdown are far less well understood in bacteria when compared with those processes in eukaryotes.
Comparative sequence analysis of the Mtb genome has revealed that it contains 250 genes encoding enzymes involved in lipid metabolism compared with only 50 enzymes in Escherichia coli, which has a genome of comparable size. Among these genes, 150 are predicted to encode proteins involved in lipid catabolism (12, 13). A family of 24 carboxyl ester hydrolases called “lip” genes (lipC to Z, except K and S) has been predicted to play a role in lipid catabolism (14). Among these, only a few have been functionally characterized and related to mycobacterial dormancy and resuscitation (15–18).
Tetrahydrolipstatin, a serine esterase inhibitor, covalently binds to and inhibits mammalian lipases and fatty acid synthase (FAS) and is marketed as “Orlistat” for the treatment of severe forms of obesity (19). THL was previously shown to inhibit both active and latent forms of mycobacteria (11, 20–22) but the bacterial target spectrum remains poorly characterized. Therefore, to (1) define the THL target spectrum in a mycobacterial species and (2) to obtain biochemical insights into regulation of lipases and esterases in different metabolic states, we employed a chemical-proteomics approach using activity-based protein profiling (ABPP) with a bait that has been described to bind to lipolytic enzymes (23–25). We identified several known lipases (as anticipated), putative lipase and esterases, and hypothetical proteins of unknown functions, thereby providing a comprehensive resource of experimentally determined THL targets in mycobacteria. Importantly, we systematically compared readouts of fluorescently tagged THL-proteins (7 bands on one-dimensional SDS-PAGE) with those of mass spectrometry-based peptide identification of enriched protein fractions (247 in growing cells). This comparison led to the identification of 14 THL targets, two of which were further validated experimentally. We furthermore provide a conceptual framework for the evaluation of this target list using both experimental as well as bioinformatics approaches in two examples, lipH and tesA. Overall, our data indicate that THL is an anti-mycobacterial drug because of its potential to (1) bind to a relatively wide range of lipolytic enzymes and (2) prevent bacilli from resuscitating from a nonreplicating persistent (NRP) state when lipid metabolism is particularly important.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
Mycobacterium bovis BCG Pasteur strain (ATCC 35734) and over-expression mutants were grown in Dubos liquid medium (Difco, USA) supplemented with 10% Dubos medium albumin (Difco), and 0.05% Tween 80, or in Middlebrook 7H11-OADC agar (Difco) at 37 °C. For precultures, 7H9-OADC-Tween 80 (Difco) was used. Where required, kanamycin (Sigma-Aldrich) (30 μg/ml) was added to the culture medium.
Culturing conditions, including logarithmic (or exponentially growing), hypoxia-induced nonreplicating persistent (NRP) and re-growth from NRP, were adapted from standardized protocols (26). Inoculums used for starting cultures were prepared from single colonies picked from 7H11 agar plates and the pre-cultures were propagated in roller bottles rotated at 50 rpm for 3 days with an initial optical density at 600 nm (OD600 nm) of 0.05. NRP BCG cultures were cultivated by subjecting bacilli to the slow withdrawal of oxygen as described elsewhere (27). Briefly, aerobic cultures (780 ml), with an initial OD600 nm of 0.005, were expanded in glass bottles with a capacity of 1000 ml (Duran, Germany), achieving an optimal headspace ratio (HSR) of air volume to liquid volume. The sealed cultures were stirred gently at 80 rpm for 18 days to allow the bacilli to enter into NRP. Subsequently, fresh oxygenated air was re-introduced to hypoxic cultures through loosening of the caps to exit from NRP for 3 days. This exposure of air allowed NRP bacilli to regrow. Oxygen depletion in these cultures was monitored by the use of the methylene blue as an oxygen indicator, as described previously (27). Growth and survival were monitored by enumeration of CFU on Middlebrook 7H11 agar after plating of appropriate dilutions. Plates were sealed with parafilm and incubated at 37 °C for 4 weeks.
Chemicals, THL Analogs and Click Reagents
Tetrahydrolipstatin (Orlistat) was purchased from Sigma-Aldrich. THL-alkyne analogs, rhodamine-azide, biotin-azide and the trifunctional probe were synthesized as described previously (25). THL and its analogs were dissolved in DMSO for all experiments.
Construction of Overexpression Plasmids
lipN (BCG2991c), lipH (BCG1460c), lipV (BCG3229) and tesA (BCG2950) were amplified (via polymerase chain reaction, PCR) from genomic DNA isolated from M. tuberculosis H37Rv using the primers listed in supplemental Table S2A. The PCR fragments were inserted into BglII and HindIII sites of an E. coli- Mycobacterium shuttle vector pMV262 (28) and placed under the control of the constitutive HSP60 promoter. The constructs were electroporated into wild-type (WT) M. bovis BCG, and transformants were selected on 7H11 plates with kanamycin. Overexpression of genes in isolated colonies from the plates was assessed using quantitative reverse transcription-PCR (qRT-PCR) using the primers listed in supplemental Table S2B.
Drug Inhibitory Concentration
The growth inhibition curve of M. bovis BCG by THL and the alkyne handle-containing analog compound was obtained in a 96-well plate assay. THL dilutions in the logarithmic concentrations were prepared in DMSO and added to 200 μl of 0.02 OD600 nm culture of BCG at different metabolic states. The final DMSO concentration was maintained at 0.5%. The plates were then incubated at 37 °C for 5 days without shaking after which the OD600 nm was recorded using SpectraMax M2 spectrophotometer. Media with 0.5% DMSO and no inoculum is recorded as blank. MIC50 (minimum inhibitory concentration required to inhibit the growth of 50% of organism) was determined by plotting the mean average slope of absorption (derived from three independent experiments) versus probe concentration and the concentration of 50% inhibition was determined using GraphPad Prism software, version 5.0a. The minimum bactericidal concentration (MBC) for THL was calculated by plating appropriate dilutions of bacterial cultures that were grown for 5 days in the presence of various THL concentrations on 7H11 agar plates and colony forming units (CFU) were counted after 3 weeks of incubation at 37 °C. Data obtained from three independent experiments was analyzed using GraphPad Prism software, version 5.0a, USA.
Huisgen Cycloaddition Reaction
M. bovis BCG total protein concentration was adjusted to 1 μg/μl using phosphate-buffered saline (PBS) and 100 μg of protein was incubated with 5 μm of THL-alkyne analogs at room temperature for 1 h. For in situ labeling, 100 ml of bacterial pellet was incubated with 10 μm of THL-alkyne for 2 h at room temperature, the pellet was washed with PBS, lysed and total protein was estimated. Subsequently, the Huisgen cycloaddition reaction was initiated by adding 20 μl of a freshly premixed solution containing 1 mm CuSO4 (Sigma-Aldrich), 1 mm tris-(Benzyltriazolylmethyl)amine (TBTA) (Sigma-Aldrich), 100 μm tris(2-carboxyethyl)phosphine (TCEP) (Sigma-Aldrich), and 100 μm rhodamine-azide (supplemental Fig. S1B) or the trifunctional probe (supplemental Fig. S1D). The reaction was stirred at room temperature for 2 h in the dark and proteins were precipitated by the addition of five volumes of ice-cold acetone followed by overnight incubation at −20 °C. Protein pellets were then washed with methanol and resuspended in 50 μl of 1× sodium dodecyl sulfate (SDS) loading dye. Approximately 10 μg of protein was separated on 12% SDS-PAGE, followed by in-gel fluorescence scanning with Typhoon 9410 Variable Mode Imager scanner (GE Amersham Biosciences, USA) and total protein was visualized using Coomassie R-250 (Bio-Rad, USA).
For target enrichment, three sets of cycloaddition reactions followed by peptide LC-MS analysis were carried out with slight modifications to control for consistency. Set 1 involved identifying individual gel bands between 60 and 20 kDa at logarithmic phase BCG culture, whereas set 2 involved identifying targets at different metabolic states (L, N and R), and for set 3 the target enrichment was performed using trifunctional probe. For all the above experiments, 2 mg protein was used to perform the Huisgen cycloaddition reaction as mentioned previously, except that biotin-azide (supplemental Fig. S1C) or the trifunctional probe (supplemental Fig. S1D) was substituted for rhodamine-azide. After acetone precipitation, proteins were resuspended in 0.2% SDS in PBS and briefly sonicated. The supernatants were then incubated with 25 μl of prewashed NeutrAvidin agarose beads (Thermo Scientific). Beads were collected by centrifugation and washed 3× with 1 ml of buffer A (8 m urea, 200 mm NaCl, 2% SDS, 100 mm Tris pH 8), buffer B (8 m urea, 1.2 m NaCl, 0.2% SDS, 100 mm Tris pH 8, 10% ethanol, 10% isopropanol), buffer C (8 m urea, 100 mm Tris pH 8), and finally with PBS. Proteins were eluted from beads with 25 μl of 2× SDS-loading dye and incubation at 96 °C for 20 min. Twenty microliters of elute was separated on a 12% SDS-PAGE and proteins were visualized by in-gel fluorescence scanning, Coomassie Blue staining or silver staining (29). Protein bands corresponding to both DMSO-treated (control) and THL-alk1 treated samples were excised as single bands (set 1) or entire lanes (set 2 and set 3) followed by in-gel trypsin digestions as described previously (30). The eluted peptides were dried under vacuum and stored at −20 °C until analysis.
