Abstract
Arginine phosphorylation is an emerging protein modification implicated in the general stress response of Gram-positive bacteria. The modification is mediated by the arginine kinase McsB, which phosphorylates and inactivates the heat shock repressor CtsR. In this study, we developed a mass spectrometric approach accounting for the peculiar chemical properties of phosphoarginine. The improved methodology was used to analyze the dynamic changes in the Bacillus subtilis arginine phosphoproteome in response to different stress situations. Quantitative analysis showed that a B. subtilis mutant lacking the YwlE arginine phosphatase accumulated a strikingly large number of arginine phosphorylations (217 sites in 134 proteins), however only a minor fraction of these sites was increasingly modified during heat shock or oxidative stress. The main targets of McsB-mediated arginine phosphorylation comprise central factors of the stress response system including the CtsR and HrcA heat shock repressors, as well as major components of the protein quality control system such as the ClpCP protease and the GroEL chaperonine. These findings highlight the impact of arginine phosphorylation in orchestrating the bacterial stress response.
Protein phosphorylation is a ubiquitous post-translational modification affecting almost every signal transduction process. Advances in speed and sensitivity of mass spectrometers, together with the higher selectivity of current phosphopeptide enrichment methods, greatly facilitated the acquisition of large scale phosphoproteomic datasets. To date, studies investigating serine, threonine and tyrosine phosphorylations in higher eukaryotes usually identify about 10,000 sites (1). Despite the technical advances in the phosphoproteomics research field, other types of protein phosphorylation, which have been reported to occur in a variety of organisms (2, 3), remain scarcely studied. Examples of such noncanonical phosphoresidues are phosphoarginine, phospholysine, and (N1- or N3-) phosphohistidine, where phosphorylation occurs at the side-chain nitrogen atom (N-phosphorylation), yielding a phosphoramidate (P-N) bond that is unstable at low pH conditions. In contrast, the phosphate moiety that is covalently attached to the hydroxyl group of serine, threonine, and tyrosine (O-phosphorylation) yields a phosphoester bond that is stable at acidic pH. Of note, low pH conditions are widely employed during sample preparation for phosphoproteomic analysis, especially when phosphopeptide enrichment and separation by strong cation exchange or reverse-phase chromatography are performed. Because of the acid lability of the P-N bond, N-phosphoryl amino acids often remain unidentified even though they have been suggested to engage in important biological functions (3–5).
Accordingly, the general occurrence and functional relevance of N-phosphorylation are vastly unknown. So far, the phosphorylation of histidine has been described as an intermediate step in the two-component and multicomponent phosphorelay signaling pathways found in bacteria, fungi, and plants. In bacteria, the corresponding histidine kinases play a major role in perception of various stimuli, allowing their hosts to rapidly adapt to changing environmental conditions, whereas in lower eukaryotes and plants, histidine kinases appear to control more specific functions (6), such as osmolarity sensing (7) and hormone signaling (8), respectively. In the case of phosphoarginine, a number of studies suggested its presence in diverse organisms, as reviewed in (3). For example, Wakim and co-workers obtained mammalian cell fractions that could arginine-phosphorylate histone H3 in vitro (9, 10), however the identity of the corresponding arginine kinase(s) was never determined. To date, the only protein arginine kinase that could be characterized in detail is the McsB protein occurring in Bacillus subtilis and closely related bacterial species (11).
The McsB kinase is part of the bacterial stress response system, where it is involved in regulating CtsR, the central transcriptional repressor of class III heat shock genes. Notably, CtsR is capable of binding to DNA in a temperature dependent manner, using the beta-wing of its winged helix-turn-helix (HTH1) as a temperature sensor. At elevated temperatures, the beta-wing induces dissociation of CtsR from DNA and consequently allowing for heat shock gene expression (12). However, high temperature is not the only stress condition where expression of class III genes is induced. It is thus likely that McsB-mediated arginine phosphorylation is an additional mechanism inhibiting the CtsR repressor under a broad range of protein folding stress situations. Consistent with this notion, it was shown that McsB phosphorylation of CtsR inhibits binding to its operator DNA in vitro (11), that McsB and CtsR interact in vivo and that the heat shock-induced degradation of CtsR depends on the presence of McsA and McsB (13). The genes regulated by CtsR encode factors belonging to the protein quality control system, such as the ClpP protease and the associated ClpC and ClpE unfoldases, as well as regulatory proteins such as CtsR, McsA, and McsB themselves (14–16). ClpCP forms an ATP-dependent protease complex, similar to the eukaryotic proteasome, that not only carries out protein quality control, by removing unfolded proteins (17, 18), but also functions as a regulatory protease in developmental processes such as sporulation (19) and competence (20). In addition, ClpCP itself appears to function as a central regulator in the bacterial stress response, by mediating CtsR degradation upon heat shock (21, 22) and inhibiting the activity of McsB, according to in vitro assays (13). Therefore, the activation of the stress response in B. subtilis is regulated by an intricate feedback loop resulting from the concerted activities of CtsR, McsB and ClpCP.
A further regulatory level is represented by the YwlE phosphatase that counteracts the activity of the McsB arginine kinase (13). Based on sequence homology, YwlE was initially annotated as a tyrosine phosphatase. However, in vitro assays using phosphopeptide substrates demonstrated that, even though phosphotyrosine phosphatase activity can occur at pH 5 (23), at physiological pH YwlE activity is highly specific toward phosphoarginine (24). By systematically analyzing phosphatase-deficient B. subtilis strains, the latter study also revealed that YwlE is the only arginine phosphatase in this bacterium. In sum, these findings indicate that arginine phosphorylation is a carefully balanced post-translational modification resulting from the antagonistic activity of very specific protein kinases and phosphatases (24).
To better understand how McsB-mediated arginine phosphorylation affects the bacterial stress response and to identify specific targets of McsB, we studied changes in the B. subtilis arginine phosphoproteome under different environmental conditions. For this purpose, we developed a quantitative MS approach adapted to the special chemical properties of phosphoarginine. Quantitative data uncovered major physiological McsB targets, which comprise the main regulators and executors of the bacterial stress response system including CtsR, HrcA, GroEL, ClpC and ClpP. Additionally, the obtained results demonstrate that the improved phosphoproteomic method reported here will be of great value for the scientific community in investigating the prevalence and the biological function of acid-labile phosphorylations, that so far escaped systematic analysis.
EXPERIMENTAL PROCEDURES
Peptide Synthesis and Phosphorylation
All model peptides for acidic hydrolysis were synthesized in house using FMOC-based coupling chemistry. Peptides were HPLC purified and identity and purity were validated by MALDI mass spectrometry. Arginine phosphorylated peptides were synthesized without the phosphomoiety. Phosphorylation was subsequently introduced in vitro using purified recombinant McsB arginine kinase from Geobacillus stearothermophilus. The resulting peptide mixture was again HPLC fractionated (Merck, Germany) on a semipreparative C18 column (GeminiC18, 15 cm × 2 mm × 5 μm, Phenomenex, Germany) using an ammonium bicarbonate buffer at pH 7 to obtain 99% pure arginine phosphorylated peptide.
Acidic Hydrolysis of Arginine-phosphorylated Peptides
Aliquots of 20 pMol/reaction of the phosphopeptide ac-K(pR)GGGGYIKIIKV were diluted in 200 mm phosphate buffer to a final concentration of 0.1 μm. All hydrolysis reactions were carried out in 200 μl scale. Time points were taken after 5, 15, 30, 60, 120, and 240 min of reaction time by neutralizing the reaction mixture with 1 m HEPES, pH 7.2. For the first replicate, samples of each time point were injected on an Ultimate plus HPLC system for C18-RP separation (PepMapAcclaim, 15 cm × 75 μm × 3 μm, 100Å, Dionex-Thermo-Fisher Scientific, United States) and detected on a LCQ DecaXP mass spectrometer (Thermo-Fisher Scientific) in data dependent mode. The second replicate was analyzed on an Ultimate 3000 beta HPLC system coupled to an LTQ FT hybrid mass spectrometer (Dionex-Thermo-Fisher Scientific) applying the same separation conditions. Mass traces corresponding to the 2+ and 3+ charge states of phosphorylated and hydrolyzed peptide were manually integrated to determine the abundance of each peptide form at the respective incubation time.
