Abstract
Human leukocyte antigen (HLA) class I molecules bind peptides derived from the intracellular degradation of endogenous proteins and present them to cytotoxic T lymphocytes, allowing the immune system to detect transformed or virally infected cells. It is known that HLA class I–associated peptides may harbor posttranslational modifications. In particular, phosphorylated ligands have raised much interest as potential targets for cancer immunotherapy. By combining affinity purification with high-resolution mass spectrometry, we identified more than 2000 unique ligands bound to HLA-B40. Sequence analysis revealed two major anchor motifs: aspartic or glutamic acid at peptide position 2 (P2) and methionine, phenylalanine, or aliphatic residues at the C terminus. The use of immobilized metal ion and TiO2 affinity chromatography allowed the characterization of 85 phosphorylated ligands. We further confirmed every sequence belonging to this subset by comparing its experimental MS2 spectrum with that obtained upon fragmentation of the corresponding synthetic peptide. Remarkably, three phospholigands lacked a canonical anchor residue at P2, containing phosphoserine instead. Binding assays showed that these peptides bound to HLA-B40 with high affinity. Together, our data demonstrate that the peptidome of a given HLA allotype can be broadened by the presentation of peptides with posttranslational modifications at major anchor positions. We suggest that ligands with phosphorylated residues at P2 might be optimal targets for T-cell-based cancer immunotherapy.
Major histocompatibility complex (MHC)1 class I molecules are cell surface glycoproteins that are expressed on almost every nucleated cell in vertebrates. They result from the noncovalent interaction of a polymorphic heavy chain, a constant light chain (β-2-microglobulin (β2m)), and a peptide ligand (1). The extracellular region of the heavy chain encompasses three domains, α1, α2, and α3, with α1 and α2 forming a groove that accommodates a peptide ligand of, typically, 8 to 11 amino acid residues. The binding of the ligand to the groove is governed by the interaction of the side chains of certain peptide residues, called anchor positions, with several pockets of the heavy chain named A to F (1, 2). The size and chemical nature of these pockets impose restrictions on the peptide repertoire that can be associated with a particular class I antigen. It is reckoned that the ligandome of a given class I allotype may comprise up to 10,000 different peptides (3), although recent reports suggest that this number may be underestimated (4).
Peptides displayed by MHC class I molecules derive from the intracellular degradation of endogenous proteins in the nucleus and cytosol and reach the lumen of the endoplasmic reticulum by means of the transporter associated with antigen processing. Inside the endoplasmic reticulum, peptides bind to the heavy chain and β2m in a multistep process involving several chaperones. Finally, if the bound peptide confers enough stability to the complex, the MHC class I molecule migrates via the Golgi network to the cell surface (5). MHC class I molecules facilitate immunological surveillance by presenting peptide ligands to CD8+ T lymphocytes. When tumor-specific peptides or peptides derived from intracellular pathogens are detected by the T cells, they exert their cytotoxic effects over the antigen-presenting cell, promoting tumor suppression or eradication of the infection.
The MHC, known as the human leukocyte antigen (HLA) system in humans, is the most polymorphic region in the entire genome (6). In particular, the IMGT/HLA database (7) currently contains about 7000 allele sequences that encode more than 5000 different human class I antigens. Most of these polymorphisms are located within the α1 and α2 domains of the heavy chain and modulate the peptide binding preferences of each allotype (8). It is thought that the great diversity of HLA class I allotypes, and of their associated ligandomes, is an adaptation to guarantee immunity against intracellular pathogens (6). In this regard, the existence of a large number of different class I molecules capable of presenting diverse peptidomes hampers immune evasion by means of viral genetic mutation.
It has been known for a long time that HLA class I molecules display posttranslationally modified peptides at the cell surface (9). Among other modifications, N-terminal acetylation (10), phosphorylation (11–13), methylation (14), and glycosylation (15) have been described in MHC class I–bound peptidomes. In this context, phosphorylated ligands have raised much interest owing to their potential as targets in T-cell-based cancer immunotherapy (12, 13), given that aberrant phosphorylation is a hallmark of malignant transformation (16, 17) and phosphorylated epitopes can be specifically recognized by CTLs (11). Therefore, the characterization of the phosphopeptidome associated with MHC class I molecules and the identification of tumor-derived phosphopeptides are necessary in order for such immunotherapeutic approaches to be implemented.
Nevertheless, the identification of HLA class I–bound phosphopeptides is difficult because of several analytical limitations. Phosphopeptides constitute only a small fraction of the peptide repertoire of a given HLA allotype. Additionally, MS analysis is hindered by the low ionization efficiency of phosphorylated species relative to their nonphosphorylated counterparts (18), which makes them more difficult to detect. Moreover, the fragmentation of phosphorylated peptides by collision-induced dissociation usually results in minimally informative MS2 spectra (19). As a consequence of the lability of the phosphate group, which is readily dissociated during fragmentation, a prominent signal corresponding to the neutral loss of phosphoric acid is often observed contrasting with poor b- and y-type ion signals. This phenomenon is especially exacerbated in the case of phosphoserine (19), which is involved in about 90% of the phosphorylation events in the human proteome (18). Thus, the unambiguous identification of phosphopeptides is usually a challenging task.
To overcome these limitations, a wide variety of proteomic approaches have been developed. Several such strategies rely on the enrichment of the phosphorylated species prior to LC-MS analysis, typically by means of IMAC or TiO2 affinity chromatography (20). These techniques have also been successfully applied to the characterization of the phosphopeptidomes of several class I molecules (11–13).
In this study, we employed IMAC and TiO2 chromatography in combination with LC-MS to enrich and identify phosphorylated peptides associated with HLA-B*40:02. This strategy allowed the identification of a large number of endogenous ligands and the fine mapping of the B*40:02 binding motif. It was also effective for the characterization of the phosphopeptidome displayed by this allotype. Among the identified phospholigands, we found some sequences lacking the canonical binding motif at peptide position 2 (P2) and carrying a phosphoserine residue instead. Thus, the presentation of ligands harboring posttranslationally modified residues at major anchor positions contributes to the increased diversity of HLA class I peptidomes. We suggest that these sorts of epitopes might be valuable in T-cell-based cancer immunotherapy.
EXPERIMENTAL PROCEDURES
Cell Lines and Monoclonal Antibodies
HMy2.C1R (C1R) is a human lymphoid cell line with low expression of its endogenous HLA class I molecules. C1R cells show reduced levels of HLA-B*35:03 and normal expression of HLA-C*04:01 (21). A full-length cDNA clone of B*40:02 was obtained from the LCL line 143.2 (22) and cloned into the RSV5neo vector. C1R-B*40:02 transfectants were generated via electroporation of 107 C1R cells at 250 mV and 960 μF. To select stable transfectants, we grew the cells in the presence of 1 mg/ml geneticin (Invitrogen), and surface expression of HLA-B*40:02 was confirmed by flow cytometry. Cells were cultured in DMEM supplemented with 7.5% FCS (both from Sigma). The mAb W6/32 (IgG2a specific for a monomorphic HLA class I determinant) has been described elsewhere (23).
Isolation of the HLA-B*40-bound Peptide Pool
B*40:02-bound peptides were isolated as described elsewhere (24), with minor variations. About 1010 C1R-B*40:02 transfectant cells were lysed in 1% Igepal CA-630 (Sigma-Aldrich), 20 mm Tris, 150 mm NaCl (pH 7.5) in the presence of a mixture of protease inhibitors (Complete-Midi, Roche). No phosphatase inhibitors were included. The lysate was subjected to differential centrifugation for 10 min at 2000 × g, 30 min at 10,000 × g, and 1 h at 100,000 × g. After ultracentrifugation, the soluble fraction was subjected to affinity chromatography using the W6/32 mAb. HLA-B40-bound peptides were eluted with 0.1% aqueous TFA at room temperature and filtered through Centricon 3 devices (Amicon, Beverly, MA). Peptides were concentrated in a SpeedVac, desalted with an OMIX C18 tip (Varian, Palo Alto, CA), and dried completely.
Phosphopeptide Enrichment
Phosphopeptide enrichment entailed the combination of two previously described approaches, namely, IMAC with Fe3+ as ligand (25) and TiO2 chromatography (26). The peptide pool, reconstituted in 200 μl of loading solution (50% acetonitrile, 0.3% TFA, pH 1.5), was incubated at room temperature with 15 μl of Phos-Select iron affinity gel (Sigma) that was subsequently packed in a homemade tip column. This column was connected to a second one packed with Oligo R3 resin (Applied Biosystems, Foster City, CA). The flow-through was recovered and both columns were washed extensively, first with loading solution, then with transfer solution (1% phosphoric acid), and finally with washing solution (2% acetonitrile, 0.1% TFA). Elution was carried out using two solutions sequentially: (i) 50% acetonitrile, 0.1% TFA; and (ii) 30% acetonitrile, 0.06 mm NH4OH. Both eluates were mixed, dried to completeness, and redissolved in 0.1% formic acid (this sample is referred to as the IMAC fraction throughout the article).