Peptide Analysis by Mass Spectrometry
Before analysis by peptide mass spectrometry (MS), the dried tryptic-digested peptides were dissolved in 1% formic acid and peptides were separated and analyzed on a ultra-fast liquid chromatography (UFLC) system (Shimadzu, Japan) coupled to an Linear ion trap-Fourier transform (LTQ-FT) Ultra MS (Thermo Electron, USA). Peptides were first enriched with a Zorbax 300SB C18 column (5 mm x 0.3 mm, Agilent Technologies) followed by elution into an integrated nanobore column (75 μm × 100 mm, New Objective, USA) packed with C18 material (5 μm particle size, 300 Å pore size, Michrom BioResources, USA). A 60 min gradient was developed using mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile). The run comprised of 45 min of 5–35% B, 8 min of 35–50% B, and 2 min of 80% B followed by re-equilibrating at 5% B for 5 min. Peptides were analyzed with an ADVANCE CaptiveSpray Source (Michrom BioResources, USA). LTQ-FT-MS was operated at electrospray potential of 1.5 kV in positive mode at a full m/z range of 350–1600. A gas-flow of 2 L/min, ion-transfer tube temperature of 180 °C and collision-gas pressure of 0.85 mTorr was used. The raw data were converted into mgf format as described previously (31).
Database Searching
Peptide and protein identification was performed using an in-house MASCOT search engine (version 2.2.07; Matrix Science, USA) with an MS tolerance of 10 ppm and MS/MS tolerance of 0.8 Da. MASCOT was set up to search Mycobacterium bovis BCG str. Pasteur 1173P2 FASTA file downloaded from NCBI (3949 entries; NCBI Reference Sequence: NC_008769.1; Accession number: PRJNA58781; ID: 58781). Two missed cleavage sites of trypsin were allowed. Parameters for both search engines were set to a fragment ion mass tolerance of 1.0 Da and a parent ion tolerance of 2.5 Da. Oxidation of methionine, iodoacetamide derivative of cysteine and phosphorylation at serine, threonine, or tyrosine were specified in Mascot as variable modifications.
Criteria for protein identification
The proteins were manually filtered from DMSO-treated (control) list and identified based on their scores, number of unique matching peptides, protein abundance and molecular weight. Protein scores greater than 80 (p < 0.05), two or more numbers of unique matching peptides and protein abundance (emPAI) score greater than 0.1 correspond to confident identifications. Respective accession numbers, COGs and their functional information were obtained from the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/), BCGList and TubercuList databases (http://tuberculist.epfl.ch). ClustalW was used for multiple sequence alignment and predictions of transmembrane helices (TMHMM Server version 2.0) and signal peptides (SignalP Server version 3.0) were carried out with the respective software at the ExPASy Proteomics server (http://ca.expasy.org/). The structural and functional annotation was obtained from Superfamily database version 1.75 (http://supfam.cs.bris.ac.uk). Cellular localization and gene essentiality information was obtained from TubercuList databases (http://tuberculist.epfl.ch).
Lipid Analysis
Fifty milligrams (dry weight) of M. bovis BCG was used for lipid extraction. For TAG lipid extraction, cells were resuspended in 3 ml of chloroform-methanol (2:1, v/v) and phases were separated with 1.5 ml of water. The lower organic phase was dried under a stream of nitrogen and stored at −20 °C until further analysis. For thin layer chromatography (TLC), the dried lipids were resuspended in 100 μl of chloroform/methanol (1:1, v/v) and 10 μl of the resuspended lipids were spotted on silica gel 60-coated high-performance TLC (HPTLC) plates (20 × 20 cm, Merck, USA). The plates were developed using hexane-diethyl ether-formic acid (45:5:1, v/v/vol) as the solvent system. Once dried the plates were sprayed uniformly with 5% phosphomolybdic acid in 95% ethanol and heated at 125 °C for 10 min to visualize total lipid.
The LC-MS analysis of TAG and short-chain esters was performed by resuspending the dried lipids in 200 μl chloroform-methanol (1:1, v/v) and 5 μl of the resuspended lipid was subject to analysis by liquid chromatography coupled with electrospray ionization mass spectrometry (LC-ESI-MS) on an LTQ-Orbitrap XL (Thermo scientific, USA). A Zorbax Eclipse XDB-C18 column (3.0 × 150 mm, 1.8 μm particle size, Agilent Technologies, USA) and a mobile phase consisting of chloroform-methanol (1:1, v/v) containing 2 mm ammonium acetate were used to separate lipids TAG from polar lipids (32). The separation was done isocratically at a flow rate of 130 μl/min. The total run-time was 20 min. The LTQ-orbitrap XL was operated with positive ionization mode with the following parameters: source voltage 3.5 kV, capillary voltage 35 V, capillary temperature 300 °C, tube lens 130 V, sheath gas flow 30.0, auxiliary gas flow 5.0. Under these conditions, the TAG ionizes as ammonium adducts and elute between 4.5 min and 19 min depending on fatty acyl chain length and unsaturation. A mixture of six TAG standards was used to control for retention time (trihexanoyl-, trioctanoyl-, tridecanoyl, tridodecanoyl-, trimyristoyl-, and tripalmitoyl-glycerol, eluting ca. 4.6 min, 5.8 min, 6.5 min, 7.5 min, 8.9 min, and 10.7 min, respectively). Deuterated tripalmitoyl-glycerol (d5-TAG, C51H972H5O6) was used as internal standard (m/z 829.8, retention time ca. 10.7 min). For MS/MS experiment, the linear ion trap was used with an isolation width of 1.0 dalton and collision induced dissociation (CID) at a normalized collision energy of 35 V.
Data Acquisition and Processing
Full scan MS data were acquired using Xcalibur software (Thermo Scientific, USA) at a resolution of 100,000. Product ion scans were acquired in ion trap mode. Lipid data were manually curated using Xcalibur as well as online LipidMaps lipid database (http://www.lipidmaps.org) with an MS tolerance of 8 ppm based on the m/z of ammonium adducts ([M+NH4]+). The identification was based on both accurate mass and retention times. Further identification by product ion scans was undertaken for selected species. Under CID, TAG ions undertake neutral losses of the fatty acyl chains, as ammonium salt.
Enzymatic Assays and IC50
The activity of recombinant LipH expressed in M. bovis BCG was measured by using p-nitrophenyl (pNP) ester (Sigma Aldrich) with butyrate chains (C4). The release of pNP was monitored at 400 nm by using a 96-well plate spectrophotometer and quantified by using a calibration curve of pNP-C4. Enzymatic reactions were performed with 2.5 mm Tris buffer (pH 8.0) containing 300 mm NaCl with 1 mm sodium tauro-deoxycholate (NaTDC) at room temperature over a period of 15 min in a final volume of 250 μl containing various amounts of enzyme and 1 mm substrate. Results are expressed as specific activities in international units/mg, corresponding to 1 μmol pNP released per minute and per mg of enzyme. IC50 was determined by plotting the mean average slope of absorption (derived from three independent experiments) versus probe concentration and the concentration of 50% inhibition of reaction was determined using GraphPad prism software, version 5.0a.
Staining and Flow Cytometry
M. bovis BCG cells were washed with PBS-T (0.05% Tween 80) and incubated with 50 ng/ml of LD540 (lipophilic dye based on the Bodipy fluorophore) for 15 min at room temperature. For esterase activity, washed cells were incubated with 1 μm of 5-Chloromethylfluorescein Diacetate (CMFDA, Invitrogen, USA) for 15 min at 37 °C. Stained cells were washed with PBS and fixed with 4% paraformaldehyde for 15 min. Cell clumps were removed by filtering the sample through polyamide 60 μm mesh fabric (Sefar, Buffalo, NY). Data was acquired using a BD LSR Fortessa cell analyzer (BD Biosciences) equipped with an air-cooled 15 mW, 500 nm argon laser. The flow rate was adjusted to 100–200 events per second. Flow cytometry data were analyzed using the FlowJo software, version 8.8.7 (TreeStar, Ashland, OR).
RNA isolation and qRT-PCR
Mycobacterial culture pellets were washed with PBS and mixed with two volumes of cell lysis buffer (Qiagen, Valencia, CA). The cell lysate was then transferred to screw cap tubes containing 0.1 mm-diameter silica beads (BioSpec Products, Bartlesville, OK). Cells were disrupted for 5 min at 50 revolutions using a bead beater at 4 °C, and unlysed bacteria and cell debris were pelleted by centrifugation. The supernatants were used to isolate total RNA with RNeasy mini kit (Qiagen, USA) according to the manufacturer's protocol. DNase-treated RNA samples (1 μg) were reverse transcribed using 50 ng random hexamer and SuperScript III RT-PCR kit (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. Quantitative PCR reactions were carried out using Taqman RT-PCR master mix kit (Applied Biosystems, Foster City, CA) containing 30 ng of cDNA template, 250 nm primers and 125 nm probe (Table S2b) with standard conditions as recommended by the manufacturer. Incubations without transcriptase/template were included as negative controls. Relative quantification was performed using the comparative threshold cycle (CT) method. Data was analyzed using GraphPad Prism software, version 5.0a.
In Silico Characterization of LipH
A homology model of the 3D structure of LipH was generated using the structural homologs of LipH which were identified using protein BLAST (http://www.ncbi.nlm.nih.gov/) against PDB database and selected as templates based on the highest sequence identity presented. The Molecular graphics software program Swiss PDB Viewer was used to thread the protein sequence on its structural homologs using MUSCLE package of Swiss PDB viewer (33) and thereafter initial structural alignments were generated after which a modeling request was submitted to Swiss model server (http://swissmodel.expasy.org/) and further refined by a fragment-guided molecular dynamic simulations FG-MD program (34), energy minimized with GROMACS 4.0.7 package (http://www.gromacs.org/) and validated using Structural Analysis and Verification Server (http://nihserver.mbi.ucla.edu/SAVES/). Docking was performed using Autodock Vina (35) (http://autodock.scripps.edu/), which uses a genetic algorithm to dock flexible ligands into flexible active site of protein. The ligand 3D structures were constructed and all-atom charges were assigned with BUILDER module of molecular modeling program Accelrys Insight-II. The active site was then analyzed using the CASTp program and molecular overlay was done using Accelrys Discovery Studio. ClustalW v2 was used for multiple sequence alignment and predictions of transmembrane helices (TMHMM Server v2.0) and signal peptides (SignalP Server v3.0) were carried out at the ExPASy Proteomics server (http://ca.expasy.org/).