Generation of the B. subtilis ΔywlE and ΔywlEΔmcsB Strains
The ywlE-deficient mutant was generated by replacing the ywlE gene with a spectinomycin resistance cassette carrying a transcriptional terminator. DNA fragments corresponding to the chromosomal DNA regions located immediately upstream and downstream of the ywlE coding sequence were generated by PCR using oligonucleotide pairs ywlE_fw_UP (5′-GCCGCCGGAGGAAGAGTGAT-3′) and ywlE_rev_UP_spec (5′-CGCTCACGAAGGGATTCGATAGACAAAAATAATATCCAT-3′); ywlE_fw_DW_spec (5′-GTTCCTTCGATAGTTTATTATTGTCAGAAAATCTGCAAAC-3′) and ywlE_rev_DW (5′-TAAACAGCACCCCTACAGGT-3′); A DNA fragment carrying the spectinomycin resistance cassette was generated using the primers reverse complementary to ywlE_rev_UP_spec and ywlE_fw_DW_spec. The three PCR fragments were ligated by PCR and then introduced into competent B. subtilis 168 [ATCC 2385] cells as described previously (25). Spectinomycin-resistant mutants that resulted from double cross-over events in which 421 bp of the ywlE coding sequence was replaced by the spectinomycin resistance cassette were selected on LB agar plates containing spectinomycin (200 μg/ml). The ΔywlEΔmcsB double mutant was generated by introducing chromosomal DNA prepared from B. subtilis ΔmcsB (26) into competent B. subtilis ΔywlE cells. Mutants were selected on LB agar plates containing spectinomycin, erythromycin (5 μg/ml), and lincomycin (20 μg/ml). The gene replacements were confirmed by PCR and sequencing analysis.
Cell Growth Conditions
B. subtilis cultures were grown in liquid Luria-Bertani (LB) medium. For the ΔywlE strain, spectinomycin (200 μg/ml) was added to the media, whereas for the ΔywlEΔmcsB strain both spectinomycin and erythromycin (5 μg/ml) were used. For phosphoproteomic experiments, cultures grown overnight at 37 °C were diluted in 200 ml of fresh medium for each condition to an A600 of 0.05 and grown at 37 °C under orbital shaking until an A600 of 0.6 was reached. For heat stress, the culture flask was transferred to a 50 °C water bath for 20 min under orbital shaking. Oxidative stress was induced by adding 1 mm diamide. The control culture remained at the initial growth conditions. After the 20 min incubation time all cells were harvested by centrifugation at 4 °C. Obtained pellets were washed once with ice cold PBS buffer and stored at −80 °C until further use.
Pervanadate Inhibition of YwlE Phosphatase
Peroxidated vanadate species have been described as potent tyrosine phosphatase inhibitors (27). Because YwlE are structurally and mechanistically related to low molecular weight tyrosine phosphatases, we reasoned that this inhibitor might also eliminate YwlE activity. Sodium pervanadate was prepared from sodium ortho-vanadate and H2O2. Briefly, 500 mm sodium ortho-vanadate in water was brought to pH 7 with concentrated HCl and boiling until the yellow color disappeared. After dilution to 10 mm concentration in 100 mm HEPES pH 7.5, 3% H2O2 in 100 mm HEPES was added and the solution was incubated at room temperature for 5 min. To remove residual H2O2, 1 μl of 1 mg/ml bovine catalase was added. The resulting pervanadate solution was added to the cell medium in a final concentration of 0.4 mm 5 min before the cells were exposed to heat shock conditions at 50 °C for 20 min. Controls in which the cells were not exposed to higher temperature were performed in parallel.
Cell Lysis and Filter-aided Sample Preparation
The pellet of a 200 ml cell culture (approx. 1 g) grown to an OD600 of 0.6 was transferred into freezer mill sample vials under liquid nitrogen. The cells were crushed in a freezer mill (Spex, New Jersey) applying the following settings: 10 cycles consisting of 2 mins of grinding (15 counts/second) followed by 3 mins of cooling per cycle. The obtained cell powder was stored at −80 °C until further use.
Samples were allowed to warm until a suspension was formed. This was transferred into 1.5 ml Eppendorf vials and one sample volume of lysis buffer (100 mm Tris, pH 7.5, 4% SDS, 200 mm dithiotreitol, 3 mm of each phosphatase inhibitor: sodium vanadate, potassium fluoride, sodium pyrophosphate, and glycerol phosphate) was added. Samples were homogenized with a sonication probe (UPH100 0.5 mm, Hielscher, Germany) to dissolve proteins and shear DNA. After centrifugation, the supernatant was incubated at 56 °C for 50 min to reduce disulfide bonds. Immediately after reduction, the protein solution was diluted with 12 ml 100 mm Tris (pH 7.5, containing 8 m Urea and 25 mm iodoacetamide) in an Amicon filter unit (Millipore, United States, MWCO 30000) for alkylation of free cysteines. After incubation for 45 min in the dark, samples were washed according to the FASP protocol (28) by centrifugation at 4000 g applying 2 × 15 ml 100 mm Tris (pH 7.5, containing 8 m Urea), 2 × 15 ml 100 mm Tris (pH 7.5, containing 4 m Urea), and 2 × 100 mm ammonium bicarbonate. The retentate was reduced to 2–3 ml in the last washing step. The protein content was determined after the last 4 m urea wash using a Bradford Protein Assay (Bio-Rad, Germany).
Proteolytic Cleavage and iTRAQ Labeling
Lyophilized Trypsin Gold (Promega, Madison, WI) was allowed to heat to room temperature before it was dissolved to 1 μg/μl with 100 mm ammonium bicarbonate solution. Tryptic digest of all B. subtilis whole cell lysates was achieved with a protein to trypsin ratio of 100:1 for 14 h at 37 °C in the filter devices. The level of digestion was monitored by retention time and UV intensity (214 nm) distribution on RP-HPLC separation of 0.25 to 0.5 μg protein on a monolithic column (Ultimate Plus equipped with a PepSwift PS-DVB column, 5 cm x 200 μm, all Dionex-Thermo-Fisher Scientific). Further, the peptide content was determined via absorbance at 280 nm in a NanoDrop device (Thermo-Fisher Scientific).
To accomplish a high degree of iTRAQ labeling, 0.5 mg of peptides from each sample were purified from ammonium bicarbonate and other reagents from the sample preparation by RP-C18 solid-phase extraction (SPE) at neutral pH (Strata-X 200 mg, 6 ml cartridge, Phenomenex, Germany). The eluent was dried under vacuum and re-dissolved in 50 μl 100 mm TEAB buffer. ITRAQ reagents (Invitrogen, New York) were dissolved in 150 μl ethanol, added to the peptide solution, and the mixture was incubated for 4h at room temperature. Labeling efficiency was monitored by LC-MS/MS analysis of each individual labeling reaction and modification of N termini and lysines of more than 97% were considered as completely labeled, otherwise fresh iTRAQ reagent was applied. Furthermore, to confirm an acid-labile arginine phosphorylation as PTM, two aliquots were prepared for the control of the first biological replicate, one was labeled with iTRAQ reagent 114 and the other one with reagent 117. The aliquot with the 114-label was incubated in 1% TFA at 60 °C for 1 h to hydrolyze all N-phosphorylations before mixing with the other aliquots. In the second replicate this treatment was omitted, because ETD fragmentation allowed sufficient phosphosite localization confidence. For quantitative protein and phosphorylation analysis, individual samples (unstressed control, acid-treated unstressed control, heat shock, and oxidative stress) were mixed in a 1:1:1:1 ratio according to their protein amount. The excess of quenched labeling reagent over peptides was removed by a second solid-phase extraction step on a 6 ml C18 cartridge Strata-X after mixing the 4 iTRAQ channels.
Determination of Protein Abundance Ratios
Apart from the regulation of phosphorylation sites, also alterations in the protein abundance are important to reveal cellular processes occurring during heat shock or oxidative stress exposure. Therefore, an aliquot of 100 μg of the final mixture was fractionated by strong cation exchange chromatography on a 1 mm Polysulfoethyl column. Solvents for the SCX were 5 mm sodium phosphate, pH 2.7 for buffers A and B and pH 6 for buffer C, respectively. For efficient elution, solvent B also contained 1 m sodium chloride. Peptides were bound to the resin in 100% solvent A. Peptide separation was achieved by a linear gradient combining increasing percentages for buffer B and C over 45 min. Fractions were collected every 5 min and analyzed by LC-MS/MS as described below.