The flow-through of the first chromatography was dried and reconstituted in 1 m glycolic acid, 80% acetonitrile, 5% TFA and applied to a microcolumn packed with Titansphere TiO2 resin (GL Sciences, Tokio, Japan). The unbound fraction was recovered, acidified with 10 μl of 0.1% formic acid, dried, and reconstituted in 0.1% TFA before desalting with a C18 ZipTip (Eppendorf, Hamburg, Germany). Afterward, the sample was dried and reconstituted in 0.1% formic acid (this sample is referred to as the flow-through fraction throughout the article). The TiO2 column was washed with 80% acetonitrile, 5% TFA and the peptides were eluted with two solutions: (i) 0.3 m NH4OH and (ii) 0.3 m NH4OH, 30% acetonitrile. Both eluates were mixed and the medium was acidified by the addition of formic acid. Then, the samples were desalted using a C18 ZipTip (Eppendorf), dried in a SpeedVac, and redissolved in 0.1% formic acid (this sample is referred to as the TiO2 fraction throughout the article).
LC-MS Analysis
Two technical replicates of each fraction (flow-through, IMAC, and TiO2) of the B*40:02-bound peptide pool were analyzed in a nano-LC Ultra HPLC (Eksigent, Framingham, MA) coupled online with a 5600 triple TOF mass spectrometer (AB Sciex, Framingham, MA) through a nanospray III source (AB Sciex). Chromatography was performed using a C18 chromXP trapping column (350 μm × 0.5 mm, 3-μm particle diameter, 120-Å pore size; Eksigent) and a C18 chromXP column (75 μm × 150 mm, 3-μm particle diameter, 120-Å pore size; Eksigent). Solvent A was 0.1% formic acid in water, and solvent B was 0.1% formic acid in acetonitrile. The loading pump was operated at isocratic conditions with buffer A at a flow rate of 2 μl/min for 10 min. The nanopump worked at 300 nl/min under gradient elution conditions as follows: 2% B for 1 min, a linear increase to 30% B in 109 min, a linear increase to 40% B in 10 min, a linear increase to 90% B in 5 min, and 90% B for 5 min. HPLC was controlled with the Eksigent Control software (version 3.12, Eksigent).
The nanospray source was equipped with a fused silica PicoTip emitter (10 μm × 12 cm, New Objective, Woburn, MA). The ion source was operated in positive ionization mode at 150 °C with a potential difference of 2800 V. Each acquisition cycle consisted of a survey scan of 250 ms between 350 and 1250 m/z units and a maximum of 50 MS2 spectra scanning between 100 and 1500 m/z units. Ions showing the highest intensities in the MS spectrum were selected for fragmentation. Singly charged ions were excluded to avoid the fragmentation of non-peptide contaminants. A dynamic exclusion window of 20 s was applied to each fragmented ion. The total cycle time was 2.8 s. The mass spectrometer was controlled with Analyst TF software (version 1.5, AB Sciex).
Synthetic peptides were dissolved in 0.5% formic acid and 20% acetonitrile at an estimated concentration of 250 fmol/μl. Peptides were directly infused at a flow rate of 3 μl/min into the 5600 triple TOF mass spectrometer. For retention time comparison of natural and synthetic phosphopeptides, a pool of 85 synthetic phosphopeptides, 100 fmol each, was prepared in 0.1% formic acid and analyzed via LC-MS/MS using the same chromatographic conditions described above. The extracted ion chromatogram of each phosphopeptide was used to determine its retention time.
MS/MS Ion Search and Peptide Identification
Raw data were processed with PeakView software (version 1.1, AB Sciex) to generate an MGF file that was used as input for a Mascot (version 2.4) MS/MS ion search. A concatenated target-decoy protein database containing 40,478 sequences was generated by combining the UniProt Homo sapiens complete proteome set (downloaded on May 23, 2011) with its corresponding reversed database generated with the DBToolKit software (version 4.1.4). Search parameters were set as follows: no enzyme; peptide tolerance, 15 ppm; MS/MS tolerance, 25 mDa; and electrospray ionization quadrupole TOF as instrument. Variable modifications included phosphorylation of serine, threonine, and tyrosine; oxidation of methionine; and pyroglutamic acid formation from N-terminal glutamine. Peptide sequences that matched the HLA-C*04:01 binding motif, Phe or Tyr at P2 and Asp at P3 (27), were discarded. Estimation of the false discovery rate (FDR) was carried out by decoy hit counting as previously described (28, 29), and only those matches with an FDR < 5% at the peptide level were considered. All the information related to the MS analysis, MS/MS ion search, and peptide identification, including raw data, MS metadata, MGF and mzIdentML files, and the corresponding MIAPE MS and MSI documents, were deposited in ProteomeXchange (PRIDE accession number 31118, ProteomeXchange accession number PXD000450). This process was aided by the MIAPE extractor tool (version 3.7.0).
For the identification of phosphorylated ligands, every match with a Mascot score greater than 25 was considered. Then, MS2 spectra were manually inspected for signals that could correspond to the neutral loss of the phosphate group (−98, −49, and −32.7 for singly, doubly, and triply charged ions, respectively). Putative phosphopeptides were validated by means of retention time comparison and fragmentation of the corresponding synthetic peptide (supplemental data S2).
Statistical Analysis
To assess preferences in residue usage, we grouped the identified peptides according to their length. The frequency of each residue at each peptide position (fobs) was compared with the frequency of the same amino acid in the database (fexp) under the null hypothesis that fobs ≤ fexp. Preliminary p values for each residue and position were calculated assuming a binomial distribution with p = fexp. Definitive P values were obtained by subjecting preliminary p values to multiple testing correction as follows:
where k is the number of residues of each peptide in the set tested (i.e. k = 9 in the nonamer set). P values less than 0.05 were considered statistically significant.
Peptide Synthesis
The stepwise solid-phase peptide synthesis was performed on an automated Multipep peptide synthesizer (Intavis, Koeln, Germany) using standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry. Peptides were purified via reversed-phase chromatography either in a Smartline HPLC instrument (Knauer, Berlin, Germany) equipped with a 218TP52 C18 column (Vydac, Deerfield, IL) or using an Oligo R3 (Applied Biosystems) microcolumn. Peptides intended for binding assays were quantified by means of amino acid analysis in a Biochrom 30 amino acid analyzer (Biochrom, Cambridge, UK). The peptide GEFGGCGSV was labeled after synthesis with 5-iodoacetamidofluoresceine (Thermo) following the manufacturer's instructions. Afterward, it was purified by means of reversed-phase HPLC and quantified based on absorbance at 491 nm as described elsewhere (30).
Peptide Binding Assays
Binding assays were performed essentially as described elsewhere (30, 31), with minor variations. In brief, C1R-B*40:02 transfectants were washed twice with PBS and acid stripped via incubation in 0.263 m citric acid, 0.123 m Na2HPO4, 1% BSA, pH 3, for 2 min at 4 °C. After two washes with ice-cold DMEM, the cells were incubated in DMEM supplemented with 5% FCS, 2 μg/ml of human β2m (Calbiochem), 400 nm fluorescent reference peptide GEFGGXGSV (where X represents fluoresceine-labeled cysteine), and different concentrations of the test peptides ranging from 50 μm to 23 nm in 3-fold dilutions. After overnight incubation at 4 °C, fluorescence was measured in an Epics XL-MCL flow cytometer (Beckman Coulter). Inhibition of reference peptide binding was plotted versus the test peptide concentration, and the IC50 (the concentration of test peptide that gives 50% inhibition) was estimated after the experimental results had been fitted to a sigmoid curve, as previously described (30).
RESULTS
Size Distribution of the B*40:02 Peptidome
HLA-B*40:02 was affinity purified and its constitutive peptidome was acid extracted and isolated via centrifugal filtration. The phosphorylated species in the peptide pool were enriched by means of IMAC and TiO2 chromatography. This strategy yielded three fractions (namely, IMAC eluate, TiO2 eluate, and flow-through) that were then analyzed via LC-MS.
Peptide matches were filtered at an FDR < 5% at the peptide level (FDR < 2.4% at the peptide spectrum match level, Mascot score > 45.73). A total of 2246 unique ligands were identified, including 337 (15%) octamers, 1131 (50%) nonamers, 445 (20%) decamers, 245 (11%) undecamers, 74 (3%) dodecamers, and 14 (1%) tridecamers (Fig. 1A and supplemental data S1). The size distribution of this set of ligands showed a Gaussian-like pattern with a mean (± S.D.) molecular weight of 1073.2 (± 130.0) Da (Fig. 1B).
Fig. 1.
Analysis of the HLA-B*40:02-associated ligandome. A, length distribution of the ligands identified from the peptide pool bound to B*40:02. B, molecular weight distribution of the same set of peptides. The mean (± S.D.) molecular weight of the identified peptides is shown. C, Venn diagram showing the number of sequences identified in the flow-through (continuous line), the IMAC (dashed line), and the TiO2 (dotted line) fractions.
Not surprisingly, the vast majority of the ligands (1935, 86%) were identified in the flow-through fraction, including 1581 sequences (70%) that were not detected in the IMAC or TiO2 samples. The numbers of ligands in the IMAC and TiO2 eluates were 399 (18%) and 371 (17%), respectively, including 79 (4%) and 203 (9%) peptides that were found in these fractions exclusively (Fig. 1C).