Statistical Analysis
Data obtained from at least three independent experiments was combined together for statistical analysis. Results were analyzed either using One-way ANOVA or Student's t test. Data are represented as the mean values either ± S.E. or ± standard deviation (n = 3) where implied. * p < 0.05, ** p < 0.01, *** p < 0.001 was considered to be significant unless otherwise noted.
Proteomics
For LC-MS based proteomics analysis, three independent biological replicate experiments were performed to identify THL targets in logarithmic culture of M. bovis BCG. Cell extracts treated only with DMSO (instead of THL-alk) were used as control incubations. Proteins present in these control experiments were removed from the target list. Next, only proteins with a score > 80 (p < 0.05), at least two or more unique matching peptides, and a protein abundance index (emPAI) of > 0.1 were considered confident identifications (supplemental Table S1, “THL targets_Log”). The % coverage for each protein on supplemental Table S1 was obtained from MASCOT server. The same approach was taken to determine THL in the other metabolic states, except that only two replicate experiments were performed (supplemental Table S1, “THL targets_NRP” and “THL targets_Regrowth”).
Growth and Enzymatic Assays
Data obtained from at least three independent experiments (total nine technical replicates) were combined together for statistical analysis. Graphical data is plotted using GraphPad Prism software (version 5.0a, USA). Dubos or 7H9 media with 0.5% DMSO was recorded as blank. IC50 was determined by plotting the mean average slope of absorption versus probe concentration and the concentration of 50% inhibition was determined using GraphPad Prism software. Statistical significance was tested using One-way ANOVA. All bar and line graphs represent mean value and error bar represents S.E. p < 0.05 was considered to be significant.
Lipidomics
For TLC lipid analysis, data obtained from at least three independent experiments was analyzed using ImageJ software (version 1.47, USA) to estimate the band intensity. The LC-MS analysis of TAG and short-chain esters was obtained from three different biological replicate and one representative data is shown.
Flow Cytometry
Flow cytometry data were analyzed using the FlowJo software, version 8.8.7 (TreeStar). Data obtained from at least three independent experiments was combined together for statistical analysis. Statistical significance was tested using One-way ANOVA and are represented as mean values ± S.E. (n = 3). p < 0.05 was considered to be significant.
qRT-PCR
Relative quantification was performed using the comparative threshold cycle (CT) method. The ΔCT value against endogenous control (sigA) was normalized against ΔCT value and the obtained ΔΔCT value was used to calculate relative quantity (RQ). Data obtained from at least three independent experiments was combined together and plotted as log2 ratios means using GraphPad Prism software. All bar and line graphs represent mean value and error bar represents standard deviation (n = 3).
RESULTS
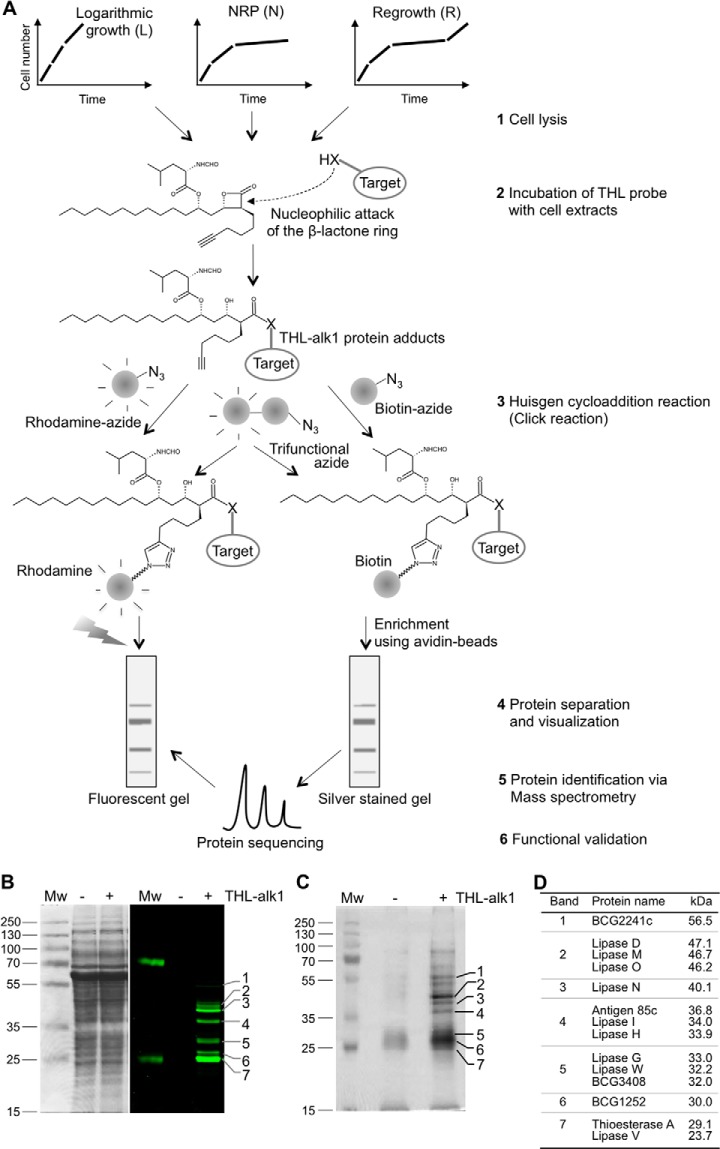
The main purpose of this study was to identify, biochemically, the targets of THL in M. bovis Bacillus Calmette-Guérin (BCG) in different metabolic states. To do so, we used recently developed probes (25) with terminal C-C triple bonds (Fig. 1A, supplemental Fig. S1A). Bacteria were grown in liquid medium and culture conditions originally developed by Wayne (3). These led to exponential growth (L) and non-replicating persistence (NRP or N) induced by limitation of oxygen. Re-growth (R) from NRP was achieved by re-introduction of fresh air into NRP cultures. Cells were harvested from these three conditions, washed and lysed (Fig. 1A, step 1) and THL probes were incubated with total cell lysates to allow binding with proteins (Fig. 1A, step 2). The β-lactone of THL is known to form covalent adducts with active site residues of target enzymes (19) via activity-based mechanisms (36). Importantly, the resulting cell lysates were next processed for (1) visualization using a fluorophore (rhodamine, supplemental Fig. S1B), or (2) enrichment via affinity chromatography using the biotin (supplemental Fig. S1C)-streptavidin system or using the trifunctional probe (supplemental Fig. S1D) that contains a biotin group in addition to its rhodamine fluorophore. The required modifications for either option were introduced by the Huisgen cycloaddition reaction (click reaction, Fig. 1A, step 3). Protein extracts were next separated by SDS-PAGE (Fig. 1A, step 4) and band patterns were compared with protein identifications obtained by mass spectrometry (Fig. 1A, step 5), followed by functional validation of some of the target enzymes (Fig. 1A, step 6).
Fig. 1.
THL target spectrum in M. bovis BCG largely comprises lipid esterases. A, Schematic representation of the workflow and steps involved in identification of THL targets. M. bovis BCG was grown in three different physiological states in vitro, cells lysed by sonication (step 1) and total cell extracts incubated with THL-alk1 (step 2). The alkyne moiety of THL-alk1 was then used to either tag (via the Huisgen cycloaddition reaction) protein-THL adducts with rhodamine-azide or biotin-azide or trifunctional-azide (step 3). Whole cell lysates (in the case of rhodamine tagged adducts) or enriched fractions (in the case of biotin tagged adducts) were separated by SDS-PAGE (step 4, B, C). Protein identities (obtained by sequencing of peptides in enriched fractions via mass spectrometry, step 5, C) were compared with visualized protein patterns (step 4) to derive the THL target list (D) from which TesA (migrating with band 7) and LipH (migrating with band 4) were further validated (step 6). B, Mycobacterial cell lysates incubated with THL-alk1 or DMSO as a control were tagged to a rhodamine-azide dye by the Huisgen cycloaddition reaction. Equal amounts of proteins were separated on SDS-PAGE and visualized by Coomassie staining (left panel) or in-gel fluorescence (right panel). C, THL-alk1 bound targets were enriched using biotin-avidin affinity chromatography. Enriched fractions were separated on SDS-PAGE and stained with silver. The bands indicated on the right side were excised and subjected to tandem MS analysis. D, THL targets in M. bovis BCG total cell extracts derived from logarithmically growing cultures. Only proteins found consistently in at least 2 out of 3 three replicate experiments, not present in control incubations (DMSO in the absence of THL-alk1), and, with matching, co-migrating fluorescent targets (B), were included in this list.
THL Target Spectrum Largely Encompasses Lipid Esterases
Typical results of such SDS-PAGE are shown in Fig. 1B (fluorescence) and Fig. 1C (silver stained gel of enriched protein fraction). Distinct bands were visible in both cases when THL-alk1 was used during incubation with little to no signal in control reactions where the probe was omitted. Bactericidal activities of three THL analogs (THL-alk1 and two additional ones which carry the C-C triple bond in different positions, supplemental Fig. S1A) were tested and their fluorescent target profiles compared. Although the inhibitory effect of THL-alk1 was indistinguishable from that of THL (i.e. the hydrated form of the “natural” inhibitor, lipstatin), THL-alk2 and THL-alk3 were slightly less effective (supplemental Fig. S2A). THL-alk1 also led to the largest number of fluorescent target proteins (supplemental Fig. S2B), which could be ablated to a large extent by THL in competition experiments (supplemental Fig. S2C). The protein target enrichment was further validated using trifunctional azide to consolidated visualization and affinity purification of labeled proteins by a combination of in-gel fluorescence and avidin chromatography procedures (supplemental Fig. S2E). We therefore chose THL-alk1 for all subsequent experiments.