TiO2 Enrichment of Phosphopeptides
The iTRAQ-labeled peptide mixture containing 0.25 mg sample per channel was lyophilized and re-dissolved in 150 μl TiO2 loading buffer (300 mg/ml lactic acid, 12.5% acetic acid (AcOH), 0.2% heptafluorobutyric acid (HFBA), 60% acetonitrile (ACN), pH 4 with NH3) immediately before incubation with the TiO2 resin (Titansphere, 5 μm, GL Sciences, Japan). After incubation for 35 min at 20 °C, unbound peptides were removed by filter centrifugation in Mobicol devices (MoBiTec, Germany) and the resin was washed with additional 150 μl of loading buffer. Further removal of unphosphorylated peptides was achieved by subsequent washing with 400 μl of solvent A (200 mg/ml lactic acid, 75% ACN, 2% trifluoroacetic acid (TFA), 2% HFBA), solvent B (200 mg/ml lactic acid, 75% ACN, 10% AcOH, 0.1% HFBA, pH 4 with NH3), and solvent C (80% ACN, 10% AcOH). The TiO2 resin was re-suspended in each washing step. For elution of phosphopeptides, the resin was incubated twice with 100 μl of 1% NH3 solution containing 30 mm ammonium phosphate for 15 min. The sample volume was reduced under low pressure and a pH of 7 was adjusted before injection onto the LC column.
LC-MS/MS Analysis
RP separation of all peptide mixtures was achieved on an Ultimate 3000 RSLC nano-flow chromatography system (Thermo-Fisher). An alternative loading solvent to 0.1% TFA was required, because already short washing periods of arginine-phosphorylated samples under these conditions lead to significant hydrolysis of the acid-labile phosphoarginine. Therefore, 0.5% AcOH (pH 4.5 with NH3) at a flow rate of 15 μl/min was used to remove salts after loading the peptide mixture onto the precolumn (PepMapAcclaim C18, 2 cm × 0.1 mm, 5 μm, Dionex-Thermo-Fisher). Peptide separation was achieved on a C18 separation column (PepMapAcclaim C18, 50 cm × 0.75 mm, 2 μm, Dionex-Thermo-Fisher) by applying a linear gradient from 2% to 40% solvent B (80% ACN, 0.1% FA) in 180 or 240 min. Solvent A was 2% ACN, 0.1% FA. Under these conditions, the hydrolysis of the phosphoarginine mark was estimated to be very low, ranging from 5 to 10%. The separation was monitored by UV detection and the outlet of the detector was directly coupled to the nano-Electrospray ionization source (Proxeon Biosystems, Denmark) for MS analysis.
The sample was infused into the LTQ Orbitrap Velos ETD mass spectrometer (Thermo-Fisher Scientific, USA) using nanoSpray emitter tips (Proxeon Biosystems, Denmark) at a voltage of 1.5 kV. Peptides were analyzed in data-dependent fashion in positive ionization mode, applying the three different fragmentation methods: collision-induced dissociation (CID), higher-energy collisional dissociation (HCD), and electron-transfer dissociation (ETD). The seven most abundant peptide signals of the survey scan at resolution 60,000 with charge state 2+ and exceeding an intensity threshold of 1500 counts were selected for fragmentation. To prevent repeated fragmentation of highly abundant peptides, selected precursors were dynamically excluded for 50 s from MS/MS analysis. CID fragmentation was achieved at normalized collision energy (NCE) of 35% with additional activation of the dephosphorylated precursor at M-49 Th, M-32.7 Th, and M-98 Th in a pseudo-MS3 method. HCD was applied in two activation steps at 34% NCE for optimal peptide identification and 48% NCE for highest iTRAQ intensity. The resolution for HCD scan was set to 7500. For ETD, peptides were incubated with fluoranthene anions allowing for charge state dependent incubation times (120 ms for 2+ charged peptides) and resulting peptide fragments were detected in the ion trap analyzer.
Peptide and Protein Identification
Raw data were extracted by the Protein Discoverer software suite (version 1.3.01, Thermo-Fisher Scientific) and searched against a combined forward/reversed database of B. subtilis Uniprot Reference Proteome with common contaminants added (10,540 entries in total) using MASCOT (version 2.2, www.matrixscience.com). Because cells were grown in LB medium, all raw data were independently searched against a combined B. subtilis and yeast (S. cerevisae) database, which did not result in a significant increase in protein hits or identification of yeast proteins. Carbamidomethylation of cysteine, iTRAQ modification of peptide N terminus and lysine ε-amino group were set as fixed modifications. Phosphorylation of serine, threonine, tyrosine, histidine, and arginine as well as methionine sulfoxidation were selected to be variable, with a maximum of four modifications per peptide. Because tryptic cleavage is impaired at phosphorylated arginine, a maximum of two missed cleavage sites was allowed, whereas fully tryptic cleavage of both termini was required. The peptide mass deviation was set to 4 ppm; fragment ions were allowed to have a mass deviation of 0.5 Da for CID and ETD data and 0.025 Da for HCD data. False discovery rates were assessed from reversed database hits including all identified peptide spectrum matches with MASCOT score above 19, rank 1, and a peptide length of at least six amino acids. This resulted in false discovery rates below 1% on the peptide spectrum match level and below 1.5% on the unique peptide level for each experiment. In the rare cases in which a peptide was mapped to more than one protein, both proteins are reported. For reliable phosphorylation site analysis, all phosphopeptide hits were automatically re-analyzed by the phosphoRS software (29) within the Protein Discoverer software suite, manually validated, and compared with known B. subtilis protein phosphorylations (30). Proteins were grouped according to their biological function assignment on the SubtiList homepage (http://genolist.pasteur.fr/SubtiList/).
Relative Quantification of Phosphorylation
Peptide quantification was achieved by the reporter ions quantifier node in Protein Discoverer. This node extracted reporter ion areas from corresponding HCD scans for all identified peptide hits (HCD, CID, and ETD) based on the precursor mass. Extracted peak areas for heat shock and oxidative stress were divided by the peak area of the control sample to give the corresponding peptide abundance ratio. Only peptides having a quantification value for all conditions (control, heat shock and oxidative stress) were included in the analysis. Systematic errors in sample mixing were corrected by dividing the individual peptide ratios by the median ratio of the correspondent sample. Regulatory cut-offs for peptide abundance ratios were determined from the distribution of unphosphorylated peptides. A 95% confidence window was applied to log2 normalized values of peptide regulations and all hits outside of this window were considered to be regulated. For the heat stress/control ratio regulatory cut-off values of 0.59 and 1.75 were determined. For the oxidative stress and control comparison peptides with abundance changes within 0.30 to 2.50 were considered not regulated. Protein regulations were the averages of all identified and quantified unphosphorylated peptides from all analyzed samples. Phosphopeptide abundances were calculated by averaging, if a phosphopeptide was identified in several replicates and/or charge states. To argument for up-regulation of phosphorylation, each phosphopeptide ratio was divided by the respective protein abundance ratio, in case the protein was identified before. For regulation of the phosphorylation, we oriented the regulatory cut-offs determined before. Peptide and protein abundances with significant regulations are shown in supplemental Table S1 with bold numbers.
Motif Analysis
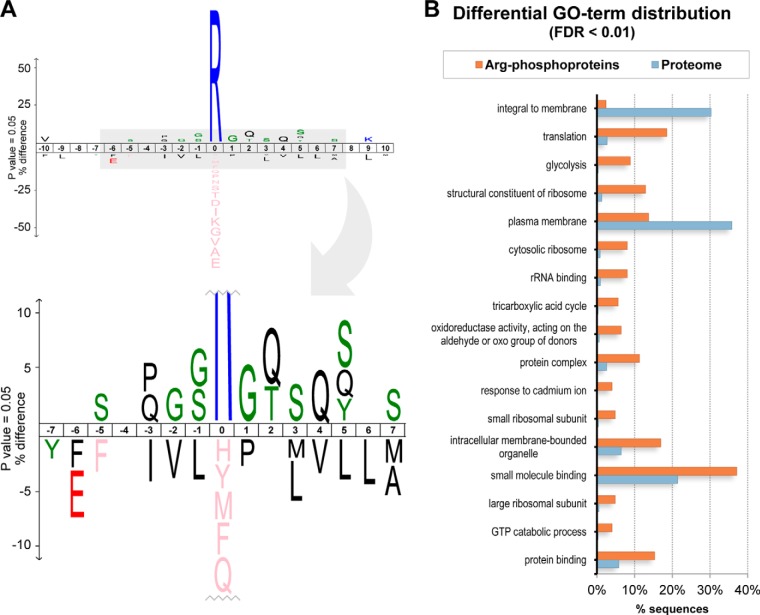
For the analysis of sequence features adjacent to the identified arginine phosphorylations, the 20 amino acids surrounding each phosphoarginine site were extracted and aligned. This alignment was used as test dataset in the IceLogo program, version 1.2 (31). To eliminate amino acid compositional bias, a reference dataset containing the full sequences of all arginine-phosphorylated proteins was used, applying random sampling. When more than one phosphorylation site per protein was found, the corresponding protein was accordingly repeated in the reference data set (i.e. when two phosphorylation sites were identified, the protein appeared twice in the reference dataset, so as to assure equal sizes in both datasets). A p value of 0.05 was as used as a cut-off for overrepresentation of amino acids (Fig. 2A).
