Abstract
Salinity is a major abiotic stress affecting plant growth and development. Understanding the molecular mechanisms of salt response and defense in plants will help in efforts to improve the salt tolerance of crops. Brachypodium distachyon is a new model plant for wheat, barley, and several potential biofuel grasses. In the current study, proteome and phosphoproteome changes induced by salt stress were the focus. The Bd21 leaves were initially treated with salt in concentrations ranging from 80 to 320 mm and then underwent a recovery process prior to proteome analysis. A total of 80 differentially expressed protein spots corresponding to 60 unique proteins were identified. The sample treated with a median salt level of 240 mm and the control were selected for phosphopeptide purification using TiO2 microcolumns and LC-MS/MS for phosphoproteome analysis to identify the phosphorylation sites and phosphoproteins. A total of 1509 phosphoproteins and 2839 phosphorylation sites were identified. Among them, 468 phosphoproteins containing 496 phosphorylation sites demonstrated significant changes at the phosphorylation level. Nine phosphorylation motifs were extracted from the 496 phosphorylation sites. Of the 60 unique differentially expressed proteins, 14 were also identified as phosphoproteins. Many proteins and phosphoproteins, as well as potential signal pathways associated with salt response and defense, were found, including three 14-3-3s (GF14A, GF14B, and 14-3-3A) for signal transduction and several ABA signal-associated proteins such as ABF2, TRAB1, and SAPK8. Finally, a schematic salt response and defense mechanism in B. distachyon was proposed.
Soil salinity is one of the severest problems in irrigated agriculture worldwide. Salt stress reduces the average yield of the major crops and severely limits agricultural production. Sustainable and equitable global food security is largely dependent on the development of crops with high salt tolerance. In addition, improved salt tolerance of perennial crop species used for fodder or fuel production has also become a key component in reducing the spread of secondary salinity in many regions of the world (1).
Under salt stress, the plant cells perceive salt stress signals and then regulate the transcription and translation of intracellular genes associated with stress response through signal transduction, which lead to a series of physiological and biochemical responses in the plant (2). Salt stress can be divided into osmotic stress, ion damage, and ion imbalance (or nutritional deficiency). Osmotic stress and ion damage are considered to be two main processes harmful to plants (3). The salt-stress-induced proteins can be divided into two categories: functional proteins and regulatory proteins (4). In recent years, proteomic approaches have been widely used to study the molecular mechanisms of plant salt response (5–9). However, little has been done to further understanding of the phosphoproteome dynamics and the comparison between proteome and phosphoproteome changes under salt stress. Experimental strategies combining different levels of salt treatments with a recovery step have seldom been adopted to investigate the proteomes.
Brachypodium distachyon L., a member of the Pooideae subfamily and a temperate wild annual grass endemic to the Mediterranean and the Middle East (10), has evolved rapidly as a model plant system, especially for economically important crops such as wheat, barley, and several potential biofuel grasses such as switchgrass. It possesses many attractive attributes such as a small diploid genome of 272 Mbp, a short growth cycle, self-fertility, and simple growth requirements (11), as well as competence to be efficiently transformed (12). Recently, genome sequencing and annotation of B. distachyon 21 (Bd21)1 was completed (10), making functional proteomic study of B. distachyon feasible. Comprehensive proteomic study analyzing both the proteome and the phosphoproteome of B. distachyon under salt stress and recovery will lead to in-depth knowledge about how cereal plants cope with salt stress.
It is known that protein post-translational modifications (PTMs) are closely related to plant growth, development, and resistance to various biotic and abiotic stresses. Protein phosphorylation is one of the most common and important modifications. A considerable amount of work has revealed that protein phosphorylation is involved in the regulation of diverse processes including metabolism, transcription/translation, protein degradation, homeostasis, cellular communication/signaling, proliferation, differentiation, and cell survival (13). In recent years, various techniques have been developed for the specific enrichment of phosphopeptides, such as immobilized metal affinity chromatography (14, 15) and TiO2 metal oxide affinity chromatography (16, 17). The metal oxide affinity chromatography procedures use a modified buffer containing a saturated solution of organic acids, such as glycolic acid or lactic acid, to decrease the nonspecific binding of acidic peptides (18, 19). TiO2 metal oxide affinity chromatography has been found to be more effective than immobilized metal affinity chromatography in the specific enrichment of phosphopeptides (18, 20).