Approximately seven distinct bands were visible in the fluorescence readout (Fig. 1B), a relatively small number given the complex nature of the protein fractions used. However, a potential complication was the overlap of multiple fluorescent protein bands. We hence implemented LC-MS-based peptide sequencing to determine the protein identity of individual bands excised from silver stained gels. The obtained target list contained 49 proteins after the removal of proteins that were also present in the control (DMSO) condition (Replicate 1, supplemental Table S1, sheet 1, column G to J). These proteins were next assigned to the bands in the enrichment as well as the fluorescent gels on the basis of the highest protein score and corresponding molecular weight. This experiment was repeated three times with slight modifications in sample processing (gel slices versus protein bands, dual tag THL probes, etc.) to control for consistency (Fig. 1C, supplemental Figs. S2D and S2E). The results varied mainly in the number of identified proteins (Replicate 2 & Replicate 3, see supplemental Table S1, sheet 1, column K to R for details); the proteins listed in Fig. 1D were identified in at least two out of three three replicate experiments (see supplemental Table S1, sheet 4 for more details). This comparison led to the identification of 14 THL targets (Fig. 1D), all α/β-hydrolases (see discussion), ten of which are reported or probable lipid esterases (LipD, LipG, LipH, LipI, LipM, LipN, LipO, LipV, LipW, and TesA), two hypothetical proteins with probable acetyl transferase activity (BCG1252, BCG3408), Antigen 85c (mycolyl transferase) and BCG2241c (probable protease). Therefore, THL-alk1 is an activity-based THL probe with high specificity for α/β-hydrolases (100% of targets in BCG) and lipid esterases (70% of the targets shown in Fig. 1D).
Esterase (Protein) Levels and (Enzymatic) Activity Correlate Inversely With Cellular TAG on Entry Into and Exit From Nonreplicative Conditions
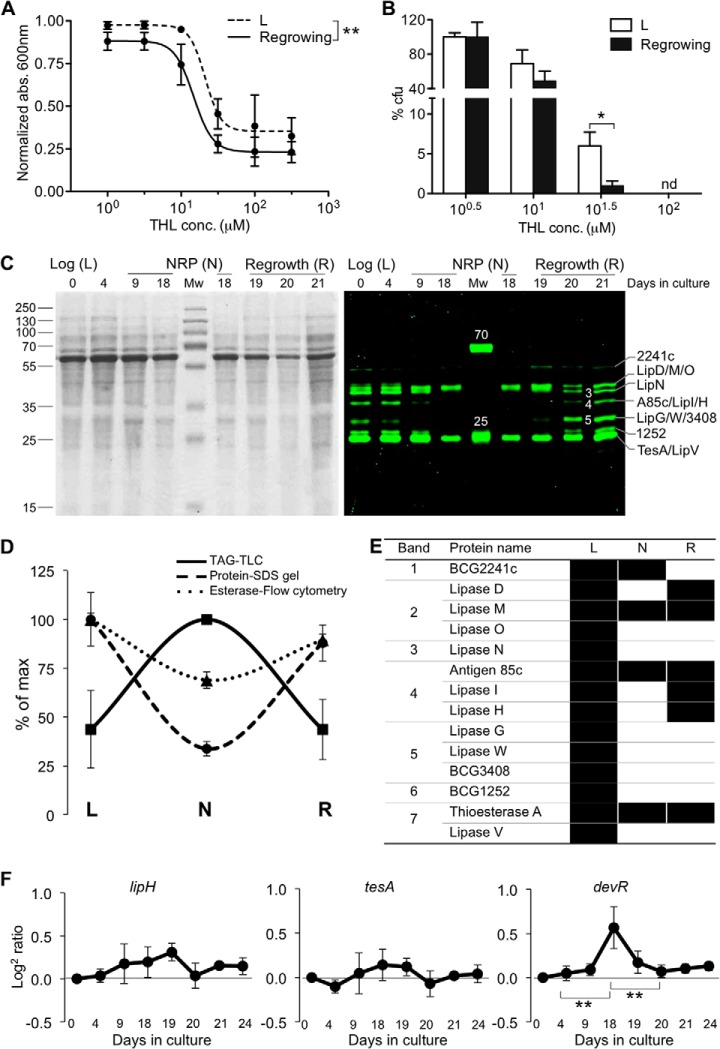
We have previously shown that THL inhibits growth of M. bovis BCG and re-growth of bacilli from hypoxic NRP (11). This is also the case for the THL analog used here (supplemental Fig. S2A). Regrowing bacilli were significantly more susceptible to THL than cells in logarithmic growth (Figs. 2A, 2B). These results are not intuitively evident given the overall similarity of the protein inventory in the different physiological states (Fig. 2C, left panel) (37, 38). We therefore used THL-alk1 to experimentally explore differences in apparently similar protein extracts. Cells were cultured in the three conditions mentioned above (Fig. 1A) and equal amounts of total cell extracts were incubated with THL-alk1 followed by SDS-PAGE of rhodamine-visualized targets (Fig. 2C, right panel). The seven distinct protein bands observed in logarithmic growth conditions (L, Fig. 2C) gave way to a very different pattern in extracts from non-replicating cells (N, Fig. 2C), with only two distinct bands and comparable overall protein composition (Fig. 2C, left panel). Re-growing cells (R) were characterized by a pattern reminiscent of the L condition, with approximately seven distinct bands in the mass range of 20–60 kDa. Therefore, using this activity-based approach, we were able to rapidly reveal profound differences for the presence or absence of lipolytic hydrolases among replicating (metabolically active) and non-replicating (metabolically inactive) conditions of BCG (see below for discussion on quantitative aspects of the fluorescent readout). One would expect a reciprocal relationship between the identified lipases if these were contributing to total cellular TAG activity in a major way. This is indeed borne out experimentally. An accumulation of large amounts of TAG was quantitatively a most prominent change in lipid composition on entry into nonreplicating conditions, and these TAG deposits were hydrolyzed during regrowth (supplemental Fig. S3A, S3B) (11). In addition, total cellular esterase activity, assessed by ester-linked fluorescent dye (FITC conjugated CMFDA, supplemental Fig. S3C), followed a trend similar to the cellular esterase activity, demonstrating that the cells in hypoxic NRP are indeed characterized by overall reduced catabolic activity. To check whether the regulation of these enzymes happens at the gene level, we performed qRT-PCR to quantify mRNA expression (Fig. 2F and supplemental Fig. S4). Our qRT-PCR data showed that M. bovis BCG carboxyl hydrolases appeared to be strongly regulated at the protein level in growing and non-replicative conditions with marginal effects at gene transcript levels. This is a second, biologically important finding and is relevant for our understanding of molecular mechanisms that lead to adaptation of mycobacteria in different environmental conditions (e.g. desiccation or TB pathogenesis).
Fig. 2.
Esterase levels and activity correlate inversely with cellular TAG upon entry into and exit from non replicative-conditions. A and B, THL inhibits M. bovis BCG cultured in liquid (A) and solid media (B) with different potencies depending on the growth condition. Cells re-growing from non-replicative persistence (“Re-growing”) are more sensitive to THL than logarithmically growing bacilli (L). Cultures were incubated with THL at 37 °C for 5 days. The data were analyzed using One-way ANOVA and are represented as the mean values ± S.E. (n = 3). * p < 0.05, ** p < 0.01. C, Cell lysates prepared from M. bovis BCG cultured in different physiological states (L, N and R) were incubated with THL-alk1 (and DMSO as a control). Huisgen cycloaddition reactions were performed using rhodamine-azide dye. Equal amounts of proteins were separated by SDS-PAGE and visualized by in-gel fluorescence (left panel), followed by coomassie staining (right panel). See also Fig. 1B for a representative fluorescent SDS-PAGE of the L condition and protein band assignments. D, Cellular TAG content (determined by TLC, supplemental Fig. S3A and S7A) accumulates in NRP (N). Esterase activity (supplemental Fig. S3C), as well as THL target intensity assessed by in gel fluorescence (C) an inverse relationship and minimum values in N. The maximum value was set as 100% for each of the three different types of measurement. The data were represented as the mean values ± S.D. (n = 3). E, THL targets in total cell extracts from M. bovis BCG derived from logarithmic (L), NRP (N) and regrowth cultures (R). Only proteins positively identified (and absent in control conditions) from three independent experiments in L and two independent experiments in N and R are shown and represented by solid bars. THL identifies lipases in growing (L and R) but not nonreplicative cells (N) except for LipM. F, qRT-PCR analysis of lipH, tesA and devR in different metabolic states. Total RNA was isolated from cells grown in different metabolic states and used for qRT-PCR to measure RNA levels. The ΔCT value against endogenous control (sigA) was normalized against ΔCT value of logarithmic culture (day 0) and the obtained ΔΔCT value is used to calculate relative quantity (RQ). Data is plotted as log2 ratios means and were represented as the mean values ± S.D. (n = 3) and were analyzed by Student's t test. ** p < 0.01 compared with day 18 (NRP).