Fig. 2.
Analysis of the arginine phosphorylations identified in the B. subtilis ΔywlE strain. A, Motif analysis of arginine-phosphorylated sites. The graph shows the difference in the frequency of amino acids surrounding the phosphoarginine mark in relation to their general frequency in the corresponding proteins. Only amino acids significantly enriched at each position (p value 0.05) are shown. B, Gene Ontology term enrichment analysis. The graph shows the frequency of significantly enriched (FDR cut-off 0.01) GO terms for the group of arginine-phosphorylated proteins in relation to the Swiss-Prot reference B. subtilis proteome.
Gene Ontology Annotation and Enrichment Analysis
GO annotations and enrichment analysis were performed using the Blast2GO software (32). Briefly, Blast2GO annotates the user provided list of proteins based on the functional information of the closest homologs found for each protein. First, the homologs were retrieved in a Blastp search of the entire B. subtilis 168 UniprotKB reference proteome against the swissprot database, with an expectation value minimum of 1 × e−3. Second, a mapping step linking BLAST Hits to functional information (GO terms) was performed using standard parameters. Third, an annotation step was performed using E-value Hit Filter 1 × e−6, Annotation cut-off 55 and GO weight 5. This resulted in a list of all B. subtilis proteins and their respectively annotated GO terms that was used as a reference dataset for the GO term enrichment analysis. As test data set, a list containing the 132 arginine-phosphorylated proteins was provided. To test the enrichment of specific GO terms, a two-tailed Fisher's Exact Test was performed using term filter value 0.05 in the FDR term filter mode.
Data Availability
The mass spectrometry data from this publication have been submitted to the ProteomeXchange Consortium http://proteomecentral.proteomexchange.org via the PRIDE partner repository (33) and assigned the identifier PXD000273.
RESULTS
Adaptation of a Phosphoproteomic Workflow for Phosphoarginine Analysis
In contrast to the phosphorylation of serine, threonine and tyrosine residues, both free and peptidic phosphoarginine have been described as acid-labile modification (34). We thus aimed to identify suitable experimental conditions that preserve the phosphoarginine modification and at the same time allow for efficient sample preparation during the phosphoproteomic analysis. For this purpose, we first characterized the chemical stability of the enzymatically generated phosphoarginine model peptide KpRGGGGYIKIIKV, at acidic, neutral and basic pH conditions at 25 °C and 60 °C. Dephosphorylation of the arginine phosphopeptide and concomitant formation of the unmodified peptide were monitored by liquid chromatography mass spectrometry (LC-MS). The recorded time course clearly revealed that acidic conditions (pH≤3) induce the rapid hydrolysis of the phosphoramidate bond, whereas the phosphorylated peptide remains stable under slightly acidic, neutral and basic pH, even at elevated temperatures (Fig. 1A).
Fig. 1.
Quantitative mass spectrometric analysis of arginine phosphorylation. A, Stability analysis of the phosphoarginine modification. The graph depicts percentage of arginine phosphorylation remaining after the incubation of the model arginine phosphopeptide KpRGGGGYIKKIKV at different pH conditions (left chart, 25 °C; right chart, 60 °C). B, Study design. The schematic presentation illustrates the methodology applied to analyze the arginine phosphoproteome of B. subtilis. A key step of this procedure (detailed information in the SI part) was to optimize phosphopeptide enrichment at mild acidic conditions to preserve the phosphoarginine modification. The iTRAQ chemistry was applied to obtain quantitative information on the abundance of phosphorylation sites under different stress conditions, as well as to report on the acid sensitivity of each identified phosphopeptide.
To perform an in-depth analysis of the phosphoproteome, the selective enrichment of phosphopeptides before MS analysis is an essential step. Most often, enrichment is achieved by performing metal oxide affinity chromatography using titanium dioxide (TiO2) (35). Of note, the TiO2-based enrichment has to be carried out under strongly acidic conditions (pH 2) to efficiently protonate carboxylate groups and prevent the co-purification of unphosphorylated, acidic peptides (36). Given the instability of the phosphoarginine mark at this pH, we adapted the TiO2 procedure accordingly by applying less acidic conditions (pH 4) during the critical binding step. Removal of unspecifically bound, unphosphorylated peptides was achieved by fast washing steps at low pH. Furthermore, peptide desalting before LC-MS analysis (both offline or online) also typically employs acidic solvents, such as 0.1% trifluoroacetic acid (TFA) or 0.5% acetic acid. Here, the acid acts as ion-pairing agent ensuring the proper binding of peptides to the chromatographic C18 material. In instances where offline peptide desalting or buffer exchange is necessary, for example before isobaric tag labeling, we thus used heptafluorobutyric acid (HFBA) buffered to pH 8 as an ion pairing agent. For online desalting, where a trap column was used in the LC-MS/MS set-up, we observed that a 0.5% acetic acid solution adjusted to pH 4.2 performs equally well to standard loading buffers (pH 1.8 - 2) and almost completely prevented phosphoarginine hydrolysis. Thus, the adjusted acetic acid solution appears to be the optimal buffer for analyzing N-phosphorylated samples by LC-MS/MS.
In addition to sample preparation, also the MS analysis per se is critical to characterize protein phosphorylation events. Recently, our group analyzed the performance of different MS fragmentation techniques in correctly identifying arginine phosphorylation events. We observed that collision-induced dissociation (CID) or higher energy collisional dissociation (HCD) data show considerably increased frequency of false localizations when compared with electron-transfer dissociation (ETD) data for N-phosphorylated samples. This is because of a high rate of H3PO4 elimination that most likely results from the intramolecular rearrangement of the N-bound phosphate in gas phase during the former collision-induced fragmentations (37). Accordingly, in the case of phosphoarginine analysis, the localization reliability has to be carefully scrutinized, as the standard phosphosite localization algorithms have been validated for serine, threonine and tyrosine phosphorylations only. To ensure the data quality of our study we thus (1) employed ETD fragmentation for the localization of phosphosites, (2) manually inspected all phosphopeptide spectra, and (3) included an acid hydrolyzed sample as an additional evidence of the nature of the underlying modifications (Fig. 1B). By including this control in the quantitative analysis, we obtained information on the acid sensitivity of every identified phosphosite, thus further distinguishing between N- and O-phosphorylations.
B. subtilis Accumulates Diverse Phosphoarginine Proteins in YwlE-deficient Cells
To evaluate the in vivo relevance of arginine phosphorylation and to characterize the interplay between the protein arginine kinase McsB and the cognate phosphatase YwlE, we employed the adapted TiO2 method to perform an in-depth arginine phosphoproteomic analysis of B. subtilis. Phosphopeptides from two biological replicates were analyzed by high resolution mass spectrometry using three different fragmentation types (ETD, HCD, and CID) in combination. Two technical replicates were recorded for each sample. Data analysis was performed using Mascot (Matrix Science) for peptide identification and PhosphoRS (29) for phosphosite localization, in a target-decoy approach. Using a B. subtilis strain lacking the arginine phosphatase YwlE (ΔywlE), which was grown under normal and stressed conditions (Fig. 1B), we identified in total 217 phosphoarginine sites distributed in 134 proteins (Table I, supplemental Table S1). Additionally, 27 serine, threonine and tyrosine phosphopeptides were identified in this dataset (supplemental Table S1). In contrast, qualitative MS analysis of wild-type B. subtilis samples solely revealed O-phosphorylation sites (supplemental Table S2), whereas phosphoarginine sites were not observed. To explore whether the identified phosphoarginine sites are exclusively introduced by the McsB kinase, we repeatedly analyzed the phosphoproteome of a ΔywlEΔmcsB double mutant strain grown both under unstressed and heat-shocked conditions. Although we could identify serine, threonine and tyrosine phosphorylations in this McsB-deficient strain (supplemental Table S3), we did not observe any phosphoarginine sites. Thus, McsB appears to be the only protein arginine kinase in B. subtilis that, together with YwlE, shapes the arginine phosphoproteome. Furthermore, based on sequence homology searches, there is no evidence that a second protein arginine kinase exists in B. subtilis. In contrast, the arginine phosphoproteomic analysis of Elsholz and coworkers indicated that four proteins (RecA, Nin, BdhA, and AroF) became arginine-phosphorylated in the ΔywlEΔmcsB mutant, and thus are not McsB products (38). Unfortunately, site localization confidence and spectral information for those phosphorylations are not reported and therefore cannot be verified. Nevertheless, under the conditions employed in our study, all identified phosphoarginine sites directly result from McsB activity.