Fine Mapping of the B*40:02 Binding Motif
To determine the B*40:02 binding motif, peptides were grouped according to their length. Only octamers to undecamers were considered because the number of sequences was large enough for proper statistical analysis only in these sets. We assumed that in peptide positions not subjected to structural constraints, residue usage should mirror the frequencies of each amino acid in the proteome. Conversely, the overrepresentation of one or more particular residues would reflect the binding preferences of HLA-B40. The frequency of each residue at each peptide position (fobs) was compared with the frequency of that particular amino acid in the database (fexp). A residue was considered to be favored at a given position if the difference between fobs and fexp was statistically significant (p < 0.05) after multiple testing correction.
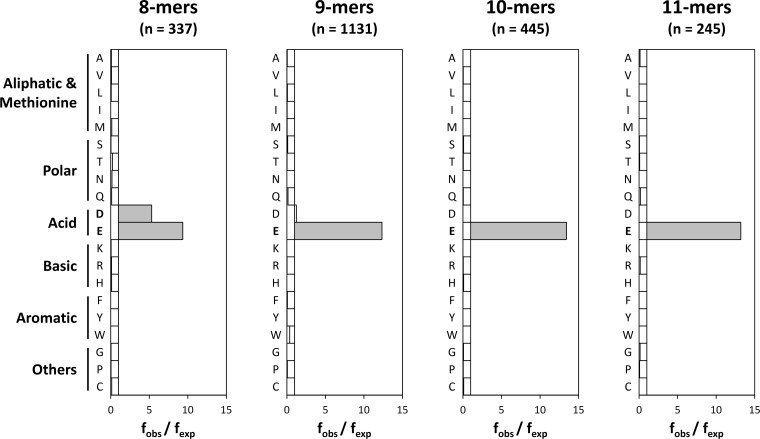
The major restriction for binding to B*40:02 involved P2. B*40:02 ligands showed an almost absolute preference for acidic residues at this position (Fig. 2). Although both Asp and Glu could be accommodated, the frequency of Asp2 was appreciably lower and declined as peptide length increased. Among octamers, 228 (68%, fobs/fexp = 9.4) and 86 (26%, fobs/fexp = 5.3) peptides showed Glu2 and Asp2 motifs, respectively. In the case of nonamers, Glu2 was present in 1010 ligands (89%, fobs/fexp = 12.3) and Asp2 was found only in 67 (6%, fobs/fexp = 1.2). Notably, no peptides containing Asp2 were observed among decamers and undecamers, where the frequency of Glu2 reached 97% (fobs/fexp = 13.4) and 96% (fobs/fexp = 13.2), respectively.
Fig. 2.
Residue usage at P2 among B*40:02 ligands. The observed frequency (fobs) of each amino acid residue at P2 is compared with the frequency of the same residue in the database (fexp). Statistically significant differences (p < 0.05 after multiple testing correction) are indicated with gray bars.
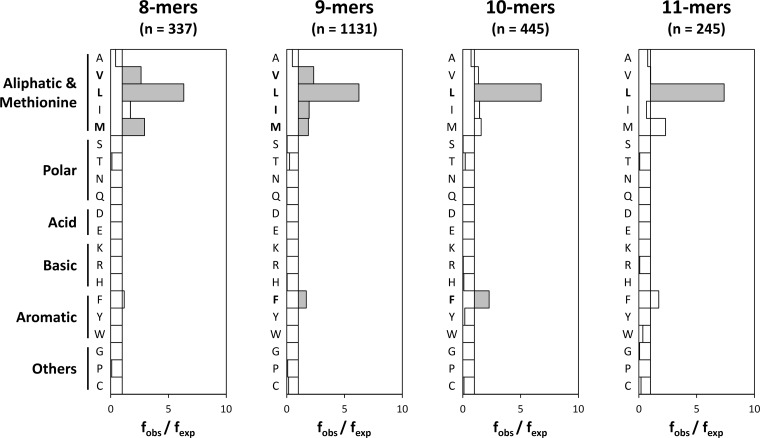
The second most relevant anchor position was the peptide C terminus (PΩ), which showed a slightly more relaxed specificity than P2 (Fig. 3). In this case, a strong selection of Leu was observed encompassing 210 octamers (62%, fobs/fexp = 6.3), 696 nonamers (62%, fobs/fexp = 6.3), 297 decamers (67%, fobs/fexp = 6.8), and 178 undecamers (73%, fobs/fexp = 7.4). Other aliphatic residues (Val, Ile), Met, and Phe were also significantly overrepresented. Finally, Ala could also be found at PΩ, albeit at a lower frequency than expected (fobs/fexp < 1).
Fig. 3.
Residue usage at the peptide C terminus among B*40:02 ligands. The observed frequency (fobs) of each amino acid residue at PΩ is compared with the frequency of the same residue in the database (fexp). Conventions are the same as in Fig. 2.
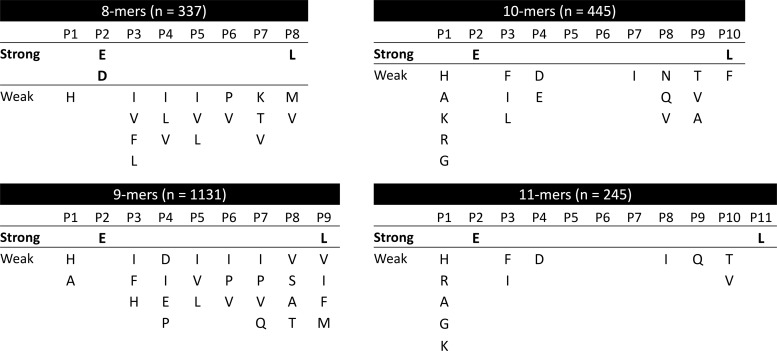
Besides these two main anchors, restrictions in residue usage were observed in most peptide positions, although much more subtle than those affecting P2 and PΩ (Fig. 4). Only the central regions of decamers (P5 and P6) and undecamers (P5, P6, and P7) showed no preference for any particular amino acid (Fig. 4).
Fig. 4.
Fine mapping of the B*40:02 binding motif. Residues statistically overrepresented (p < 0.05 after multiple testing correction) at any peptide position are shown ordered downward according to their fobs/fexp ratio. Binding motifs were arbitrary classified as strong (fobs/fexp > 5) or weak (fobs/fexp < 5).
Characterization of the B*40:02-bound Phosphopeptidome
In order to gain insight into the B*40:02 phosphopeptidome, an enrichment strategy was deployed. Two tandem affinity steps (namely, IMAC with Fe3+ and TiO2 affinity chromatography) were used to capture the putative phosphorylated species present in the B*40:02 peptide pool. Afterward, LC-MS analysis of the bound material and database searching were performed.
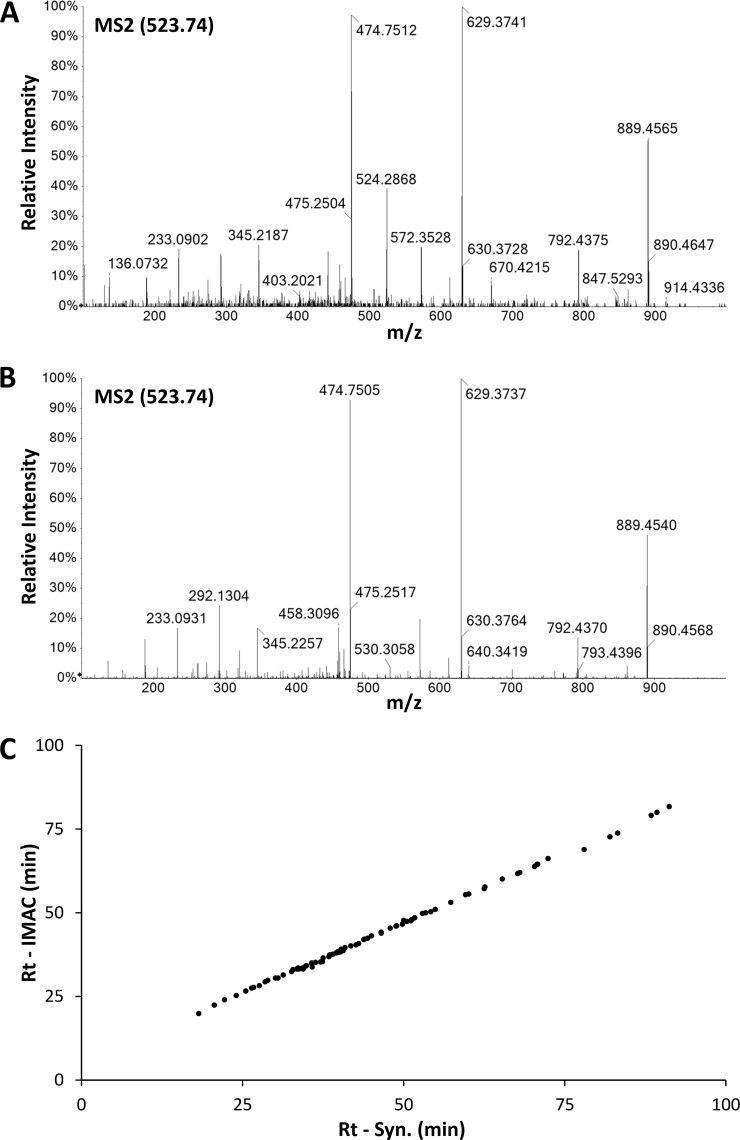
To bypass the limitations related to the identification of phosphopeptides via MS, every peptide match with a score greater than 25 in the Mascot search was considered. Then, MS2 spectra were manually inspected for signals that could derive from the neutral loss of phosphoric acid (see “Experimental Procedures”). Finally, putative phosphorylated sequences were further confirmed through comparison of the retention times and the MS2 spectra of the endogenous and the synthetic peptides (Fig. 5 and supplemental data S2).