These results prompted us to identify THL target proteins by LC-MS-based peptide analysis after enrichment and tryptic protein digestion (Fig. 1A, step 5). In contrast to the previous experiment, we excised the entire lane between 60–20 kDa in order to gather more information on the proteins with low abundance. Approximately 250 proteins with an ion score of ≥80, emPAI of >0.1 and peptide match of ≥2 were identified in total cell extracts from cultures harvested in logarithmic growth conditions (supplemental Table S1, sheet 1, column K to N). On the other hand, a dramatic reduction in the total protein in NRP (total proteins 27, supplemental Table S1, sheet 2, column G to J) was observed, which doubled during 3 days of regrowth (supplemental Table S1, sheet 3, column G to J). This was consistent with the pattern observed in the fluorescent gel. To strengthen and relate fluorescence visualization with affinity enrichment of tagged proteins, we performed cycloaddition with a trifunctional probe, which has the advantage of fluorescent visualization followed by separation of enriched proteins on SDS-polyacrylamide gels (supplemental Fig. S2E), but with low affinity; this is evident from the number of proteins obtained, ion score and peptide matches (see supplemental Table S1, sheet 1, column O to R and sheet 2 and 3, column K to N for details).
Functional Validation: Overexpression of Target Proteins Leads to Reduced Susceptibility to THL
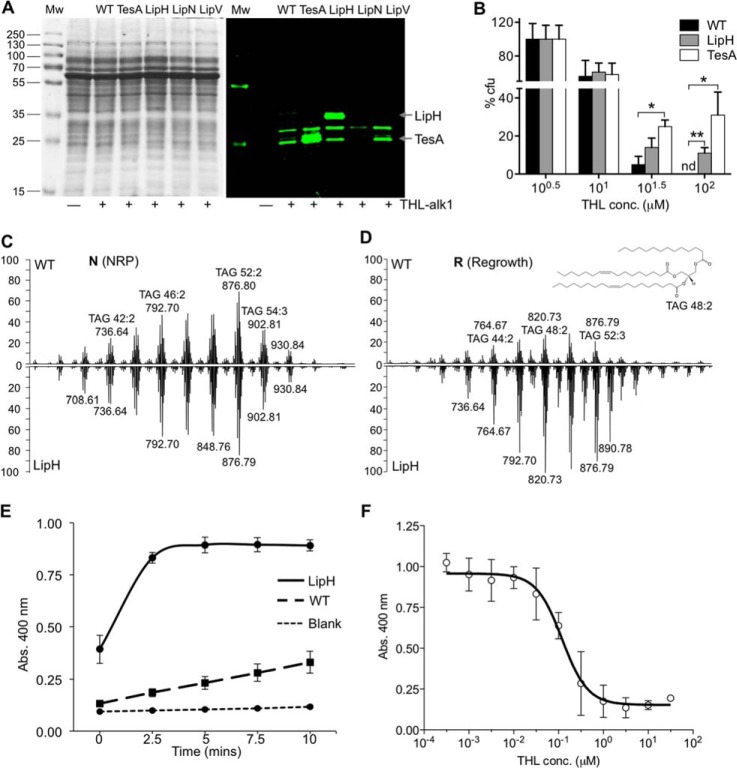
We next cloned and overexpressed several of the genes listed in Fig. 1D (see supplemental Table S2A for primer details) but only successfully managed to achieve >10-fold expression of transcript levels for lipH (migrating with band 4 in Figs. 1B and 2C and with very variable fluorescent intensity between growth stages) and tesA (migrating with band 7 in Figs. 1B and 2C and with relatively constant fluorescent intensity between growth stages). Overexpression of tesA and lipH led to ∼50- and 10-fold increase in transcript levels (assessed by qRT-PCR, supplemental Fig. S5A) and ∼60- and 80-fold increase in fluorescence intensity for TesA and LipH, respectively, when reacted with THL-alk1 (supplemental Fig. S5B). These results demonstrate that the fluorescence levels are reflective of protein levels (Fig. 3A) and cellular enzymatic activity (Fig. 3E). However, elucidation of the precise quantitative relationship would require additional calibrations which was not the aim of this study (see also Discussion). Instead, we assayed for THL susceptibility and found that overexpression of lipH and tesA rendered these strains substantially more resistant to THL on solid (Fig. 3B), liquid media (supplemental Fig. S5C), as well as in an enzymatic inhibition assay (Fig. 3F), therefore functionally validating our biochemical results.
Fig. 3.
Functional validation of THL targets (lipH and tesA). A, Mycobacterial lysates from wild type (WT) and cells overexpressing THL targets (tesA, lipH, lipN and lipV) were incubated with THL-alk1 and processed as described in Fig. 1. The molecular weight of LipH and TesA are indicated. B, Cells expressing tesA and lipH were significantly less susceptible to THL on solid growth media. The data were analyzed using One-way ANOVA and are represented as mean values ± S.E. (n = 3). * p < 0.05, ** p < 0.01, nd: value not determined. C and D, LC-MS of lipid extracts from M. bovis BCG WT (upward trace) and strain over-expressing lipH (downward trace on same scale as WT) during NRP (N) and re-growth from NRP (R). Individual TAG species were further characterized using tandem MS. A possible structure of TAG 48:2 (m/820.73) is shown as an inset. The spectra are calibrated using d5-TAG m/z 829.8 as an internal standard. E, Whole cell lysate from LipH overproducing cells have higher esterase activity in a biochemical assay using p-nitrophenyl-butyrate (pNP-C4) as a substrate. The data were represented as the mean values ± S.D. (n = 3). F, IC50 study of THL in LipH over-expressing cell lysate were performed by incubating lysates with different THL log scale concentration. Product formation using substrate pNP-C4 was recorded at 400 nm and IC50 was determined as the concentration of THL that inhibits enzymatic activity by 50%. The data were represented as the mean values ± S.D. (n = 3).
Biochemical Validation: Molecular Examination of Natural Substrate Recognition and Mimicry by THL
LipH is an enzyme belonging to the hormone sensitive lipase (HSL) family. Its cell-free substrate specificity has been characterized (39) but relatively little is known about its activities in vitro (culture) and in vivo (infection models and human tissue). Here we evaluated the effect of lipH overexpression on TAG dynamics in the Wayne NRP model. Overexpression of lipH did not affect accumulation of TAG (supplemental Figs. S7A, S7B), or the profiles in N or L (Fig. 3C, supplemental Fig. S7C) but, surprisingly, led to less effective hydrolysis of TAG upon resuscitation (Fig. 3D). Instead, the short-chain ester p-nitrophenyl-butyrate (pNP-C4) was rapidly hydrolyzed in lysates from cells overexpressing LipH (Fig. 3E). LC-ESI-MS findings of cellular lipid extracts were consistent with these results and furthermore provided a detailed chemical composition of TAG species, which showed that qualitatively the TAG compositions were comparable with respect to fatty acyl chain length (mostly C14-C18) and unsaturation (mostly 0–2 double bonds per fatty acyl, Fig. 3D). This data suggests that LipH utilizes substrates other than the long-chain TAGs (fatty acyl chain length >C14, Fig. 3C), consistent with cell-free enzymology (Fig. 3E) (39). It cannot be ruled out however, that LipH might inhibit TAG breakdown, for example, by interfering with other lipases.
Crystallographic determination of the thioesterase domain of human fatty acid synthase (FAS) in complex with THL has provided experimental insight into the molecular basis of inhibition (40). Furthermore, together with biochemical studies (41), this technique has provided mechanistic explanations for the predominant product of human FAS, namely palmitic acid, C16, which is determined, at least in part, by a “specificity channel” accommodating the aliphatic moiety. Thioesterase A, a type II thioesterase potentially involved in the biosynthesis of phthiocerol dimycocerosate (PDIM) and a THL target in all physiological conditions tested here (Fig. 2E), is enzymatically similar to the thioesterase domain in FAS. Since no structural data of TesA is currently available, a homology model of the three-dimensional (3D) structure of TesA was generated using the thioesterase structures available in the protein data bank (PDB) for RifR from Amycolactopsis mediterranei, a type II thioesterase in the biosynthetic pathway of rifampicin (42) (PDB ID: 3FLB, 31% sequence identity to TesA) and thioesterase RedJ (43) (PDB ID: 3QMV, 33% sequence identity) as templates (supplemental Fig. S8A). Docking of the natural substrate of TesA, phthiocerol, the common lipid core in mycocerosates, was next simulated using a force field-based algorithm that minimizes the binding energies of flexible substrates and active sites of enzymes (supplemental Fig. S9A). The steric orientation of the active site amino acids, the catalytic triad, Ser104, His236, and Asp208, was recorded and compared with the orientation when THL was docked instead of the natural ligand using the same algorithm (supplemental Fig. S9B). Both docking of the natural substrate ligand (phthiocerol) as well as the inhibitor (THL) led to binding energies of −7.2 Kcal/Mol and −6.4 Kcal/Mol, respectively, which are within the expected range (44). Importantly, the virtual superimposition of the catalytic triad (supplemental Fig. S9C) provides in silico validation of the experimental results of the THL-TesA interaction and complements the functional validation in cellular overexpression.