In the current study, for the first time, we conducted a comprehensive proteome analysis on Bd21 leaves under various levels of salt stress plus a recovery step. Phosphoproteome characterization subject to salt stress was also conducted. A suite of technologies including TiO2 metal oxide affinity chromatography, MALDI-TOF-MS/MS, and LC-MS/MS was used. Large-scale bioinformatic analyses including gene ontology (GO) enrichment, KEGG pathway analysis, phosphorylation motif enrichment, and protein–protein interaction analysis were conducted. Our results provide new insights into the mechanisms of salt response in plants.
EXPERIMENTAL PROCEDURES
Plant Material and NaCl Treatments
Seeds of Bd21 were kindly provided by Dr. John Vogel from the U.S. Department of Agriculture Agricultural Research Service. The Bd21 seeds were surface sterilized by 5% sodium hypochlorite for 5 min and rinsed four times in sterile distilled water. Seeds were submerged in water for 12 h at room temperature and then transferred to wet filter paper to germinate at room temperature (22 °C to 25 °C) for 24 h. The uniformly germinated seeds were selected to grow in plastic pots containing Hoagland solution that was changed every 2 days. At the three-leaf stage, the seedlings were treated with NaCl at five different concentrations, 0 mm, 80 mm, 160 mm, 240 mm, and 320 mm. For each concentration, three biological replicates were conducted. After 48 h of treatment, half of the seedling leaves were harvested for analyses. Some of the sampled leaves were used to measure the relative water content (RWC) and chlorophyll content immediately after sampling, and the remaining leaves were kept frozen at −80 °C for later use. After the stress treatment, the seedlings were transferred to Hoagland solution without NaCl to recover for 48 h. After recovery, leaves were collected and analyzed as above.
Measurement of Leaf RWC and Chlorophyll Content
Leaf RWC was measured immediately after sampling according to the method outlined by Gao et al. (21). RWC was calculated using the following formula: RWC = (FW − DW)/(TW − DW) × 100, where FW represents the weight of freshly collected material, TW represents the weight after rehydration for 24 h at 4 °C in the dark, and DW represents the weight after drying in an oven at 60 °C for 48 h. The final leaf RWC was the mean value taken from four individual seedlings. Chlorophyll contents (chlorophyll a and b) were determined according to the method of Arnon (22). Chlorophyll was extracted from fresh leaves (50 mg) ground in a tissue homogenizer together with 5 ml of ice-cold 80% acetone. The homogenate was centrifuged at 3000 rpm for 2 min. The supernatant was saved and the pellet was re-extracted with 5 ml of 80% acetone. All supernatants were pooled together to a final volume of 12.5 ml. The absorbance of the extract was taken at 663 nm and 645 nm for chlorophyll a and b measurement, respectively, using an Ultrospec 3100 Pro (GE Healthcare). The concentrations of chlorophyll a, chlorophyll b, and total chlorophyll were calculated using Arnon's equations (22). Three biological replicates were used to minimize experimental error.