Fig. 5.
Sequence confirmation of phosphorylated ligands. The identity of each reported phosphopeptide was confirmed by fragmentation of the corresponding synthetic peptide. A, experimental MS2 spectrum of a molecular species at m/z = 523.74 fragmented during LC-MS analysis of the B*40:02-bound peptidome. B, MS2 spectrum of the synthetic peptide S[pS]YGNIRAV. C, the retention time of each phosphorylated peptide in the IMAC fraction (Rt - IMAC) is plotted against the retention time of the corresponding synthetic peptide (Rt - Syn.) in an independent LC-MS run. The data fit a linear function (R2 = 0.9991).
A total of 85 unique phosphopeptides were sequenced using this approach (Table I and supplemental data S2). Of them, 69 (81%) were identified only in the IMAC eluate and 16 (19%) were found in both the IMAC and the TiO2 samples. No single peptide belonged exclusively to the TiO2 fraction. In our dataset, phosphorylation occurred exclusively at serine (77 sequences, 91%) and threonine residues (8 sequences, 9%) and peptides containing phosphotyrosine could not be identified. Notably, in 60 ligands (71%) phosphorylation involved SP or TP sites (i.e. phosphorylation occurred before a proline residue). Finally, 48 out of the 85 phosphorylated positions described (56%) had been previously annotated in either the HPRD or the UniProt database (Table I). Thus, the remaining 44% were novel phosphosites described here for the first time.
Table I. Phosphorylated ligands displayed by HLA-B*40:02.
| N | Sequence | AN | Protein name | IMAC | TiO2 | Reference |
|---|---|---|---|---|---|---|
| 1 | AEL[pS]PVEQKL | Q9Y5P8 | Protein phosphatase 2A 48 kDa regulatory subunit | + | + | This study |
| 2 | REL[pS]PQRNL | Q96F44 | E3 ubiquitin-protein ligase TRIM11 | + | − | This study |
| 3 | TER[pS]PGPRL | O14978 | Zinc finger protein 263 | + | + | This study |
| 4 | IES[pS]PQYRL | Q8WWM7 | Ataxin-2-like protein | + | + | (53) |
| 5 | KEKDF[pS]PEAL | O15372 | Eukaryotic translation initiation factor 3 subunit H | + | − | (54) |
| 6 | GEW[pS]ASLPHRF | O43164 | E3 ubiquitin-protein ligase Praja-2 | + | − | This study |
| 7 | SEA[pS]PSREAI | Q13111 | Chromatin assembly factor 1 subunit A | + | + | (55) |
| 8 | YEG[pS]PIKVTL | P06748 | Nucleophosmin | + | + | (56) |
| 9 | HERSI[pS]PLL | Q9UBT6 | DNA polymerase κ | + | + | This study |
| 10 | VEKKKIR[pS]L | O75394 | 39S ribosomal protein L33, mitochondrial | + | − | This study |
| 11 | AEN[pS]PTRQQF | Q86XP3 | ATP-dependent RNA helicase DDX42 | + | − | (56) |
| 12 | QEV[pT]PSRSTL | P49454 | Centromere protein F | + | + | (57) |
| 13 | RE[pS]PPPSAL | Q86VE0 | Myb-related transcription factor, partner of profilin | + | − | This study |
| 14 | REK[pT]PSEDKL | Q8NFC6 | Biorientation of chromosomes in cell division protein 1-like 1 | + | − | This study |
| 15 | GEDR[pS]PPTAL | Q9BUB5 | MAP kinase-interacting serine/threonine-protein kinase 1 | + | − | (51) |
| 16 | REV[pS]PAPAVA | O60292 | Signal-induced proliferation-associated 1-like protein 3 | + | − | (50) |
| 17 | KEV[pT]PKPVL | P17544 | Cyclic AMP-dependent transcription factor ATF-7 | + | − | This study |
| 18 | HEL[pS]PPQKRM | Q9BXP5 | Serrate RNA effector molecule homolog | + | − | (55) |
| 19 | RE[pT]PSPRPM | Q9UQ35 | Serine/arginine repetitive matrix protein 2 | + | − | (58) |
| 20 | REYG[pS]PLKA | Q9UQ88 | Cyclin-dependent kinase 11A | + | − | (51) |
| 21 | KEM[pS]PTRQL | Q4G0N7 | Protein FAM229B | + | + | This study |
| 22 | GENK[pS]PLLL | Q8IZD2 | Histone-lysine N-methyltransferase MLL5 | + | + | This study |
| 23 | VEFPH[pS]PEI | Q9BR61 | Acyl-CoA-binding domain-containing protein 6 | + | + | This study |
| 24 | YEKAN[pT]PEL | Q9H8V3 | Protein ECT2 | + | + | (55) |
| 25 | VEK[pS]PRPPL | Q7L590 | Protein MCM10 homolog | + | − | This study |
| 26 | QEK[pS]PKQAL | O43683 | Mitotic checkpoint serine/threonine-protein kinase BUB1 | + | − | (51) |
| 27 | GEI[pS]PQREV | Q8WWI1 | LIM domain only protein 7 | + | − | (56) |
| 28 | HEFH[pS]QENL | Q9Y3M8 | StAR-related lipid transfer protein 13 | + | − | (59) |
| 29 | REG[pS]GRFSL | P57764 | Gasdermin-D | + | − | This study |
| 30 | GEN[pS]GIGKL | Q6P0N0 | Mis18-binding protein 1 | + | − | (51) |
| 31 | SEI[pS]PIKGSV | Q7Z5K2 | Wings apart-like protein homolog | + | − | (56) |
| 32 | SEK[pS]LENETL | P23588 | Eukaryotic translation initiation factor 4B | + | − | (56) |
| 33 | RED[pS]QRPGAHL | Q32P51 | Heterogeneous nuclear ribonucleoprotein A1-like 2 | + | − | (60) |
| 34 | RED[pS]GTFSL | O75569 | Protein activator of the interferon-induced protein kinase | + | − | (51) |
| 35 | REA[pS]PSRLSV | O75122 | CLIP-associating protein 2 | + | + | This study |
| 36 | SEQ[pS]LGQKL | P33991 | DNA replication licensing factor MCM4 | + | − | (55) |
| 37 | REV[pS]PAPAV | O60292 | Signal-induced proliferation-associated 1-like protein 3 | + | − | (50) |
| 38 | SERM[pS]LPLL | Q15398 | Disks large-associated protein 5 | + | − | (61) |
| 39 | SES[pS]PPKAM | Q9UHB6 | LIM domain and actin-binding protein 1 | + | − | (51) |
| 40 | SEV[pS]PSGVGF | Q6ULP2 | Aftiphilin | + | − | This study |
| 41 | REK[pS]PGRML | O14578 | Citron Rho-interacting kinase | + | − | (50) |
| 42 | SEF[pS]DVDKL | O75665 | Oral-facial-digital syndrome 1 protein | + | − | This study |
| 43 | REA[pS]PLSSNKL | Q9Y462 | Zinc finger protein 711 | + | − | This study |
| 44 | REIES[pS]PQYRL | Q8WWM7 | Ataxin-2-like protein | + | − | (53) |
| 45 | REY[pS]PPYAP | Q9UPT8 | Zinc finger CCCH domain-containing protein 4 | + | − | (51) |
| 46 | KEKEV[pS]QEL | A8MX76 | Calpain-14 | + | + | This study |
| 47 | KEL[pS]PPPGL | Q7Z4S6 | Kinesin-like protein KIF21A | + | + | (51) |
| 48 | RER[pS]PSPSF | P49585 | Choline-phosphate cytidylyltransferase A | + | − | (56) |
| 49 | GET[pS]LMRTL | O96017 | Serine/threonine-protein kinase Chk2 | + | − | (62) |
| 50 | AEK[pS]PGPIV | Q86WB0 | Nuclear-interacting partner of ALK | + | − | (51) |
| 51 | REN[pS]PVQSI | Q7Z2Z1 | Treslin | + | − | This study |
| 52 | RET[pS]PNRIGL | Q7Z460 | CLIP-associating protein 1 | + | + | This study |
| 53 | REG[pS]FRVTTA | P49757 | Protein numb homolog | + | − | This study |
| 54 | PEV[pS]PRPAL | Q96L73 | Lysine N-methyltransferase 3B | + | − | This study |
| 55 | KEL[pS]PAGSI | O95453 | Poly(A)-specific ribonuclease PARN | + | − | (55) |
| 56 | REG[pS]YDRYL | Q9HCD5 | Nuclear receptor coactivator 5 | + | − | (63) |
| 57 | S[pS]YGNIRAV | Q8N122 | Regulatory-associated protein of mTOR | + | − | (64) |
| 58 | GEPHS[pS]PEL | Q9NUA8 | Zinc finger and BTB domain-containing protein 40 | + | − | This study |
| 59 | REK[pS]PGRMLST | O14578 | Citron Rho-interacting kinase | + | − | This study |
| 60 | SEL[pS]PGRSV | Q8NFF5 | FAD synthase | + | − | (50) |
| 61 | HEFS[pS]PSHL | Q9H3D4 | Tumor protein 63 | + | − | This study |
| 62 | QEK[pS]PEERL | Q7RTP6 | Protein-methionine sulfoxide oxidase MICAL3 | + | − | (51) |
| 63 | KEQ[pS]PEPHL | Q5VV52 | Zinc finger protein 691 | + | − | (65) |
| 64 | NEA[pS]PVKRPRL | Q9H8U3 | AN1-type zinc finger protein 3 | + | − | (66) |
| 65 | G[pS]FSRFYSL | P22695 | Cytochrome b-c1 complex subunit 2, mitochondrial | + | − | This study |
| 66 | REF[pS]KEPEL | Q9NQS7 | Inner centromere protein | + | − | (57) |
| 67 | SEKPV[pS]PKSGTL | Q14155 | Rho guanine nucleotide exchange factor 7 | + | − | (63) |
| 68 | REL[pS]PLISL | P51825 | AF4/FMR2 family member 1 | + | − | (50) |
| 69 | RE[pT]PVPPAM | Q9Y2F5 | Uncharacterized protein KIAA0947 | + | − | (56) |
| 70 | REP[pS]PSSSSL | O60336 | Mitogen-activated protein kinase-binding protein 1 | + | − | This study |
| 71 | LEKRT[pS]PQVL | Q7Z417 | Nuclear fragile X mental retardation-interacting protein 2 | + | − | (51) |
| 72 | REL[pS]GTIKEI | P30050 | 60S ribosomal protein L12 | + | − | This study |
| 73 | GEA[pS]PSHII | Q9ULL5-3 | Proline-rich protein 12 | + | − | This study |