Table I. ••••.

Next, we asked whether there is any recurring sequence motif in the sites phosphorylated by McsB. For this purpose, the amino acid sequences comprising ± 10 residues flanking each phosphoarginine site were submitted for consensus sequence analysis using the IceLogo program (31). The corresponding sequences of full-length proteins were used as reference dataset to eliminate any amino acid frequency bias. As shown in Fig. 2A, some amino acids located in the phosphoarginine vicinity appear to be significantly overrepresented as judged by probability analysis. However, the relative frequency of those residues compared with the reference dataset was always low (below 10%). Accordingly, McsB appears not to target a specific sequence motif. The only tendency that could be derived from these data is that glycine, serine and glutamine residues are slightly overrepresented in the mapped phosphoarginine sites, suggesting that many McsB phosphorylation sites are located in flexible regions. However, it has to be noted that the observed sequence frequencies may also result from a possible variation in stability of the phosphoarginine modification in distinct peptide contexts rather than reflecting the substrate preference of the McsB kinase. Taken together, our data suggest that McsB relies on additional regulatory mechanisms such as temporal activation, cellular localization and/or collaboration with certain adaptor proteins such as McsA to target substrates in a specific manner.
To evaluate whether McsB has a preference for distinct classes of proteins, we performed a gene ontology (GO) enrichment analysis of the phosphoarginine-containing proteins identified in the ΔywlE strain. Blast2GO (32) was used to assign GO terms for all B. subtilis proteins, based on the closest GO-annotated homologs present in the Swiss-Prot database. Analysis of the enrichment of GO terms in the arginine-phosphorylated protein dataset, compared with the overall proteome, was performed by applying the Fisher's exact test in the Blast2GO interface. A full report of enriched GO terms using term filter value 0.05 at FDR mode is provided in supplemental Table S4. The most specific GO terms (cut-off 0.01) are shown in Fig. 2B. According to these data, membrane proteins are clearly underrepresented, reflecting the fact that McsB is a cytosolic kinase. The process “translation” is strongly enriched, together with the related functional terms “structural constituent of ribosome” and “rRNA binding” and the cellular component terms “cytosolic ribosome,” “small ribosomal subunit” and “large ribosomal subunit.” Additionally, the processes “glycolysis” and “tricarboxylic acid cycle,” “response to cadmium ion” and “GTP catabolic process” are also enriched. In general, it seems that a broad variety of proteins become arginine-phosphorylated in the phosphatase-deficient strain, many of them connected with protein synthesis and metabolic functions.
Stress Induces Arginine Phosphorylation of Key Components of the Protein Quality Control System
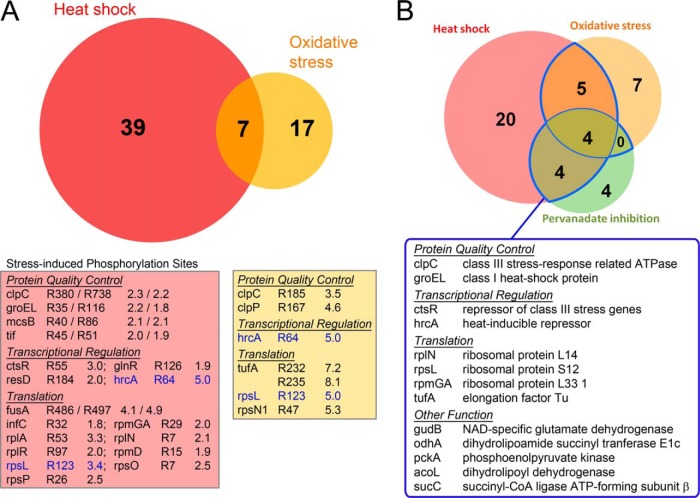
As the McsB arginine kinase is a member of the bacterial stress response, which is known to phosphorylate and inhibit the heat shock response regulator CtsR (11), we next evaluated how different stress situations affect the arginine phosphoproteome. For this purpose, cell lysates of B. subtilis grown under physiological, heat shock (20 min at 50 °C) and oxidative stress conditions (20 min incubation in the presence of 1 mm diamide) (Fig. 1B) were prepared for MS analysis. An isobaric, isotope-coded tag (iTRAQ) was incorporated into the peptides to deduce the relative abundance of arginine phosphorylations under different growth conditions from the corresponding MS/MS spectra (39). To distinguish if the observed changes result from distinct protein amounts or higher phosphorylation rates, protein ratios were also measured (supplemental Table S5) and used to normalize phosphopeptide ratios. Comparison of the individual iTRAQ ratios revealed that, even though the abundance of the majority of the arginine-phosphorylated proteins did not significantly change (supplemental Table S1), 41 phosphosites were up-regulated during heat shock and 24 phosphosites during oxidative stress, as summarized in Fig. 3A (see also supplemental Table S1). Interestingly, not only CtsR but also its immediate targets McsB, ClpC, and ClpP showed an up-regulation of phosphorylation upon stress. In this context, it is remarkable that another heat shock repressor, HrcA, also exhibits elevated arginine phosphorylation during stress situations. This finding strongly suggests that arginine phosphorylation is an active mechanism inducing expression of HrcA-repressed heat shock genes (dnaK and groE operons). In fact, the chaperone GroEL is also one of the regulated targets of McsB. As illustrated in Fig. 3A, the overlap between the regulated sites in heat shock and in oxidative stress is relatively low (11%), indicating that the mounted stress response, as characterized by its phosphoarginine pattern, is highly dependent on the environmental conditions.
Fig. 3.
Comparative analysis of bacterial arginine phosphoproteome on different stress situations. A, Quantitative analysis of arginine phosphorylation on different growth conditions. The Venn diagram shows the heat shock (red) and oxidative stress (yellow) induced phosphoarginine sites in B. subtilis ΔywlE. Overlapping sites are shown in orange. Individual regulatory ratios for proteins involved in protein quality control, transcriptional and translational regulation are listed below. The highlighted proteins HrcA and ribosomal protein S12 (rpsL) exhibit increased phosphorylation levels under both stress conditions. B, Analysis of arginine phosphorylation of heat-shocked wild-type cells subjected to YwlE inactivation by pervanadate. The Venn diagram illustrates the overlap of all arginine-phosphorylated proteins identified in the heat-shocked, pervanadate-treated wild-type cells (green) with proteins showing stress-increased arginine phosphorylation in the quantitative ΔywlE strain analysis (red: heat shock, yellow: oxidative stress). Among the 44 arginine-phosphorylated proteins identified, 13 proteins occur under at least two distinct conditions (blue border, and depicted in the table below) and should represent the most prominent physiological targets of McsB during the bacterial stress response.
To further confirm the most prominent phosphorylation sites that are specifically targeted by McsB, we analyzed the arginine phosphoproteome of wild-type cells under different conditions. To block the in vivo activity of the YwlE arginine phosphatase during heat stress exposure, sodium pervanadate, a cell membrane permeable phosphatase inhibitor oxidizing the active site cysteine, was employed. Although the number of detected arginine phosphorylations (15 sites in 12 proteins, Fig. 3B and supplemental Table S6) was significantly lower in comparison to those detected in ΔywlE cells (supplemental Table S1), several targets showing a pronounced up-regulation on folding stress were also identified in the wild-type background, notably ClpC, GroEL, CtsR, and HrcA. Moreover, these sites were not identified in the unstressed control. Taken together, the quantitative analyses of the B. subtilis ΔywlE arginine phosphoproteome and the precisely timed inhibition of the YwlE arginine phosphatase in wild-type cells markedly reduced the list of potential McsB substrates. Among the 134 candidates, the transcriptional repressors CtsR and HrcA, the chaperone GroEL, the unfoldase ClpC, together with several ribosomal subunits, should represent the main physiological targets of McsB under the analyzed stress conditions.