Protein Preparation
Leaf total protein was extracted according to the method of Wang et al. (23), with minor modifications. Approximately 400 mg of fresh leaves of each sample were ground into fine power in liquid nitrogen. The powder of each sample was resuspended in 4 ml of SDS buffer (30% sucrose, 2% SDS, 100 mm Tris-HCl, pH 8.0, 50 mm EDTA-Na2, 20 mm DTT) and 4 ml of phenol (Tris-buffered, pH 8.0) in a 10-ml tube. 1 mm phenylmethanesulfonyl fluoride and PhosSTOP Phosphatase Inhibitor Mixture (Roche) were added to inhibit the activity of proteases and phosphatases. The mixture was vortexed thoroughly for 30 s and the phenol phase was separated via centrifugation at 14,000 × g for 15 min at 4 °C. The upper phenol phase was pipetted into fresh 10-ml tubes. Four volumes of cold methanol plus 100 mm ammonium acetate were added to the phenol phase and the mixture was stored at −20 °C for at least 30 min. After centrifugation at 14,000 × g for 15 min at 4 °C, the supernatant was discarded carefully and then the precipitated proteins were washed with cold methanolic ammonium acetate twice and ice-cold 80% acetone twice. Finally, the pellet was vacuum-dried and then dissolved in lysis buffer (7 m urea, 2 m thiourea, 4% w/v CHAPS, 65 mm DTT) over 3 h at 4 °C. The protein mixtures were harvested via centrifugation at 14,000 × g for 15 min at 4 °C to remove insoluble material. Then, the concentration of the extracted protein mixtures was determined with a 2-D Quant Kit (Amersham Biosciences) using BSA (2 mg/ml) as the standard. The final protein solution was stored at −80 °C for later use. Three biological replicates were conducted for the protein extraction of each sample.
Two-dimensional Gel Electrophoresis, Image Acquisition, and Data Analysis
Each sample including 1 mg of total protein in 360 μl of rehydration buffer (7 m urea, 2 m thiourea, 2% w/v CHAPS, 0.2% bromphenol blue) containing 65 mm DTT and 0.5% immobilized pH gradient buffer (pH 4–7) (GE Healthcare) was loaded onto an 18-cm pH 4–7 linear immobilized pH gradient strip (GE Healthcare). Isoelectric focusing was performed using a PROTEAN IEF cell (Bio-Rad) according to the manufacturer's instructions. Briefly, active rehydration was carried out at 30 V for 12 h, followed by 300 V for 1 h, 500 V for 1 h, 1000 V for 1 h, 3000 V for 1 h, and then focusing at 8000 V until 80,000 Vh at 20 °C. After isoelectric focusing, the strips were equilibrated with an equilibration solution (50 mm Tris-HCl pH 8.8, 6 m urea, 30% glycerol, 2% SDS, 0.2% bromphenol blue) containing 1% DTT for 15 min, with a second equilibration step of 15 min with the same equilibration buffer containing 2.5% w/v iodoacetamide. The equilibrated strips were loaded on the top of 12% SDS-polyacrylamide gels and sealed with 0.5% w/v agarose. The SDS-PAGE step was performed at 15 °C in an Ettan Dalt Twelve (Amersham Biosciences) electrophoresis system at a constant current setting of 15 mA/gel for 1 h, followed by 20 mA/gel until the bromphenol blue tracking dye arrived at the bottom edge of the gel. The two-dimensional gel electrophoresis (2-DE) experiments were repeated three times for error control. After electrophoresis, all gels were stained with Coomassie Brilliant Blue. Image analysis was performed with ImageMaster 2D Platinum Software Version 5.0 (Amersham Biosciences). The experimental Mr (kDa) of each protein was estimated by means of comparison with the protein marker, and the experimental pI was determined by its migration on the immobilized pH gradient strip. The abundance of each spot was estimated based on the percentage volume. Only those with significant and biological reproducible changes (abundance variation at least 2-fold, Student's t test, p < 0.05) were considered as differentially expressed protein (DEP) spots.