| 74 | GEQ[pS]PNVSL | Q58WW2 | DDB1- and CUL4-associated factor 6 | + | − | (51) |
| 75 | KE[pT]PDKVEL | Q13610 | Periodic tryptophan protein 1 homolog | + | − | (57) |
| 76 | NEFH[pS]PIGL | Q06413 | Myocyte-specific enhancer factor 2C | + | + | This study |
| 77 | REV[pS]PEPIV | Q8N103 | T-cell activation Rho GTPase-activating protein | + | − | This study |
| 78 | NEK[pS]PYASL | Q6IQ21 | Zinc finger protein 770 | + | − | This study |
| 79 | REL[pS]GTIKEIL | P30050 | 60S ribosomal protein L12 | + | − | This study |
| 80 | SERIMQL[pS]L | Q81IY63 | Angiomotin-like protein 1 | + | − | This study |
| 81 | R[pS]FPTLPTL | Q13761 | Runt-related transcription factor 3 | + | − | This study |
| 82 | KES[pS]PIPSP | Q01082 | Spectrin β chain, non-erythrocytic 1 | + | − | (51) |
| 83 | SE[pS]GSNLHAL | Q6P4F7 | Rho GTPase-activating protein 11A | + | − | This study |
| 84 | KELARQI[pS]F | Q9Y385 | Ubiquitin-conjugating enzyme E2 J1 | + | − | This study |
| 85 | KEN[pT]PPAL | P31350 | Ribonucleoside-diphosphate reductase subunit M2 | + | − | (50) |
The sequence, accession number (AN), and putative parental protein are indicated. + and − symbols specify whether the peptide was sequenced from the IMAC or the TiO2 fraction. If the described phosphorylation was annotated in the HPRDB and/or the UniProt database, a reference is given. Peptides phosphorylated at P2 (n + 57, 65, and 81) are shown in bold letters. Phosphoserine and phosphothreonine are represented as [pS] and [pT], respectively.
B*40:02 Displays Peptides with Phosphorylated Residues at Position 2
Interestingly, 3 out of the 85 phosphorylated ligands identified in this study lacked Asp or Glu at P2 and contained phosphoserine (pSer) instead: S[pS]YGNIRAV, G[pS]FSRFYSL, and R[pS]FPTLPTL (peptides 57, 65, and 81 in Table I and supplemental data S2). It is worth noting that pSer and Glu share structural similarities. Both residues hold side chains of comparable length with a net negative charge. As a consequence, we reasoned that the interaction of pSer with the B pocket could confer enough stability to the complex to allow B*40:02 to display ligands with phosphorylated residues at P2.
To test this hypothesis, the natural ligands S[pS]YGNIRAV and G[pS]FSRFYSL and the related mutant peptides SEYGNIRAV and GEFSRFYSL were tested for binding to B*40:02. In this assay, C1R-B*40:02 cells were acid stripped to dissociate surface HLA class I complexes. Then, a reference peptide that bound specifically to HLA-40 (Fig. 6A) was added to the cells together with human β2m and different concentrations of the test peptides. The amount of fluorescent peptide bound to B*40:02 was determined via flow cytometry, and the binding affinity of the test peptides was inferred from the concentration-dependent inhibition of the binding of the reference peptide.
Fig. 6.

Competitive binding assay of peptides with phosphoserine at P2 to HLA-B*40:02. C1R-B*40:02 cells were acid stripped to denature surface HLA class I complexes. After extensive washing, β2m, the reference peptide GEFGGXGSV (X = fluoresceine-labeled cysteine), and different concentrations of the indicated test peptides were added. HLA class I complexes were left to reassociate overnight at 4 °C. A, binding of the reference peptide to C1R (blue line) and C1R-B40 (red line). The percentage of the maximum fluorescence is plotted against the peptide concentration. The mean ± S.D. of three independent experiments is shown. B, the inhibition of the binding of the reference peptide is plotted against the concentration of the test peptides (colored dots). The mean of three independent experiments is shown. Experimental data were fitted to sigmoid curves (colored lines) to allow the estimation of the IC50 values. C, estimated IC50 values of the test peptides.
Both phosphopeptides bound to B*40:02 with high affinity. The IC50 value, defined as the concentration of test peptide that yielded 50% inhibition, was 1.8 and 0.3 μm for S[pS]YGNIRAV and G[pS]FSRFYSL, respectively (Fig. 6). Likewise, the mutant peptides SEYGNIRAV and GEFSRFYSL showed affinity similar to or slightly higher than that of their phosphorylated counterparts (IC50 = 1.3 and 0.2 μm, respectively). This further confirmed that S[pS]YGNIRAV and G[pS]FSRFYSL are true B*40:02 ligands and indicated that the substitution of Glu by pSer has little or no effect on binding affinity. The peptide GRIDKPILK, a known HLA-B*27:05 ligand, was included as a negative control, as it lacks proper motifs at P2 and PΩ to fit the B*40:02 groove. As seen in Fig. 6, this ligand failed to inhibit the binding of the reference peptide, demonstrating that the rest of the inhibition curves actually reflect specific binding of the test peptides.
DISCUSSION
Modern mass spectrometers in combination with database searching strategies allow high-throughput identification of peptides and proteins on a routine basis. The same techniques devised to identify proteins have been applied to the characterization of class I–bound peptide repertoires (4, 32–34). The identification of HLA class I–bound peptidomes, however, is usually a more complex task because of the relatively low amount of sample typically available. We estimate that after affinity purification, only about 2 to 4 μg of peptides are obtained from 1010 cells transfected with the allotype of interest (data not shown). Despite this drawback, the high-throughput identification of HLA ligands is now feasible, and some authors have even proposed the staging of a Human Immunopeptidome Project, analogous to the Human Proteome Project (35), to systematically characterize the ligandomes of HLA antigens (36).
In this work, we focused on the characterization of the peptidome and phosphopeptidome presented by HLA-B*40:02, a member of the B44 supertype (8). B*40:02 has been reported to predispose to adult T-cell leukemia, a non-Hodgkin's lymphoma caused by human T-lymphotropic virus type 1 (37). Apparently, this association is explained by the limited capability of HLA-B*40:02 to present epitopes derived from human T-lymphotropic virus type 1 and to trigger a strong CTL response. If tumor-specific epitopes were described, it is conceivable that B40+ adult T-cell leukemia patients could benefit from T-cell-based immunotherapy.
The HLA-B40-associated peptide pool was affinity purified and its constitutive peptide repertoire was acid extracted. Afterward, phosphorylated ligands were enriched sequentially by means of IMAC and TiO2 affinity chromatography, yielding three different fractions, IMAC, TiO2, and flow-through, that were then analyzed via LC-MS. An MS/MS ion search allowed the identification of more than 2000 B*40:02 ligands. Most of them were identified in the flow-through, as expected, but both the IMAC and the TiO2 fractions contributed significantly to the number of detected ligands, providing more than 300 additional sequences. The size distribution of this peptide pool was that expected for a class I ligandome, with nonamers being by far the most abundant species and accounting for 50% of the peptide repertoire. Additionally, a relatively high frequency of octamers (15%) was detected. Although not common, this feature is shared by other HLA class I antigens such as B37 or B18 (38).
For fine mapping of the B40 binding motif, peptides were grouped according to their lengths. This grouping is required because class I ligands are anchored through their N and C termini (1) to the heavy chain. Consequently, short peptides are bound in an extended conformation, whereas the central region of longer ligands protrudes from the groove (39–42). For this reason, the proper alignment of peptides of different lengths is not straightforward, especially regarding peptide positions P4 to PΩ-2.