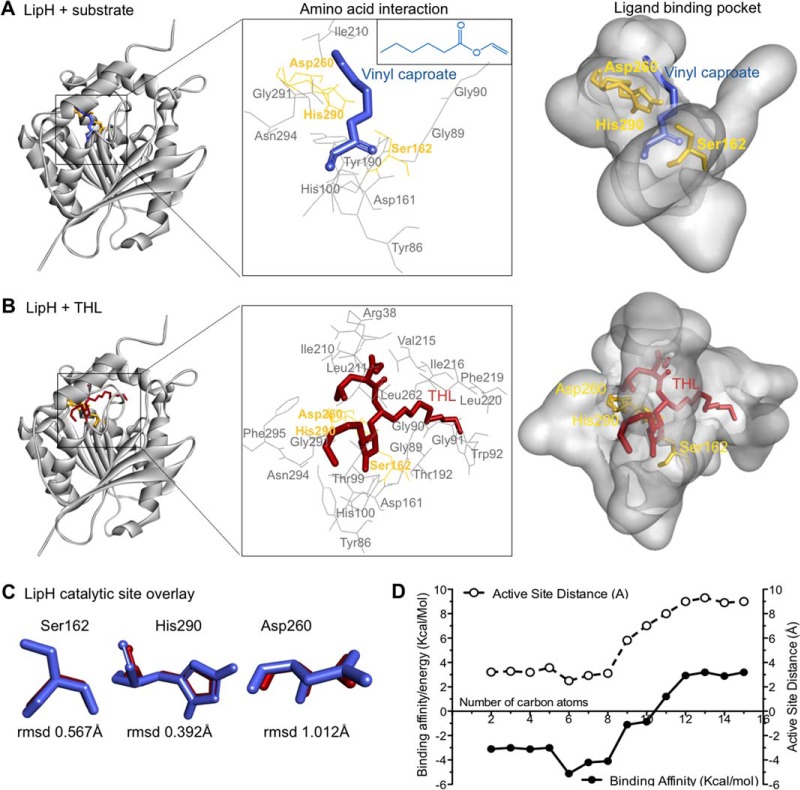
We next extended this approach to the study of LipH where natural substrates are less well characterized (39, 45). Three-dimensional models of LipH (Fig. 4A) were generated based on homology with experimentally determined lipase structures. The top-ranked amino acid sequence homology hits in the PDB for LipH included carboxyesterases from Alicyclobacillus acidocaldarius (Est2, PDB ID: 1U4N, 44% sequence identity with LipH) and Alicyclobacillus fulgidus (AFEST, PDB ID: 1JJI, sequence similarity 41%), and bacterial acetyl esterase (HerE, PDB ID: 1LZL, sequence similarity 40%) (supplemental Fig. S8B). Vinyl caproate (Fig. 4A) and short-chain TAGs were used as potential substrates for LipH using the same in silico docking approach described above. THL bound within the LipH active site (Fig. 4B) with a binding energy of −7.10 Kcal/mol, which was comparable to −5.11 Kcal/mol of THL with virtually identical orientation of the catalytic triad (Fig. 4C). Using this approach for TAGs with increasing fatty acyl chain lengths, we found that species with FA carbon chain lengths of 6–8 are predicted to be energetically preferred substrates (minimal binding energies) whereas chain lengths >10 do not lead to any docking (Fig. 4D); this is consistent with the biochemical results with synthetic substrates (Fig. 3E, 3F) (39, 46). This approach therefore provides a computational basis for LipH substrate preference and demonstrates the utility of THL as a tool compound to probe enzymatic activities of lipid esterases. THL-alk1 (and the closely related derivatives, supplemental Fig. S1A) is a useful experimental probe for the identification of such enzymes.
Fig. 4.
Molecular examination of natural substrate recognition and mimicry by THL. A and B, Three-dimensional model of the LipH structure depicting the core α/β-hydrolase fold with catalytic triad (yellow). Vinyl caproate (blue, insert E), a described substrate of LipH in vitro, and THL (red) are docked to the active site using computational methods. The insert (middle) depicts the catalytic triad and other amino acids interacting with docked ligands. C, Overlay of catalytic triad shows the individual amino acid conformation when LipH is docked with vinyl caproate (blue) and with THL (red), respectively. The root mean square deviations (rmsd) of the differences between vinyl caproate and THL predictions are indicated at the bottom. D, Plot depicting the change in binding energy (Kcal/mol) and distance to active site serine (Å) when vinyl esters of different chain lengths are docked with the LipH 3D-model.
DISCUSSION
Target identification using affinity-based protein profiling is often primarily (and solely) centered on affinity enrichment (“pull down,” Fig. 1C). Here, we defined the target spectrum of THL as the common overlap between fluorescent tagging without enrichment (Fig. 1B) and mass spectrometry-based identification after affinity enrichment (Fig. 1C, supplemental Table S1). This stringent selection led to a substantially refined short-list of 14 THL targets in total cell extracts from M. bovis BCG (Fig. 1D), reduced from 261 protein IDs (supplemental Table S1, sheet 1), to provide (1) a clearer picture of specificity and (2) allow for a more informed selection for validation (Fig. 3 for two select targets). This workflow feature has important implications for the applicability of the THL probes characterized in detail here, both in general terms and in the context of M. bovis BCG, a widely used model organism with very broad metabolic capabilities. In the following sections we discuss how our analysis exemplifies the broad utility of this lipase tag and the future applications for this method.
The value of this advance in the analysis of ABPP readouts is nicely illustrated by the correlation of target intensity (Fig. 2C, right panel) to enzymatic activity in whole cell extracts (Fig. 2D). Growing stages (L, R) are characterized by high levels of THL-alk1-targeted esterases and high cellular esterase and TAG lipase activity (supplemental Fig. S3). The opposite is true for the nonreplicative condition. Therefore, THL-alk1 is a suitable probe for “facile” (i.e. not requiring sophisticated proteomics to quantify lipases) monitoring of lipid esterases using a fluorescent readout. Although the quantitative relationship between fluorescence and protein levels has not been determined in detail in this study, imaging of THL-alk1 adducts in cells with larger dimensions (such a eukaryotic cells) will help understand better the spatial distribution of enzymes involved in the dynamics of lipid droplets (47–49). In addition, THL-alk1 and its closely related derivatives could be useful in other innovative approaches for the systematic determination of protein-small molecule interactions (50–52).
In Mycobacterium tuberculosis, the dormancy survival regulator regulon (dosR, also called devR) comprises about 50 genes and is critical for persistence (53, 54). It is controlled by a two-component system, uses oxygen levels as key input, and allows the bacteria to survive in hypoxic conditions for extended periods of time. Quantitative real time PCR analyses showed that dosR transcripts are elevated during nonreplicative conditions (day 18, Fig. 2F); however, these studies have not provided any conclusive explanation for the regulation of THL targets, which are not part of dosR (Fig. 2F and supplemental Fig. S4). The transcript levels of lipH and tesA are relatively constant throughout all growth conditions (Fig. 2F) whereas the fluorescent signal of hydrolases/lipases (Fig. 2C, right panel) and total cell esterase activity (Fig. 2D) were substantially reduced in the nonreplicative condition when cells are metabolically much less active. The easiest explanation for this observation is that these enzymes are regulated at the post-translational level, such as modifications that lead to the re-localization and/or degradation of enzymes, or alterations that interfere with THL binding and activity. These examples further demonstrate how THL-alk1 can be used as a tool to study lipases at the proteomic level via widely established SDS-PAGE separation.
M. bovis BCG is most susceptible to THL during resuscitation when carbon sources are needed to revive cellular growth (Fig. 2B). The in vitro conditions used here provide sufficient substrate sources during all growth conditions, including the resuscitation phase. Tween 80, polyoxyethylene sorbitan oleate, added as a surfactant to avoid aggregation in liquid cultures, is metabolized by the bacteria thus providing oleic acid (C18:1). In addition, intracellular TAG deposits and, possibly wax esters (50) serve as an extra fatty acyl/carbon source during resuscitation. On the other hand, mycobacteria growing inside host cells (e.g. in macrophage cultures) are not exposed to such an abundance of nutrients and thus are forced to adapt their metabolism. It is therefore likely that carbon originates from multiple sources in vivo (i.e. macrophages and lung tissue/granuloma) consistent with recent studies on carbon utilization and metabolism under such conditions (55, 56). Probes with a specific affinity to lipid esterases, such as THL-alk1, will therefore be of great value for studies on “carbon sourcing” which, in addition to mycobacterial pathogenicity, is intensively studied in biofilm formation and hydrocarbon deposition.
For future applications, it will be necessary to prepare suitable starting fractions. Here, we used total cell lysate as starting material, a crude and relatively complex mixture. The bulk of THL targets are soluble after ultracentrifugation (supplemental Fig. S10). However, it can be expected that further separation into more defined biochemical fractions (e.g. cell wall, lipid droplets, and secreted fractions) will provide additional biologically important information. For example, four of the factors shown in Fig. 2E are potential secretory enzymes (BCG2241c, LipM, A85c and LipH); the lipid metabolic surface and secretory proteins are likely preferential targets for the immune system early in infection. As anticipated, we found many lipases in our total cell lysate; the enzymes shown in supplemental Table S1 fall mainly into the categories of hydrolases (16%), transferases (20%), and oxidoreductases (17%), collectively representing >50% of all identified proteins, when categorized according to the enzyme commission (E.C.) classification. This is expected for conditions where the proteomic composition of the different test extracts is comparably large and diverse (Figs. 1B, 2C). Despite this, the results are consistent with recent findings in other biological species (57), which supports the validity of our approach, and are consistent with the reported inhibitory action of THL on intestinal lipases (19, 23).
It is becoming increasingly evident that particularities of major lipid metabolic pathways are unique even among closely related bacteria such as the Corynebacterium-Mycobacterium-Nocardia (CMN) group of Actinomycetes. This is true for both the chemical diversity of lipids (e.g. carbon chain lengths and side chain functionalities of mycolic acids) as well as the protein machinery involved in their metabolism (e.g. lipid droplet associated factors) (51). THL-alk1 will provide a starting point for biochemical identification of such related lipid esterases in different biological species where sequence similarity is often low (50) and insufficient for identification. This is supported by the findings of this study.