The ClpC Unfoldase is a Prominent Target of McsB
ClpC stands out in our analysis because of the large number of identified phosphoarginine sites (12 in total). However, only R380, R783, and R185 exhibited increased levels under stress conditions. Indeed, co-immunoprecipitation experiments have shown that McsB forms a complex with McsA and ClpCP both in physiological and high temperature conditions (22). Given the anticipated close contact of McsB and ClpC, such high number of constitutive phosphorylations is not unexpected. The active form of ClpC is a hexameric complex that only forms in the presence of its adaptor protein, MecA (18). This hexameric complex is capable of binding to the heptameric ClpP protease complex, and is responsible for ATP-dependent unfolding and translocation of substrate proteins into the ClpP proteolytic chamber (40). Inspection of the available ClpC-MecA crystal structure (PDB ID 3PXI, (41)) revealed that the majority of the phosphoarginine sites are buried in the hexameric complex (8 from the 11 residues defined in the structure), whereas almost all identified phosphoarginine sites are solvent-accessible in the monomeric ClpC form (Fig. 4). Given the distinct accessibility of the phosphoarginine sites, McsB appears to have a clear preference for targeting the monomeric, inactive state of ClpC. As most of the targeted sites are close to protein interfaces critical for hexamer assembly, McsB-mediated phosphorylation is expected to impair formation of the functional form of ClpC and thus to have a rather inhibitory effect on the ClpCP proteolytic system. Notably, McsB has also been proposed to function as an adaptor protein targeting substrates to ClpCP and promoting the assembly of the functional ClpCP protease (22). Elsholz and co-workers identified five phosphoarginine sites in ClpC, three of those—R5, R254, and R443—were not observed in our study. The authors report that the ATPase activity of recombinant ClpCR5A and ClpCR254A variants fail to get activated by McsB, and that CtsR is not degraded under high temperature when those variants are introduced in vivo. Therefore, the authors conclude that the R5 and R254 phosphorylations are necessary for McsB mediated ClpCP activation and regulated CtsR degradation (12). It seems, though, that such phosphorylations are not prominent, because we never observed them in repeated analyses. In conclusion, it appears that both effects—activation and inhibition—can result from McsB-mediated arginine phosphorylation of the ClpC unfoldase, evidencing the complexity of the regulation of this highly energy demanding molecular machine. Clearly, deciphering the interplay between McsB and ClpC is an interesting topic for further investigation.
Fig. 4.
Structural localization of arginine phosphorylation sites in the ClpC ATPase/unfoldase. Crystal structure of the ClpC-MecA hexameric complex (left) and the corresponding ClpC monomer (right), according to PDB 3PXI. MecA is distinguished in a darker shade of gray. Arginine residues phosphorylated in the B. subtilis ΔywlE strain are marked in spheres (red, accessible; orange, inaccessible sites). To indicate the orientation of the arginine side chains, nitrogen atoms are colored blue. Accordingly, in the hexameric complex (A, side and top views) most arginine residues are inaccessible. Therefore, the ClpC monomer (B, side and top views) should be the primary target of the McsB kinase.
DISCUSSION
The B. subtilis Arginine Phosphoproteome
In this work, we identified 231 arginine-phosphorylated peptides representing 217 sites in 134 distinct proteins of the Gram-positive model bacterium B. subtilis (Fig. 2, supplemental Table S1). Notably, this phosphoarginine dataset contains the highest number of phosphorylation sites reported for a bacterial species, thus reflecting the high efficiency of our MS-based analysis of phosphoarginine, which is considered a technically challenging modification, given its acid lability (Fig. 1A). Recently, Elsholz and co-workers published the identification of 121 arginine phosphorylation sites in 87 proteins, also employing a B. subtilis ΔywlE strain, using standard protocols for phosphoproteomic analysis (38). However, to obtain this result 20 mg of starting material was used, whereas we used about 1 mg of cell extract to obtain almost twice the number of phosphopeptides. Most importantly, all but one of the phosphosites reported in our study have been localized based on ETD fragmentation data, whereas the Elsholz study employed CID fragmentation. As we have recently demonstrated, ETD provides considerably more accurate phosphosite localizations of N-phosphorylated samples than CID (37). Furthermore, we demonstrated that the identified in vivo phosphoarginine sites are acid-labile, thus further evidencing the nature of the underlying phosphorylations. Taking all this into consideration, our dataset represents a more reliable and comprehensive view of the arginine phosphoproteome of this bacterium.
Comparison of the two distinct phosphoarginine datasets reveals a rather low overlap: only 15% of the arginine-phosphorylated proteins were identified in both studies. The observed discrepancy could be explained by the different experimental conditions, as Elsholz and coworkers analyzed exponential and stationary B. subtilis cultures grown under unstressed conditions only, but also by the different sample preparation and analysis methods, as discussed above. Most importantly, however, the low overlap suggests that McsB is capable of phosphorylating a strikingly large number of proteins, a fact that is consistent with the lack of a clear sequence motif for the kinase (Fig. 2A). An in-depth phosphoproteomic study analyzing O-phosphorylations in B. subtilis identified only 78 sites (30), most likely representing the combined activity of a number of protein serine/threonine and tyrosine kinases. This comparison suggests that the phosphorylation of such a large number of substrates is not a common feature of bacterial protein kinases, but a particular feature of McsB. Given the number and the functional diversity of the arginine-phosphorylated proteins, it seems unlikely that all those phosphorylations are biologically relevant. We assume that a single kinase would not be participating in so many processes in the cell, especially when this kinase is part of a specific stress-regulated operon. It is important to mention, though, that McsB activity is not entirely promiscuous, because a number of phosphorylations were found to be specifically regulated according to distinct environmental conditions. Consistent with this notion, a previous study using McsB-YFP fusions in B. subtilis revealed that the kinase is equally distributed in the cytoplasm, but assumes a polar position on the development of competence. This localization overlapped with the localization of the competence protein ComGA-CFP, which is known to be peripherally associated with the inner face of the membrane, accumulating at the cell poles when cells become transformable (42). Therefore, in addition to the reported McsB targets, other regulatory arginine phosphorylations may occur during the development of competence, when McsB is concentrated to a specific cellular location. This indicates that localization may be an important substrate targeting mechanism for McsB.
The Role of McsB in the Bacterial Stress Response
Given the large number of arginine-phosphorylated proteins, it is difficult to propose a mechanistic model of how McsB promotes the adaptation of bacteria toward stress situations. To address this question, we performed a quantitative phosphoproteomic analysis of the B. subtilis ΔywlE strain grown under different situations. Importantly, heat shock proteins belonging to the protein quality control system showed increased levels of arginine phosphorylation during stress conditions (Fig. 3A). The favored McsB targets included the class III heat shock proteins CtsR, McsB, ClpC and ClpP and the class I proteins HrcA and GroEL. Also the Trigger factor chaperone showed increased levels of phosphorylation upon heat shock.
Most remarkable is the finding that the repressor HrcA becomes arginine-phosphorylated in a regulated manner. Phosphorylation of R64, located within the winged helix-turn-helix (HTH) DNA-binding domain, showed a 5-fold increase upon both heat shock and oxidative stress. HrcA phosphorylation was also identified in pervanadate-treated wild-type cells exposed to high temperature. Remarkably, the HrcA repressor was shown to control the expression of two operons in B. subtilis: heptacistronic dnaK and the bicistronic groE operon (43). The first one encodes for HrcA itself, the DnaK chaperone complex and 3 uncharacterized proteins, whereas the second operon includes the GroEL/ES chaperonine. Previous studies revealed that the activity of the HrcA repressor is modulated by the GroEL chaperonine system. According to the current model, de novo synthesized HrcA is present in the inactive conformation and requires remodeling by GroEL/ES to adopt its active state. This functional switch is transiently prevented at high temperatures, when the accumulating misfolded proteins bind to GroEL, thereby decreasing the levels of activated (DNA-binding competent) HrcA (44, 45). This titration model is supported by the observations that GroEL and HrcA interact in vitro, that purified HrcA relies on the presence of GroEL to bind to its operator DNA (46, 47) and that deletion of the GroES and GroEL proteins led to a high constitutive expression of the dnaK operon at low temperature, whereas overexpression of GroEL led to lower dnaK levels (46). As pointed out by Schumann (44), a remaining open question is whether titration of GroE immediately after a heat shock is sufficient to turn on the HrcA regulon or whether an additional mechanism accounts for the rapid increase in transcription of the dnaK and groE operons. Our findings strongly suggest that McsB-mediated arginine phosphorylation is this additional mechanism inducing the rapid deactivation of HrcA upon folding stress. Therefore, our data represent the first direct link between the regulation of class I (HrcA regulon) and class III (CtsR regulon) heat shock genes of B. subtilis. In some organisms, including the pathogenic bacterium Staphylococcus aureus, CtsR further controls the expression of the general chaperones GroEL and DnaK together with the HrcA repressor (48). Accordingly, the coordination of the activities of CtsR and HrcA, as revealed by our study, seems to be important for the proper mounting of the bacterial protein quality control system in response to stress situations.