The major constraint for binding to B40 was found at P2, where the overwhelming majority of ligands contained acidic residues. Indeed, only 4.7% of the identified peptides showed alternative amino acids at this position, which is consistent with the estimated FDR of the whole set (<5%). This indicates that the presence of Glu2 or Asp2 is mandatory for binding to B*40:02 and suggests that the sequences without this motif are probably random matches. It is also possible that contaminating peptides bound to HLA-B*35:03 were present in our dataset because, in contrast to C*04:01, B35 ligands were not filtered out during data analysis. However, only 8 out of the 2246 reported sequences (0.36%) matched the HLA-B*35:03 binding motif (43). This is consistent with the very low expression level of HLA-B35 in C1R cells caused by a point mutation in its translation initiation codon (21).
Although the three-dimensional structure of HLA-B*40:02 has not yet been elucidated, other members of the B44 supertype with an identical B pocket, such as B41, show a similar restriction at P2 (39). Analysis of the crystal structures of HLA-B*41:03 and B*41:04 reveals the presence of hydrogen bonds between the residue at P2 with Tyr99 and Glu63, van der Waals interactions with Tyr7, and potential salt bridges with His9 and Lys45 (39). As a general rule, position 45 is critical for the specificity of the B pocket. In this regard, allotypes with Lys45 such as B41 (39) or B44 (44) bind peptides with acidic residues at P2, whereas other molecules with Glu45, such as B27, show an almost absolute preference for Arg at this position (33, 45, 46).
Intriguingly, a size-dependent modulation of residue usage at P2 was found. Whereas B40 bound octamers with Glu and Asp at this position (68% and 26% of the sequences, respectively), among nonamers, ligands with Asp2 accounted for only 6% of the peptide set. Furthermore, no decamers or undecamers containing Asp2 were identified. At present, we have no structural explanation for this finding, and probably the determination of the three-dimensional structure of B40 will be required in order for light to be shed on this issue.
As in other HLA class I antigens, the second most influential position for binding to B*40:02 was found at the peptide C terminus. Residues with hydrophobic side chains at PΩ were found in most cases (Fig. 3 and supplemental data S1). By far, Leu was the preferred C-terminal residue and was present in about 65% of the identified ligands. Phe, Val, Ile, and Met were statistically overrepresented, at least in the nonamer set. Finally, Ala was also found in a number of ligands, although its frequency, lower than expected by chance, suggests that it is a suboptimal anchor motif. The molecular basis for this preference can be inferred from the crystal structure of B41, which shares with B40 most of the residues that shape the F pocket, including the key amino acids Leu95, Tyr116, Tyr123, and Trp147. In B*41:03 and B*41:04, the side chain of the residue at PΩ is deeply buried in a hydrophobic F pocket in contact with the abovementioned residues (39). Regarding the main anchor positions, a similar binding motif has been described for the closely related allotype HLA-B*40:01 (38). However, some differences in residue usage exist. B*40:01 is more restrictive at P2; only Glu is found at this position, and at PΩ, where Phe is particularly disfavored. The structural basis for this discrepancy is not clear, as both allotypes share the same B pocket and a nearly identical F pocket. Perhaps the limited number of B*40:01 ligands identified to date—56 in the abovementioned study (38)—or indirect effects involving secondary anchor residues could explain this divergence.
Finally, most peptide positions showed some bias in residue usage, with the exception of P5 and P6 in decamers and P5, P6, and P7 in undecamers (Fig. 4). This lack of selection is probably a consequence of the bulged conformation that long peptides adopt to fit the binding groove (39–42). As a result, the central region of the peptide establishes no contact with the heavy chain, and thus there are no structural constraints that drive the selection of particular motifs.
One of the main goals of this work was the characterization of the phosphopeptidome displayed by HLA-B40. The identification of phosphorylated species associated with HLA class I molecules has gained considerable attention since they were proposed as putative targets for cancer immunotherapy (12, 13). Nevertheless, the identification of phosphopeptides via mass spectrometry is challenging because of several analytical limitations, such as their low stoichiometry, their inefficient ionization, or the poorly informative MS2 spectra obtained upon their fragmentation by collision-induced dissociation (18, 19). To circumvent these pitfalls, two strategies were adopted: (i) perform phosphopeptide enrichment prior to LC-MS analysis, and (ii) validate every single identification by comparing both the retention times and the MS2 spectra of the natural and the synthetic peptides.
Enrichment of phosphopeptides is mandatory for the mapping of phosphorylation events in classical bottom-up workflows (18). In the same way, identification of HLA class I–associated phospholigands has benefited from the implementation of these approaches (11–13). We combined IMAC and TiO2 affinity chromatography before LC-MS analysis, resulting in the identification of 85 B40-bound phosphopeptides. To our knowledge, this is the largest set of MHC class I phosphorylated ligands reported to date. All of them were found in the IMAC fraction, and 16 (19%) were also observed in the TiO2 sample. No single identification belonged exclusively to the TiO2 set, indicating that, in terms of the identification of phosphopeptides, the contribution of TiO2 affinity chromatography after IMAC was not really valuable. However, as stated above, about 200 nonphosphorylated endogenous ligands were sequenced from this fraction exclusively, meaning that fractionating the B40-bound peptide pool using TiO2 columns had a positive effect on the sensitivity of the LC-MS analysis.
To overcome the low quality of their MS2 spectra, all the phosphorylated sequences with Mascot scores greater than 25 were manually analyzed, and those showing signals compatible with a neutral phosphate loss were compared with the fragmentation spectra of the equivalent synthetic peptide to remove false positives. In addition, the retention times of the natural and the synthetic phosphopeptides were found to correlate closely. This approach guaranteed that the set of phosphorylated sequences presented in this study was highly curated.
In the 85 sequences reported, phosphorylation occurred in serine (91%) and threonine (9%) but not in tyrosine residues. This distribution parallels the relative frequencies of these posttranslational modifications in the proteome, namely, 90%, 10%, and <0.05% for phosphoserine, phosphothreonine, and phosphotyrosine, respectively (18). In a relatively high number of cases, phosphorylation occurred before a proline residue, probably reflecting the substrate specificity of proline-directed serine/threonine kinases, such as the MAP or the cyclin-dependent protein kinase families, which recognize and phosphorylate SP or TP sites (47). About half of the phosphorylation events described in this study had been previously reported, though not in the context of HLA-class peptide repertoires, supporting the accuracy of the identifications. 37 sequences revealed novel phosphorylation sites described here for the first time. This proves that HLA peptidomics may also contribute to the characterization and annotation of posttranslational modifications in proteins.
The main finding of this work was the identification of three ligands that lacked the canonical B40 binding motif at P2. These sequences were not false positives, as the MS2 spectra of the corresponding synthetic peptides were identical to the experimental ones. One of them derives from residues 721 to 729 of the regulatory-associated protein of mTOR (Raptor), a component of the mammalian target of rapamycin complex I, which regulates cell growth and autophagy in response to starvation (48, 49). Phosphorylation of Raptor at Ser722 has been previously described (50, 51). The other two sequences correspond to novel phosphorylation sites of cytochrome b-c1 complex subunit 2, a member of respiratory chain complex III, and runt-related transcription factor 3.
The three sequences harbored a pSer residue at the main anchor position instead of Asp or Glu. Furthermore, S[pS]YGNIRAV and G[pS]FSRFYSL were shown to bind to B40 with high affinity, confirming that they were bona fide ligands. Given the structural similarities between pSer and Glu in terms of size and charge distribution, we hypothesized that pSer and acidic residues would interact with the B pocket in a similar way, leading to the formation of stable complexes. Indeed, these two ligands behaved similarly, in terms of binding affinities, to the related mutant sequences SEYGNIRAV and GEFSRFYSL.
The identification of HLA class I ligands harboring phosphorylated residues at their major anchor position might be relevant in the design of immunotherapeutic approaches for cancer treatment. Given that abnormal phosphorylation is frequently observed in transformed cells, tumor-specific phosphopeptides are obvious candidates for T-cell-based therapies. However, besides a target epitope, a specific CTL response is required in order to eradicate transformed cells. When phosphorylation occurs at nonanchor residues, both the phosphorylated and the nonphosphorylated species will likely be displayed. If this is the case, a T-cell might recognize both species due to cross-reactivity. Although specific CTLs can be raised against phosphopeptides (11), some cytotoxic activity against their nonphosphorylated counterparts may be present (52). Thus, even if a CTL recognizing a phosphorylated ligand could escape negative selection in the thymus, cross-reactivity with the nonphosphorylated epitope would hamper its use as a therapeutic agent. In contrast, peptides phosphorylated at P2 can bypass this limitation, as the posttranslational modification of the residue is essential for binding to the class I molecule. In this scenario, cross-reactivity with the unmodified epitope is not possible, guaranteeing the specificity of the CTL response. Therefore, an important goal of future work will be the identification of HLA class I ligands phosphorylated at P2 in tumor samples.