All of the 14 targets shown in Fig. 1D belong to the α/β-hydrolase fold family of enzymes, one of the largest groups of structurally related enzymes (58). Their structural characteristics are eight β-sheets (mostly in parallel alignment), which are connected via α-helices. Despite this structural similarity, α/β-hydrolases catalyze a wide range of reactions, all via a nucleophilic His residue in the catalytic triad (59), His290 (Fig. 4C) and His236 (supplemental Fig. S9C) in the case of LipH and TesA, respectively. Lipases (E.C. 3.1.1.3) constitute the largest enzyme class within the α/β-hydrolases which should make the findings reported here of broad general interest beyond the specific biological example of mycobacteria.
THL has been successfully used to study molecular recognition at the atomic level (40). We illustrate how substrate-docking studies (computational) in combination with the (experimental) THL target list can be extended to learn more about orientation of the natural product inhibitor in enzymatic cavities. The basis for this analysis is (1) the assumption that the amino acids comprising the catalytic triad are oriented sterically comparable; (2) that the binding energies are minimal between known substrates (phthiocerol and vinyl ester for TesA and LipH, respectively) and the inhibitor (THL for both TesA and LipH); and (3) with a minimal distance to active site (serine OH group) which describes a near-to-attack conformation of the ligand (substrate/inhibitor) in the active site (60). The minimum distance from the serine OH group to the carbonyl carbon of docked ligands (“active site distance” in Fig. 4D) is 2.4 Å (vinyl caproate docked to LipH) and co-incides with a minimal binding energy of −5.11 kcal/mol (Fig. 4D). The lactone ring distance for the computed LipH-THL complex is 1.82 Å with a minimal binding energy of −7.1 kcal/mol; this is thus very comparable. Note the strikingly different positional orientations of the short, long aliphatic side chains and amino-terminal of THL in these docked predictions (Fig. 4B and supplemental Fig. S9B) and their relation to the respective substrates. Further exploration using THL-alk1–3 (supplemental Fig. S1A), additional THL derivatives (61), and the vast pool of related compounds of plant and microbial origin (59) are likely to provide interesting new findings on substrate recognition, based on subtle differences between the probes, as demonstrated by the preliminary results shown in supplemental Fig. S2B. This detailed characterization shows that THL, with its spatially small “privileged structure” and trifunctional diversity ends is able to accommodate active sites of different hydrolases with almost identical steric orientation of catalytic triad residues expected for natural substrate binding, as demonstrated for LipH in comparison with TesA.
In summary, the main result of this work is a list of experimentally determined and partially validated targets of the lipase inhibitor THL in M. bovis BCG. It therefore provides a resource for future functional studies of THL targets in M. bovis BCG and related mycobacteria where lipid metabolism is now intensively studied for various reasons. In addition, we demonstrate the utility of this THL probe for the tagging of lipases with a convenient fluorescent readout. This should be of substantial general interest to the growing community of investigators interested in correlating lipase functions with physiological changes such as growth, differentiation and development in biological systems other than mycobacteria.
Supplementary Material
Acknowledgments
We thank Chionh Yok Hian and Peter C. Dedon for helpful scientific discussions. We are grateful to Paul Huchinson for technical help with flow cytometry and Newman Sze (Nanyang Technical University) for support with peptide sequencing.
Footnotes
Author contributions: M.R., S.P.R., X.C., A.C., S.Q.Y., and M.R.W. designed research; M.R., S.P.R., X.C., A.S., and A.C. performed research; S.P.R., X.C., and S.Q.Y. contributed new reagents or analytic tools; M.R., X.C., A.S., A.C., and M.R.W. analyzed data; M.R., S.Q.Y., and M.R.W. wrote the paper.
* This work was supported by grants from the National University of Singapore and the Singapore National Research Foundation under CRP Award No. 2007-04.
 This article contains supplemental Figs. S1 to S10 and Tables S1 and S2.
This article contains supplemental Figs. S1 to S10 and Tables S1 and S2.
1 The abbreviations used are:
- ABPP
- activity-based protein profiling
- BCG
- Bacillus Calmette-Guérin
- CFU
- colony forming units
- CMFDA
- 5-Chloromethylfluorescein diacetate
- ESI-MS
- electrospray ionization-mass spectrometry
- FAS
- fatty acid synthase
- HSL
- hormone sensitive lipase
- LTQ-FT
- linear ion trap-fourier transform
- MBC
- minimum bactericidal concentration
- MIC
- minimum inhibitory concentration
- NRP
- non-replicating persistent
- PDIM
- Phthiocerol dimycocerosate
- qRT-PCR
- quantitative reverse transcription-PCR
- TAG
- triacylglycerol
- THL
- tetrahydrolipstatin.
REFERENCES
- 1. Gengenbacher M., Kaufmann S. H. (2012) Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol. Rev. 36, 514–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Russell D. G., Cardona P. J., Kim M. J., Allain S., Altare F. (2009) Foamy macrophages and the progression of the human tuberculosis granuloma. Nat. Immunol. 10, 943–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wayne L. G. (1994) Dormancy of Mycobacterium tuberculosis and latency of disease. Eur. J. Clin. Microbiol. Inf. Dis. 13, 908–914 [DOI] [PubMed] [Google Scholar]
- 4. Chao M. C., Rubin E. J. (2010) Letting sleeping dos lie: does dormancy play a role in tuberculosis? Ann. Rev. Microbiol. 64, 293–311 [DOI] [PubMed] [Google Scholar]
- 5. Bishai W. (2000) Lipid lunch for persistent pathogen. Nature 406, 683–685 [DOI] [PubMed] [Google Scholar]
- 6. Russell D. G. (2003) Phagosomes, fatty acids and tuberculosis. Nat. Cell Biol. 5, 776–778 [DOI] [PubMed] [Google Scholar]
- 7. Garton N. J., Christensen H., Minnikin D. E., Adegbola R. A., Barer M. R. (2002) Intracellular lipophilic inclusions of mycobacteria in vitro and in sputum. Microbiology 148, 2951–2958 [DOI] [PubMed] [Google Scholar]
- 8. Daniel J., Deb C., Dubey V. S., Sirakova T. D., Abomoelak B., Morbidoni H. R., Kolattukudy P. E. (2004) Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J. Bacteriol. 186, 5017–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wayne L. G., Sohaskey C. D. (2001) Nonreplicating persistence of mycobacterium tuberculosis. Ann. Rev. Microbiol. 55, 139–163 [DOI] [PubMed] [Google Scholar]
- 10. Deb C., Lee C. M., Dubey V. S., Daniel J., Abomoelak B., Sirakova T. D., Pawar S., Rogers L., Kolattukudy P. E. (2009) A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PloS One 4, e6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Low K. L., Rao P. S., Shui G., Bendt A. K., Pethe K., Dick T., Wenk M. R. (2009) Triacylglycerol utilization is required for regrowth of in vitro hypoxic nonreplicating Mycobacterium bovis bacillus Calmette-Guerin. J. Bacteriol. 191, 5037–5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 13. Camus J. C., Pryor M. J., Medigue C., Cole S. T. (2002) Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology 148, 2967–2973 [DOI] [PubMed] [Google Scholar]
- 14. Singh G., Singh G., Jadeja D., Kaur J. (2010) Lipid hydrolizing enzymes in virulence: Mycobacterium tuberculosis as a model system. Critical Rev. Microbiol. 36, 259–269 [DOI] [PubMed] [Google Scholar]
- 15. Low K. L., Shui G., Natter K., Yeo W. K., Kohlwein S. D., Dick T., Rao S. P., Wenk M. R. (2010) Lipid droplet-associated proteins are involved in the biosynthesis and hydrolysis of triacylglycerol in Mycobacterium bovis bacillus Calmette-Guerin. J. Biol. Chem. 285, 21662–21670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deb C., Daniel J., Sirakova T. D., Abomoelak B., Dubey V. S., Kolattukudy P. E. (2006) A novel lipase belonging to the hormone-sensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. J. Biol. Chem. 281, 3866–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shen G., Singh K., Chandra D., Serveau-Avesque C., Maurin D., Canaan S., Singla R., Behera D., Laal S. (2012) LipC (Rv0220) is an immunogenic cell surface esterase of Mycobacterium tuberculosis. Infection Immunity 80, 243–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Richter L., Tai W., Felton J., Saviola B. (2007) Determination of the minimal acid-inducible promoter region of the lipF gene from Mycobacterium tuberculosis. Gene 395, 22–28 [DOI] [PubMed] [Google Scholar]
- 19. Hadvary P., Sidler W., Meister W., Vetter W., Wolfer H. (1991) The lipase inhibitor tetrahydrolipstatin binds covalently to the putative active site serine of pancreatic lipase. J. Biol. Chem. 266, 2021–2027 [PubMed] [Google Scholar]
- 20. Kremer L., de Chastellier C., Dobson G., Gibson K. J., Bifani P., Balor S., Gorvel J. P., Locht C., Minnikin D. E., Besra G. S. (2005) Identification and structural characterization of an unusual mycobacterial monomeromycolyl-diacylglycerol. Mol. Microbiol. 57, 1113–1126 [DOI] [PubMed] [Google Scholar]