Regarding the molecular mechanism of McsB mediated CtsR inhibition, gel shift experiments showed that arginine phosphorylation of CtsR induces the displacement of the repressor from DNA (11). An analogous effect can be expected for the structurally similar HrcA. Noteworthy, the observed up-regulated phosphoarginine site (R64) is located in the beta-wing of the HTH domain, that in the case of CtsR has been shown to be essential for DNA binding (11). Indeed, basic residues such as arginine are often critical for interaction with nucleic acids; adding a negative charge to such positions by phosphorylation can potentially serve as a universal mechanism for regulating protein-DNA interactions. This idea is supported by our results, as a considerable number of transcriptional factors were found to be arginine-phosphorylated (Table I). It will be interesting to see in future analysis to what extent arginine phosphorylation modulates transcriptional regulation in different systems.
Arginine Phosphorylation in Protein Homeostasis
Finally, it is important to mention that many proteins important for protein synthesis, i.e. nine ribosomal proteins, the translation initiation factor IF-3 the elongation factors Tu and G exhibited increased levels of arginine phosphorylation under stress situations (Fig. 3A). This re-affirms the notion, supported by the GO term enrichment analysis (Fig. 2B), that the process “protein translation” is regulated by arginine phosphorylation. The protein quality control machinery is both acting on the folding of newly synthesized proteins as well as the repair and degradation of damaged ones. When environmental conditions are unfavorable, levels of unfolded proteins increase and, to cope with the higher load, protein synthesis has to be down-regulated. Accordingly, recent reports highlight the impact of different stress situations on the mRNA selectivity and subunit assembly of the ribosome. In B. subtilis, for example, the ribosome plays an important role as stress sensor mediating the activation of σB, the stress response sigma factor (49). Similarly, generation of a modified E. coli ribosome in response to the MazF toxin emphasizes the importance of ribosomal re-assembly to adapt to stress situations (50). Hence, phosphoarginine modification of selected ribosomal subunits could represent another molecular signal modulating the overall efficiency and/or selectivity of protein translation in response to external signals, in pace with cellular protein degradation pathways. According to our data, arginine phosphorylation appears to be of great importance to the control of protein homeostasis in B. subtilis.
CONCLUSIONS
In sum, the substantial improvements in the analysis of protein arginine phosphorylation by mass spectrometry reported here enabled us to obtain a detailed and reliable view on the arginine phosphoproteome of B. subtilis. A quantitative analysis was used to identify major physiological targets of McsB in the bacterial stress response. We demonstrate that McsB is a highly active protein arginine kinase that can target a large number of substrates in the absence of the cognate phosphatase. Despite this promiscuity, specific factors belonging to the protein quality control system, such as the ClpCP protease complex and the CtsR and HrcA transcriptional repressors, are targeted in a regulated manner under stress conditions. This suggests that arginine phosphorylation acts not only at the transcriptional level by regulating expression of class I and class III heat shock genes, but also by modulating activity of relevant proteins such as chaperones, proteases and the translation machinery. In conclusion, these findings support the notion that arginine phosphorylation is an important mechanism controlling protein homeostasis in B. subtilis.
Supplementary Material
Acknowledgments
We thank Dr. N. Ogasawara for the B. subtilis ΔmcsB strain, T. Taus, Dr. P. Pichler and Dr. W. Straube for assistance with MS data analysis. The IMP is funded by Boehringer Ingelheim.
Footnotes
Author contributions: A.S., D.B.T., S.S., G.A., K.M., and T.C. designed research; A.S., D.B.T., S.S., and J.F. performed research; A.S. and D.B.T. analyzed data; A.S., D.B.T., and T.C. wrote the paper.
* This work was supported by the Christian Doppler Research Association, the European Community's Seventh Framework Programme (FP7/2007–2013)/grant agreement n° 241548 - MeioSys and 262067 - PRIME-XS, and from the Austrian Science Fund SFB F3402, P 22570-B09 (DBT) and TRP 308-N15.
 This article contains supplemental Tables S1 to S6.
This article contains supplemental Tables S1 to S6.
1 The abbreviations used are:
- GO
- gene ontology
- HFBA
- heptafluorobutyric acid
- HTH
- helix-turn-helix
- iTRAQ
- isobaric tag for relative and absolute quantitation
- RP
- reverse-phase
- SPE
- solid-phase extraction
- TiO2
- titanium dioxide.
REFERENCES
- 1. Lemeer S., Heck A. (2009) The phosphoproteomics data explosion. Curr. Opin. Chem. Biol. 13, 414–420 [DOI] [PubMed] [Google Scholar]
- 2. Attwood P. V., Piggott M. J., Zu X. L., Besant P. G. (2007) Focus on phosphohistidine. Amino Acids 32, 145–156 [DOI] [PubMed] [Google Scholar]
- 3. Besant P. G., Attwood P. V., Piggott M. J. (2009) Focus on phosphoarginine and phospholysine. Curr. Protein Pept. Sci. 10, 536–550 [DOI] [PubMed] [Google Scholar]
- 4. Cieœla J., Frączyk T., Rode W. (2011) Phosphorylation of basic amino acid residues in proteins: important but easily missed. Acta Biochim. Pol. 58, 137–148 [PubMed] [Google Scholar]
- 5. Matthews H. R. (1995) Protein kinases and phosphatases that act on histidine, lysine, or arginine residues in eukaryotic proteins: a possible regulator of the mitogen-activated protein kinase cascade. Pharmacol. Ther. 67, 323–350 [DOI] [PubMed] [Google Scholar]
- 6. Wuichet K., Cantwell B., Zhulin I. (2010) Evolution and phyletic distribution of two-component signal transduction systems. Curr. Opin. Microbiol. 13, 219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maeda T., Wurgler-Murphy S. M., Saito H. (1994) A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369, 242–245 [DOI] [PubMed] [Google Scholar]
- 8. Nongpiur R., Soni P., Karan R., Singla-Pareek S., Pareek A. (2012) Histidine kinases in plants: cross talk between hormone and stress responses. Plant Signal. Behav. 7, 1230–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wakim B. T., Aswad G. D. (1994) Ca(2+)-calmodulin-dependent phosphorylation of arginine in histone 3 by a nuclear kinase from mouse leukemia cells. J. Biol. Chem. 269, 2722–2727 [PubMed] [Google Scholar]
- 10. Wakim B., Grutkoski P., Vaughan A., Engelmann G. (1995) Stimulation of a Ca2+-Calmodulin-activated Histone 3 Arginine Kinase in Quiescent Rat Heart Endothelial Cells Compared to Actively Dividing Cells. J. Biol. Chem. 270, 23155–23158 [DOI] [PubMed] [Google Scholar]
- 11. Fuhrmann J., Schmidt A., Spiess S., Lehner A., Turgay K. R., Mechtler K., Charpentier E., Clausen T. (2009) McsB is a protein arginine kinase that phosphorylates and inhibits the heat-shock regulator CtsR. Science 324, 1323–1327 [DOI] [PubMed] [Google Scholar]
- 12. Elsholz A., Michalik S., Zühlke D., Hecker M., Gerth U. (2010) CtsR, the Gram-positive master regulator of protein quality control, feels the heat. EMBO J. 29, 3621–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kirstein J., Zühlke D., Gerth U., Turgay K., Hecker M. (2005) A tyrosine kinase and its activator control the activity of the CtsR heat shock repressor in B. subtilis. EMBO J. 24, 3435–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Derré I., Rapoport G., Msadek T. (1999) CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol. Microbiol. 31, 117–131 [DOI] [PubMed] [Google Scholar]
- 15. Krüger E., Hecker M. (1998) The first gene of the Bacillus subtilis clpC operon, ctsR, encodes a negative regulator of its own operon and other class III heat shock genes. J. Bacteriol. 180, 6681–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Derré I., Rapoport G., Devine K., Rose M., Msadek T. (1999) ClpE, a novel type of HSP100 ATPase, is part of the CtsR heat shock regulon of Bacillus subtilis. Mol. Microbiol. 32, 581–593 [DOI] [PubMed] [Google Scholar]