Supplementary Material
Acknowledgments
We specially thank José Antonio López de Castro (Centro de Biología Molecular Severo Ochoa, Madrid, Spain) for his dedicated help in the generation of the C1R-B*40:02 transfectant cell line. We also thank Iñaki Álvarez (Institut de Biotecnonologia i Biomedicina, Barcelona, Spain), Alberto Paradela, and Severine Gharbi (both from the CNB, Madrid, Spain) for critical comments on the manuscript; Salvador Martínez-Bartolomé (CNB, Madrid, Spain) for his expert support during data submission to ProteomeXchange; and the flow cytometry facility of the CNB for their technical assistance. The mass spectrometry proteomics data have been deposited at the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD000450 and the PRIDE accession number 31118. The Proteomics Unit of the Centro Nacional de Biotecnología is a member of the Spanish National Institute for Proteomics (ProteoRed-ISCIII).
Footnotes
Author contributions: M.M. and J.P.A. designed research; M.M., A.A., M.L., and M.R. performed research; M.M., A.A., and A.R. analyzed data; M.M. wrote the paper.
* M.M. and A.A. were funded by the JAE-Doc 2009 and JAE-Pre 2011 CSIC programs, respectively.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- MHC
- major histocompatibility complex
- β2m
- β-2-microglobulin
- C1R
- HMy2.C1R
- CTL
- cytotoxic T lymphocyte
- FDR
- false discovery rate
- HLA
- human leukocyte antigen
- IMAC
- immobilized metal ion affinity chromatography
- LC-MS
- liquid chromatography coupled to mass spectrometry
- P2
- peptide position 2
- pSer
- phosphoserine.
REFERENCES
- 1. Madden D. R. (1995) The three-dimensional structure of peptide-MHC complexes. Annu. Rev. Immunol. 13, 587–622 [DOI] [PubMed] [Google Scholar]
- 2. Garrett T. P., Saper M. A., Bjorkman P. J., Strominger J. L., Wiley D. C. (1989) Specificity pockets for the side chains of peptide antigens in HLA-Aw68. Nature 342, 692–696 [DOI] [PubMed] [Google Scholar]
- 3. Brusic V., Bajic V. B., Petrovsky N. (2004) Computational methods for prediction of T-cell epitopes—a framework for modelling, testing, and applications. Methods 34, 436–443 [DOI] [PubMed] [Google Scholar]
- 4. Hassan C., Kester M. G., de Ru A. H., Hombrink P., Drijfhout J. W., Nijveen H., Leunissen J. A., Heemskerk M. H., Falkenburg J. H., van Veelen P. A. (2013) The human leukocyte antigen-presented ligandome of B lymphocytes. Mol. Cell. Proteomics 12, 1829–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blum J. S., Wearsch P. A., Cresswell P. (2013) Pathways of antigen processing. Annu. Rev. Immunol. 31, 443–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gras S., Burrows S. R., Turner S. J., Sewell A. K., McCluskey J., Rossjohn J. (2012) A structural voyage toward an understanding of the MHC-I-restricted immune response: lessons learned and much to be learned. Immunol. Rev. 250, 61–81 [DOI] [PubMed] [Google Scholar]
- 7. Robinson J., Halliwell J. A., McWilliam H., Lopez R., Parham P., Marsh S. G. (2013) The IMGT/HLA database. Nucleic Acids Res. 41, D1222–D1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sidney J., Peters B., Frahm N., Brander C., Sette A. (2008) HLA class I supertypes: a revised and updated classification. BMC Immunol. 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Engelhard V. H., Altrich-Vanlith M., Ostankovitch M., Zarling A. L. (2006) Post-translational modifications of naturally processed MHC-binding epitopes. Curr. Opin. Immunol. 18, 92–97 [DOI] [PubMed] [Google Scholar]
- 10. Yague J., Alvarez I., Rognan D., Ramos M., Vazquez J., de Castro J. A. (2000) An N-acetylated natural ligand of human histocompatibility leukocyte antigen (HLA)-B39. Classical major histocompatibility complex class I proteins bind peptides with a blocked NH(2) terminus in vivo. J. Exp. Med. 191, 2083–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zarling A. L., Ficarro S. B., White F. M., Shabanowitz J., Hunt D. F., Engelhard V. H. (2000) Phosphorylated peptides are naturally processed and presented by major histocompatibility complex class I molecules in vivo. J. Exp. Med. 192, 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zarling A. L., Polefrone J. M., Evans A. M., Mikesh L. M., Shabanowitz J., Lewis S. T., Engelhard V. H., Hunt D. F. (2006) Identification of class I MHC-associated phosphopeptides as targets for cancer immunotherapy. Proc. Natl. Acad. Sci. U.S.A. 103, 14889–14894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meyer V. S., Drews O., Gunder M., Hennenlotter J., Rammensee H. G., Stevanovic S. (2009) Identification of natural MHC class II presented phosphopeptides and tumor-derived MHC class I phospholigands. J. Proteome Res. 8, 3666–3674 [DOI] [PubMed] [Google Scholar]
- 14. Yague J., Vazquez J., Lopez de Castro J. A. (2000) A post-translational modification of nuclear proteins, N(G),N(G)-dimethyl-Arg, found in a natural HLA class I peptide ligand. Protein Sci. 9, 2210–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haurum J. S., Hoier I. B., Arsequell G., Neisig A., Valencia G., Zeuthen J., Neefjes J., Elliott T. (1999) Presentation of cytosolic glycosylated peptides by human class I major histocompatibility complex molecules in vivo. J. Exp. Med. 190, 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Evan G. I., Vousden K. H. (2001) Proliferation, cell cycle and apoptosis in cancer. Nature 411, 342–348 [DOI] [PubMed] [Google Scholar]
- 17. Blume-Jensen P., Hunter T. (2001) Oncogenic kinase signalling. Nature 411, 355–365 [DOI] [PubMed] [Google Scholar]
- 18. Paradela A., Albar J. P. (2008) Advances in the analysis of protein phosphorylation. J. Proteome Res. 7, 1809–1818 [DOI] [PubMed] [Google Scholar]
- 19. Boersema P. J., Mohammed S., Heck A. J. (2009) Phosphopeptide fragmentation and analysis by mass spectrometry. J. Mass Spectrom. 44, 861–878 [DOI] [PubMed] [Google Scholar]
- 20. Larsen M. R., Trelle M. B., Thingholm T. E., Jensen O. N. (2006) Analysis of posttranslational modifications of proteins by tandem mass spectrometry. BioTechniques 40, 790–798 [DOI] [PubMed] [Google Scholar]
- 21. Zemmour J., Little A. M., Schendel D. J., Parham P. (1992) The HLA-A,B “negative” mutant cell line C1R expresses a novel HLA-B35 allele, which also has a point mutation in the translation initiation codon. J. Immunol. 148, 1941–1948 [PubMed] [Google Scholar]
- 22. Ramos M., Postigo J. M., Vilches C., Layrisse Z., Lopez de Castro J. A. (1995) Primary structure of a novel HLA-B39 allele (B*3909) from the Warao Indians of Venezuela. Further evidence for local HLA-B diversification in South America. Tissue Antigens 46, 401–404 [DOI] [PubMed] [Google Scholar]
- 23. Barnstable C. J., Bodmer W. F., Brown G., Galfre G., Milstein C., Williams A. F., Ziegler A. (1978) Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens—new tools for genetic analysis. Cell 14, 9–20 [DOI] [PubMed] [Google Scholar]
- 24. Marcilla M., Cragnolini J. J., Lopez de Castro J. A. (2007) Proteasome-independent HLA-B27 ligands arise mainly from small basic proteins. Mol. Cell. Proteomics 6, 923–938 [DOI] [PubMed] [Google Scholar]
- 25. Navajas R., Paradela A., Albar J. P. (2011) Immobilized metal affinity chromatography/reversed-phase enrichment of phosphopeptides and analysis by CID/ETD tandem mass spectrometry. Methods Mol. Biol. 681, 337–348 [DOI] [PubMed] [Google Scholar]
- 26. Thingholm T. E., Jensen O. N., Robinson P. J., Larsen M. R. (2008) SIMAC (sequential elution from IMAC), a phosphoproteomics strategy for the rapid separation of monophosphorylated from multiply phosphorylated peptides. Mol. Cell. Proteomics 7, 661–671 [DOI] [PubMed] [Google Scholar]
- 27. Buchsbaum S., Barnea E., Dassau L., Beer I., Milner E., Admon A. (2003) Large-scale analysis of HLA peptides presented by HLA-Cw4. Immunogenetics 55, 172–176 [DOI] [PubMed] [Google Scholar]
- 28. Elias J. E., Gygi S. P. (2010) Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol. Biol. 604, 55–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 30. Kessler J. H., Benckhuijsen W. E., Mutis T., Melief C. J., van der Burg S. H., Drijfhout J. W. (2004) Competition-based cellular peptide binding assay for HLA class I. Curr. Protoc. Immunol. Chapter 18, Unit 18.12 [DOI] [PubMed] [Google Scholar]
- 31. van der Burg S. H., Ras E., Drijfhout J. W., Benckhuijsen W. E., Bremers A. J., Melief C. J., Kast W. M. (1995) An HLA class I peptide-binding assay based on competition for binding to class I molecules on intact human B cells. Identification of conserved HIV-1 polymerase peptides binding to HLA-A*0301. Hum. Immunol. 44, 189–198 [DOI] [PubMed] [Google Scholar]