- 21. Waddell S. J., Stabler R. A., Laing K., Kremer L., Reynolds R. C., Besra G. S. (2004) The use of microarray analysis to determine the gene expression profiles of Mycobacterium tuberculosis in response to anti-bacterial compounds. Tuberculosis 84, 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Parker S. K., Barkley R. M., Rino J. G., Vasil M. L. (2009) Mycobacterium tuberculosis Rv3802c encodes a phospholipase/thioesterase and is inhibited by the antimycobacterial agent tetrahydrolipstatin. PloS One 4, e4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Borgstrom B. (1988) Mode of action of tetrahydrolipstatin: a derivative of the naturally occurring lipase inhibitor lipstatin. Biochim. Biophys. Acta 962, 308–316 [DOI] [PubMed] [Google Scholar]
- 24. Haalck L., Spener F. (1997) On the inhibition of microbial lipases by tetrahydrolipstatin. Methods Enzymol. 286, 252–263 [DOI] [PubMed] [Google Scholar]
- 25. Yang P. Y., Liu K., Ngai M. H., Lear M. J., Wenk M. R., Yao S. Q. (2010) Activity-based proteome profiling of potential cellular targets of Orlistat–an FDA-approved drug with anti-tumor activities. J. Am. Chem. Soc. 132, 656–666 [DOI] [PubMed] [Google Scholar]
- 26. Wayne L. G. (2001) In Vitro Model of Hypoxically Induced Nonreplicating Persistence of Mycobacterium tuberculosis. Methods Mol. Med. 54, 247–269 [DOI] [PubMed] [Google Scholar]
- 27. Wayne L. G., Hayes L. G. (1996) An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infection Immunity 64, 2062–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stover C. K., de la Cruz V. F., Fuerst T. R., Burlein J. E., Benson L. A., Bennett L. T., Bansal G. P., Young J. F., Lee M. H., Hatfull G. F., Snapper S. B., Barletta R. G., Jacobs W. R., Jr, Bloom B. R. (1991) New use of BCG for recombinant vaccines. Nature 351, 456–460 [DOI] [PubMed] [Google Scholar]
- 29. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 30. Yang P. Y., Liu K., Zhang C., Chen G. Y., Shen Y., Ngai M. H., Lear M. J., Yao S. Q. (2011) Chemical modification and organelle-specific localization of orlistat-like natural-product-based probes. Chemistry 6, 2762–2775 [DOI] [PubMed] [Google Scholar]
- 31. Gan C. S., Guo T., Zhang H., Lim S. K., Sze S. K. (2008) A comparative study of electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) versus SCX-IMAC-based methods for phosphopeptide isolation/enrichment. J. Proteome Res. 7, 4869–4877 [DOI] [PubMed] [Google Scholar]
- 32. Shui G., Guan X. L., Low C. P., Chua G. H., Goh J. .S, Yang H., Wenk. M. R. (2010) Toward one step analysis of cellular lipidomes using liquid chromatography coupled with mass spectrometry: application to Saccharomyces cerevisiae and Schizosaccharomyces pombe lipidomics. Mol. Biosystems 6, 1008–1017 [DOI] [PubMed] [Google Scholar]
- 33. Guex N., Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 34. Zhang J., Liang Y., Zhang Y. (2011) Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure 19, 1784–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trott O., Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Computational Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li N., Overkleeft H. S., Florea B. I. (2012) Activity-based protein profiling: an enabling technology in chemical biology research. Current Opinion Chem. Biol. 16, 227–233 [DOI] [PubMed] [Google Scholar]
- 37. Boon C., Li R., Qi R., Dick T. (2001) Proteins of Mycobacterium bovis BCG induced in the Wayne dormancy model. J. Bacteriol. 183, 2672–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gumber S., Taylor D. L., Marsh I. B., Whittington R. J. (2009) Growth pattern and partial proteome of Mycobacterium avium subsp. paratuberculosis during the stress response to hypoxia and nutrient starvation. Veterinary Microbiol. 133, 344–357 [DOI] [PubMed] [Google Scholar]
- 39. Canaan S., Maurin D., Chahinian H., Pouilly B., Durousseau C., Frassinetti F., Scappuccini-Calvo L., Cambillau C., Bourne Y. (2004) Expression and characterization of the protein Rv1399c from Mycobacterium tuberculosis. A novel carboxyl esterase structurally related to the HSL family. Eur. J. Biochem. 271, 3953–3961 [DOI] [PubMed] [Google Scholar]
- 40. Pemble C. W., IV, Johnson L. C., Kridel S. J., Lowther W.T. (2007) Crystal structure of the thioesterase domain of human fatty acid synthase inhibited by Orlistat. Nat. Structural Mol. Biol. 14, 704–709 [DOI] [PubMed] [Google Scholar]
- 41. Singh N., Wakil S. J., Stoops J. K. (1984) On the question of half- or full-site reactivity of animal fatty acid synthetase. J. Biol. Chem. 259, 3605–3611 [PubMed] [Google Scholar]
- 42. Claxton H. B., Akey D. L., Silver M. K., Admiraal S. J., Smith J. L. (2009) Structure and functional analysis of RifR, the type II thioesterase from the rifamycin biosynthetic pathway. J. Biol. Chem. 284, 5021–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Whicher J. R., Florova G., Sydor P. K., Singh R., Alhamadsheh M., Challis G. L., Reynolds K. A., Smith J. L. (2011) Structure and function of the RedJ protein, a thioesterase from the prodiginine biosynthetic pathway in Streptomyces coelicolor. J. Biol. Chem. 286, 22558–22569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cheng F., Wang Q., Chen M., Quiocho F. A., Ma J. (2008) Molecular docking study of the interactions between the thioesterase domain of human fatty acid synthase and its ligands. Proteins 70, 1228–1234 [DOI] [PubMed] [Google Scholar]
- 45. Dedieu L., Serveau-Avesque C., Kremer L., Canaan S. (2012) Mycobacterial lipolytic enzymes: A gold mine for tuberculosis research. Biochimie 95, 66–73 [DOI] [PubMed] [Google Scholar]
- 46. Chahinian H., Ali Y. B., Abousalham A., Petry S., Mandrich L., Manco G., Canaan S., Sarda L. (2005) Substrate specificity and kinetic properties of enzymes belonging to the hormone-sensitive lipase family: comparison with non-lipolytic and lipolytic carboxylesterases. Biochim. Biophys. Acta 1738, 29–36 [DOI] [PubMed] [Google Scholar]
- 47. Spandl J., White D. J., Peychl J, Thiele C. (2009) Live cell multicolor imaging of lipid droplets with a new dye, LD540. Traffic 10, 1579–1584 [DOI] [PubMed] [Google Scholar]
- 48. Sherer N. M., Lehmann M. J., Jimenez-Soto L. F., Ingmundson A., Horner S. M., Cicchetti G., Allen P. G., Pypaert M., Cunningham J. M., Mothes W. (2003) Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4, 785–801 [DOI] [PubMed] [Google Scholar]
- 49. Fei W., Shui G., Zhang Y., Krahmer N., Ferguson C., Kapterian T. S., Lin R. C., Dawes I. W., Brown A. J., Li P., Huang X., Parton R. G., Wenk M. R., Walther T. C., Yang H. (2011) A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genetics 7, e1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sirakova T. D., Deb C., Daniel J., Singh H. D., Maamar H., Dubey V. S., Kolattukudy P. E. (2012) Wax ester synthesis is required for Mycobacterium tuberculosis to enter in vitro dormancy. PloS One 7, e51641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li X., Wang X., Snyder M. (2013) Systematic investigation of protein-small molecule interactions. IUBMB life 65, 2–8 [DOI] [PubMed] [Google Scholar]
- 52. Gallego O., Betts M. J., Gvozdenovic-Jeremic J., Maeda K., Matetzki C., Aguilar-Gurrieri C., Beltran-Alvarez P., Bonn S., Fernandez-Tornero C., Jensen L. J., Kuhn M., Trott J., Rybin V., Muller C. W., Bork P., Kaksonen M., Russell R. B., Gavin A. C. (2010) A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae. Molecular Systems Biol. 6, 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Flores Valdez M. A., Schoolnik G. K. (2010) DosR-regulon genes induction in Mycobacterium bovis BCG under aerobic conditions. Tuberculosis 90, 197–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Muttucumaru D. G., Roberts G., Hinds J., Stabler R. A., Parish T. (2004) Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis 84, 239–246 [DOI] [PubMed] [Google Scholar]
- 55. Shi L., Sohaskey C. D., Pfeiffer C., Datta P., Parks M., McFadden J., North R. J., Gennaro M. L. (2010) Carbon flux rerouting during Mycobacterium tuberculosis growth arrest. Mol. Microbiol. 78, 1199–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rohde K. H., Veiga D. F., Caldwell S., Balazsi G., Russell D. G. (2012) Linking the transcriptional profiles and the physiological states of Mycobacterium tuberculosis during an extended intracellular infection. PLOS Pathogens 8, e1002769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yang P. Y., Wang M., Liu K., Ngai M. H., Sheriff O., Lear M. J., Sze S. K., He C. Y., Yao S. Q. (2012) Parasite-based screening and proteome profiling reveal orlistat, an FDA-approved drug, as a potential anti Trypanosoma brucei agent. Chemistry 18, 8403–8413 [DOI] [PubMed] [Google Scholar]
- 58. Schrag J. D., Cygler M. (1997) Lipases and alpha/beta hydrolase fold. Methods Enzymol. 284, 85–107 [DOI] [PubMed] [Google Scholar]
- 59. Birari R. B., Bhutani K. K. (2007) Pancreatic lipase inhibitors from natural sources: unexplored potential. Drug Discovery Today 12, 879–889 [DOI] [PubMed] [Google Scholar]
- 60. Suplatov D. A., Besenmatter W., Svedas V. K., Svendsen A. (2012) Bioinformatic analysis of alpha/beta-hydrolase fold enzymes reveals subfamily-specific positions responsible for discrimination of amidase and lipase activities. Protein Engineering, Design Selection 25, 689–697 [DOI] [PubMed] [Google Scholar]
- 61. Ngai M. H., Yang P. Y., Liu K., Shen Y., Wenk M. R., Yao S. Q., Lear M. J. (2010) Click-based synthesis and proteomic profiling of lipstatin analogues. Chem. Commun. 46, 8335–8337 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.