- 17. Krüger E., Witt E., Ohlmeier S., Hanschke R., Hecker M. (2000) The clp proteases of Bacillus subtilis are directly involved in degradation of misfolded proteins. J. Bacteriol. 182, 3259–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schlothauer T., Mogk A., Dougan D., Bukau B., Turgay K. R. (2003) MecA, an adaptor protein necessary for ClpC chaperone activity. Proc. Natl. Acad. Sci. U.S.A. 100, 2306–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pan Q., Losick R. (2003) Unique degradation signal for ClpCP in Bacillus subtilis. J. Bacteriol. 185, 5275–5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Turgay K., Hahn J., Burghoorn J., Dubnau D. (1998) Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 17, 6730–6738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krüger E., Zühlke D., Witt E., Ludwig H., Hecker M. (2001) Clp-mediated proteolysis in Gram-positive bacteria is autoregulated by the stability of a repressor. EMBO J. 20, 852–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kirstein J., Dougan D., Gerth U., Hecker M., Turgay K. R. (2007) The tyrosine kinase McsB is a regulated adaptor protein for ClpCP. EMBO J. 26, 2061–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Musumeci L., Bongiorni C., Tautz L., Edwards R., Osterman A., Perego M., Mustelin T., Bottini N. (2005) Low-molecular-weight protein tyrosine phosphatases of Bacillus subtilis. J. Bacteriol. 187, 4945–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fuhrmann J., Mierzwa B., Trentini D. B., Spiess S., Lehner A., Charpentier E., Clausen T. (2013) Structural Basis for Recognizing Phosphoarginine and Evolving Residue-Specific Protein Phosphatases in Gram-Positive Bacteria. Cell Rep. 3, 1832–1839 [DOI] [PubMed] [Google Scholar]
- 25. Anagnostopoulos C., Spizizen J. (1961) Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81, 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kobayashi K., Ehrlich S. D., Albertini A., Amati G., Andersen K. K., Arnaud M., Asai K., Ashikaga S., Aymerich S., Bessieres P., Boland F., Brignell S. C., Bron S., Bunai K., Chapuis J., Christiansen L. C., Danchin A., Débarbouillé M., Dervyn E., Deuerling E., Devine K., Devine S. K., Dreesen O., Errington J., Fillinger S., Foster S. J., Fujita Y., Galizzi A., Gardan R., Eschevins C., Fukushima T., Haga K., Harwood C. R., Hecker M., Hosoya D., Hullo M. F., Kakeshita H., Karamata D., Kasahara Y., Kawamura F., Koga K., Koski P., Kuwana R., Imamura D., Ishimaru M., Ishikawa S., Ishio I., Le Coq D., Masson A., Mauëlx C., Meima R., Mellado R. P., Moir A., Moriya S., Nagakawa E., Nanamiya H., Nakai S., Nygaard P., Ogura M., Ohanan T., O'Reilly M., O'Rourke M., Pragai Z., Pooley H. M., Rapoport G., Rawlins J. P., Rivas L. A., Rivolta C., Sadaie A., Sadaie Y., Sarvas M., Sato T., Saxild H. H., Scanlan E., Schumann W., Seegers J. F. M. L., Sekiguchi J., Sekowska A., Séror S. J., Simon M., Stragier P., Studer R., Takamatsu H., Tanaka T., Takeuchi M., Thomaides H. B., Vagner V., van Dijl J. M., Watabe K., Wipat A., Yamamoto H., Yamamoto M., Yamamoto Y., Yamane K., Yata K., Yoshida K., Yoshikawa H., Zuber U., Ogasawara N. (2003) Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U.S.A. 100, 4678–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pumiglia K. M., Lau L. F., Huang C. K., Burroughs S., Feinstein M. B. (1992) Activation of signal transduction in platelets by the tyrosine phosphatase inhibitor pervanadate (vanadyl hydroperoxide). Biochem. J. 286(Pt 2), 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wis′niewski J., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 29. Taus T., Köcher T., Pichler P., Paschke C., Schmidt A., Henrich C., Mechtler K. (2011) Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354–5362 [DOI] [PubMed] [Google Scholar]
- 30. Macek B., Mijakovic I., Olsen J., Gnad F., Kumar C., Jensen P., Mann M. (2007) The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6, 697–707 [DOI] [PubMed] [Google Scholar]
- 31. Colaert N., Helsens K., Martens L., Vandekerckhove J. l., Gevaert K. (2009) Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6, 786–787 [DOI] [PubMed] [Google Scholar]
- 32. Conesa A., Götz S., García-Gómez J. M., Terol J., Talón M., Robles M. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 [DOI] [PubMed] [Google Scholar]
- 33. Vizcaíno J. A., Côté R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Pérez-Riverol Y., Reisinger F., Ríos D., Wang R., Hermjakob H. (2013) The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sickmann A., Meyer H. E. (2001) Phosphoamino acid analysis. Proteomics 1, 200–206 [DOI] [PubMed] [Google Scholar]
- 35. Pinkse M., Uitto P., Hilhorst M., Ooms B., Heck A. (2004) Selective Isolation at the Femtomole Level of Phosphopeptides from Proteolytic Digests Using 2D-NanoLC-ESI-MS/MS and Titanium Oxide Precolumns. Anal. Chem. 76, 3935–3943 [DOI] [PubMed] [Google Scholar]
- 36. Beltran L., Cutillas P. (2012) Advances in phosphopeptide enrichment techniques for phosphoproteomics. Amino Acids 43, 1009–1024 [DOI] [PubMed] [Google Scholar]
- 37. Schmidt A., Ammerer G., Mechtler K. (2013) Studying the fragmentation behavior of peptides with arginine phosphorylation and its influence on phospho-site localization. Proteomics 13, 945–954 [DOI] [PubMed] [Google Scholar]
- 38. Elsholz A., Turgay K., Michalik S., Hessling B., Gronau K., Oertel D., Mäder U., Bernhardt J., Becher D., Hecker M., Gerth U. (2012) Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 109, 7451–7456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ross P., Huang Y., Marchese J., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 40. Kirstein J., Schlothauer T., Dougan D., Lilie H., Tischendorf G., Mogk A., Bukau B., Turgay K. R. (2006) Adaptor protein controlled oligomerization activates the AAA+ protein ClpC. EMBO J. 25, 1481–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang F., Mei Z., Qi Y., Yan C., Hu Q., Wang J., Shi Y. (2011) Structure and mechanism of the hexameric MecA-ClpC molecular machine. Nature 471, 331–335 [DOI] [PubMed] [Google Scholar]
- 42. Hahn J., Kramer N., Briley K., Dubnau D. (2009) McsA and B mediate the delocalization of competence proteins from the cell poles of Bacillus subtilis. Mol. Microbiol. 72, 202–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yuan G., Wong S. L. (1995) Isolation and characterization of Bacillus subtilis groE regulatory mutants: evidence for orf39 in the dnaK operon as a repressor gene in regulating the expression of both groE and dnaK. J. Bacteriol. 177, 6462–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schumann W. (2003) The Bacillus subtilis heat shock stimulon. Cell Stress Chaperones 8, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schumann W. (2009) Temperature sensors of eubacteria. In: Sariaslani S., Gadd G. M., eds. Advances in Applied Microbiology, Elsevier; [DOI] [PubMed] [Google Scholar]
- 46. Mogk A., Homuth G., Scholz C., Kim L., Schmid F. X., Schumann W. (1997) The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 16, 4579–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reischl S., Wiegert T., Schumann W. (2002) Isolation and analysis of mutant alleles of the Bacillus subtilis HrcA repressor with reduced dependency on GroE function. J. Biol. Chem. 277, 32659–32667 [DOI] [PubMed] [Google Scholar]
- 48. Chastanet A., Fert J., Msadek T. (2003) Comparative genomics reveal novel heat shock regulatory mechanisms in Staphylococcus aureus and other Gram-positive bacteria. Mol. Microbiol. 47, 1061–1073 [DOI] [PubMed] [Google Scholar]
- 49. Zhang S., Scott J. M., Haldenwang W. G. (2001) Loss of ribosomal protein L11 blocks stress activation of the Bacillus subtilis transcription factor sigma(B). J. Bacteriol. 183, 2316–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vesper O., Amitai S., Belitsky M., Byrgazov K., Kaberdina A. C., Engelberg-Kulka H., Moll I. (2011) Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 147, 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data from this publication have been submitted to the ProteomeXchange Consortium http://proteomecentral.proteomexchange.org via the PRIDE partner repository (33) and assigned the identifier PXD000273.