- 32. Hunt D. F., Henderson R. A., Shabanowitz J., Sakaguchi K., Michel H., Sevilir N., Cox A. L., Appella E., Engelhard V. H. (1992) Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 255, 1261–1263 [DOI] [PubMed] [Google Scholar]
- 33. Ben Dror L., Barnea E., Beer I., Mann M., Admon A. (2010) The HLA-B*2705 peptidome. Arthritis Rheum. 62, 420–429 [DOI] [PubMed] [Google Scholar]
- 34. Mester G., Hoffmann V., Stevanovic S. (2011) Insights into MHC class I antigen processing gained from large-scale analysis of class I ligands. Cell. Mol. Life Sci. 68, 1521–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Legrain P., Aebersold R., Archakov A., Bairoch A., Bala K., Beretta L., Bergeron J., Borchers C., Corthals G. L., Costello C. E., Deutsch E. W., Domon B., Hancock W., He F., Hochstrasser D., Marko-Varga G., Salekdeh G. H., Sechi S., Snyder M., Srivastava S., Uhlen M., Hu C. H., Yamamoto T., Paik Y. K., Omenn G. S. (2011) The human proteome project: current state and future direction. Mol. Cell. Proteomics. 10, M111.009993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Admon A., Bassani-Sternberg M. (2011) The Human Immunopeptidome Project, a suggestion for yet another postgenome next big thing. Mol. Cell. Proteomics 10, O111.011833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yashiki S., Fujiyoshi T., Arima N., Osame M., Yoshinaga M., Nagata Y., Tara M., Nomura K., Utsunomiya A., Hanada S., Tajima K., Sonoda S. (2001) HLA-A*26, HLA-B*4002, HLA-B*4006, and HLA-B*4801 alleles predispose to adult T cell leukemia: the limited recognition of HTLV type 1 tax peptide anchor motifs and epitopes to generate anti-HTLV type 1 tax CD8(+) cytotoxic T lymphocytes. AIDS Res. Hum. Retroviruses 17, 1047–1061 [DOI] [PubMed] [Google Scholar]
- 38. Hillen N., Mester G., Lemmel C., Weinzierl A. O., Muller M., Wernet D., Hennenlotter J., Stenzl A., Rammensee H. G., Stevanovic S. (2008) Essential differences in ligand presentation and T cell epitope recognition among HLA molecules of the HLA-B44 supertype. Eur. J. Immunol. 38, 2993–3003 [DOI] [PubMed] [Google Scholar]
- 39. Bade-Doding C., Theodossis A., Gras S., Kjer-Nielsen L., Eiz-Vesper B., Seltsam A., Huyton T., Rossjohn J., McCluskey J., Blasczyk R. (2011) The impact of human leukocyte antigen (HLA) micropolymorphism on ligand specificity within the HLA-B*41 allotypic family. Haematologica 96, 110–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Probst-Kepper M., Hecht H. J., Herrmann H., Janke V., Ocklenburg F., Klempnauer J., van den Eynde B. J., Weiss S. (2004) Conformational restraints and flexibility of 14-meric peptides in complex with HLA-B*3501. J. Immunol. 173, 5610–5616 [DOI] [PubMed] [Google Scholar]
- 41. Tynan F. E., Reid H. H., Kjer-Nielsen L., Miles J. J., Wilce M. C., Kostenko L., Borg N. A., Williamson N. A., Beddoe T., Purcell A. W., Burrows S. R., McCluskey J., Rossjohn J. (2007) A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat. Immunol. 8, 268–276 [DOI] [PubMed] [Google Scholar]
- 42. Guo H. C., Jardetzky T. S., Garrett T. P., Lane W. S., Strominger J. L., Wiley D. C. (1992) Different length peptides bind to HLA-Aw68 similarly at their ends but bulge out in the middle. Nature 360, 364–366 [DOI] [PubMed] [Google Scholar]
- 43. Steinle A., Falk K., Rotzschke O., Gnau V., Stevanovic S., Jung G., Schendel D. J., Rammensee H. G. (1996) Motif of HLA-B*3503 peptide ligands. Immunogenetics 43, 105–107 [DOI] [PubMed] [Google Scholar]
- 44. Macdonald W. A., Purcell A. W., Mifsud N. A., Ely L. K., Williams D. S., Chang L., Gorman J. J., Clements C. S., Kjer-Nielsen L., Koelle D. M., Burrows S. R., Tait B. D., Holdsworth R., Brooks A. G., Lovrecz G. O., Lu L., Rossjohn J., McCluskey J. (2003) A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition. J. Exp. Med. 198, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Madden D. R., Gorga J. C., Strominger J. L., Wiley D. C. (1992) The three-dimensional structure of HLA-B27 at 2.1 A resolution suggests a general mechanism for tight peptide binding to MHC. Cell 70, 1035–1048 [DOI] [PubMed] [Google Scholar]
- 46. Lopez de Castro J. A., Alvarez I., Marcilla M., Paradela A., Ramos M., Sesma L., Vazquez M. (2004) HLA-B27: a registry of constitutive peptide ligands. Tissue Antigens 63, 424–445 [DOI] [PubMed] [Google Scholar]
- 47. Weigel N. L., Moore N. L. (2007) Kinases and protein phosphorylation as regulators of steroid hormone action. Nucl. Recept. Signal. 5, e005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 49. Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. (2002) Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110, 177–189 [DOI] [PubMed] [Google Scholar]
- 50. Daub H., Olsen J. V., Bairlein M., Gnad F., Oppermann F. S., Korner R., Greff Z., Keri G., Stemmann O., Mann M. (2008) Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell 31, 438–448 [DOI] [PubMed] [Google Scholar]
- 51. Dephoure N., Zhou C., Villen J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Andersen M. H., Bonfill J. E., Neisig A., Arsequell G., Sondergaard I., Valencia G., Neefjes J., Zeuthen J., Elliott T., Haurum J. S. (1999) Phosphorylated peptides can be transported by TAP molecules, presented by class I MHC molecules, and recognized by phosphopeptide-specific CTL. J. Immunol. 163, 3812–3818 [PubMed] [Google Scholar]
- 53. Sugiyama N., Masuda T., Shinoda K., Nakamura A., Tomita M., Ishihama Y. (2007) Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol. Cell. Proteomics 6, 1103–1109 [DOI] [PubMed] [Google Scholar]
- 54. Damoc E., Fraser C. S., Zhou M., Videler H., Mayeur G. L., Hershey J. W., Doudna J. A., Robinson C. V., Leary J. A. (2007) Structural characterization of the human eukaryotic initiation factor 3 protein complex by mass spectrometry. Mol. Cell. Proteomics 6, 1135–1146 [DOI] [PubMed] [Google Scholar]
- 55. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3. [DOI] [PubMed] [Google Scholar]
- 56. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 57. Wang B., Malik R., Nigg E. A., Korner R. (2008) Evaluation of the low-specificity protease elastase for large-scale phosphoproteome analysis. Anal. Chem. 80, 9526–9533 [DOI] [PubMed] [Google Scholar]
- 58. Kyono Y., Sugiyama N., Imami K., Tomita M., Ishihama Y. (2008) Successive and selective release of phosphorylated peptides captured by hydroxy acid-modified metal oxide chromatography. J. Proteome Res. 7, 4585–4593 [DOI] [PubMed] [Google Scholar]
- 59. Stokes M. P., Rush J., Macneill J., Ren J. M., Sprott K., Nardone J., Yang V., Beausoleil S. A., Gygi S. P., Livingstone M., Zhang H., Polakiewicz R. D., Comb M. J. (2007) Profiling of UV-induced ATM/ATR signaling pathways. Proc. Natl. Acad. Sci. U.S.A. 104, 19855–19860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Molina H., Horn D. M., Tang N., Mathivanan S., Pandey A. (2007) Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 104, 2199–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yu C. T., Hsu J. M., Lee Y. C., Tsou A. P., Chou C. K., Huang C. Y. (2005) Phosphorylation and stabilization of HURP by Aurora-A: implication of HURP as a transforming target of Aurora-A. Mol. Cell. Biol. 25, 5789–5800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lovly C. M., Yan L., Ryan C. E., Takada S., Piwnica-Worms H. (2008) Regulation of Chk2 ubiquitination and signaling through autophosphorylation of serine 379. Mol. Cell. Biol. 28, 5874–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rigbolt K. T., Prokhorova T. A., Akimov V., Henningsen J., Johansen P. T., Kratchmarova I., Kassem M., Mann M., Olsen J. V., Blagoev B. (2011) System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 4, rs3. [DOI] [PubMed] [Google Scholar]
- 64. Carriere A., Cargnello M., Julien L. A., Gao H., Bonneil E., Thibault P., Roux P. P. (2008) Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr. Biol. 18, 1269–1277 [DOI] [PubMed] [Google Scholar]
- 65. Ruse C. I., McClatchy D. B., Lu B., Cociorva D., Motoyama A., Park S. K., Yates J. R., 3rd (2008) Motif-specific sampling of phosphoproteomes. J. Proteome Res. 7, 2140–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brill L. M., Xiong W., Lee K. B., Ficarro S. B., Crain A., Xu Y., Terskikh A., Snyder E. Y., Ding S. (2009) Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell 5, 204–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.