Protein Identification Using MALDI-TOF/TOF-MS
The selected protein spots were excised from preparative gels, destained with 100 mm NH4HCO3 in 30% acetonitrile (ACN). After the destaining buffer had been removed, the gel pieces were lyophilized and rehydrated in 30 μl of 50 mm NH4HCO3 containing 50 ng of trypsin (sequencing grade; Promega, Madison, WI). After overnight digestion at 37 °C, the peptides were extracted three times with 0.1% TFA in 60% ACN. Extracts were pooled together and lyophilized. The resulting lyophilized tryptic peptides were kept at −80 °C until mass spectrometric analysis. A protein-free gel piece was treated as above and used as a control to identify autoproteolysis products derived from trypsin. Identification of the spots was performed via MALDI-TOF/TOF-MS. MS and MS/MS spectra were obtained using an ABI 4800 Proteomics Analyzer MALDI-TOF/TOF (Applied Biosystems, Foster City, CA) operating in a result-dependent acquisition mode. Peptide mass maps were acquired in positive ion reflector mode (20-kV accelerating voltage) with 1000 laser shots per spectrum. Monoisotopic peak masses were automatically determined within the mass range of 800–4000 Da with a signal-to-noise ratio minimum of 10 and a local noise window width of m/z 250. Up to five of the most intense ions with a minimum signal-to-noise ratio of 50 were selected as precursors for MS/MS acquisition, excluding common trypsin autolysis peaks and matrix ion signals. In MS/MS positive ion mode, spectra were averaged, the collision energy was 2 kV, and default calibration was set. Monoisotopic peak masses were automatically determined with a minimum signal-to-noise ratio of 5 and a local noise window width of m/z 250. The MS and MS/MS spectra were searched against the NCBI Brachypodium protein database (26,035 entries in total; downloaded on July 16, 2011) using MASCOT version 2.1 (Matrix Science, London, UK) with the following parameter settings: trypsin cleavage; one missed cleavage allowed; carbamidomethylation as fixed modification; oxidation of methionines and phosphorylation of serine, threonine, and tyrosine allowed as variable modifications; peptide mass tolerance of 100 ppm; and fragment tolerance of ±0.3 Da. All searches were evaluated based on the significant scores obtained from MASCOT. The protein score confidence interval percentage and total ion score confidence interval percentage were both set above 95%, and the significance threshold was p < 0.05 for the MS/MS.
Quantitative RT-PCR Analysis
Total RNA was isolated from frozen samples using TRIZOL Reagent (Invitrogen). Then genomic DNA was removed and cDNA was synthesized using the PrimeScript® RT Reagent Kit with gDNA Eraser (TaKaRa, Shiga, Japan). Gene-specific primers of proteins of interest were designed using online Primer3Plus according to the procedure described by Untergasser et al. (24). The sequence of each protein of interest was first used as a BLASTP search term against the B. distachyon genome. The best-aligned gene was used for primer design. The specificity of the primers was checked by observing the melting curve of the quantitative RT-PCR products and the specific band on the agarose gel. The B. distachyon constitutively expressed S-adenosylmethionine decarboxylase gene was used as a reference for normalization (25). The primers were as follows: sense, 5′-TGCTAATCTGCTCCAATGGC-3′; antisense, 5′-GACGCAGCTGACCACCTAGA-3′. Quantitative RT-PCR was performed in a 20-μl volume containing 10 μl of 2×SYBR® Premix Ex Taq™ (TaKaRa, Shiga, Japan), 2 μl of 50-fold diluted cDNA, 0.15 μl of each gene-specific primer, and 7.7 μl of double distilled H2O. The PCR conditions were as follows: 95 °C for 3 min, 40 cycles of 15 s at 95 °C, and 61 °C for 30 s. Three biological replicates were used for each sample. Reaction was conducted on a CFX96 Real-Time PCR Detection System (Bio-Rad). All data were analyzed using CFX Manager software (Bio-Rad).
Phosphopeptide Enrichment
The extracted protein mixtures were directly reduced with DTT, alkylated with iodoacetamide, and subsequently digested with endoproteinase Lys-C and trypsin as described by Olsen et al. (26). The enrichment for phosphopeptides was performed according to a previously reported procedure (17), with some modifications. In detail, the TiO2 beads (GL Sciences, Tokyo, Japan) were incubated in 400 μl of loading buffer containing 65% ACN/2% TFA saturated by glutamic acid. A total of 2 mg of tryptic peptides were dissolved in 600 μl of loading buffer and incubated with an appropriate amount (tryptic peptide:TiO2 = 1:1, w/w) of TiO2 beads. After being washed with 600 μl of wash buffer (65% ACN/0.1% TFA), phosphopeptides were eluted twice with 300 μl of elution buffer (500 mm NH4OH/60% ACN) and the eluates were dried down and reconstituted in 0.1% formic acid/H2O for MS analysis.
Phosphopeptide Identification and Phosphorylation Site Localization
The enriched phosphopeptides were separated on a self-packed C18 reverse-phase column (75-μm inner diameter, 150-mm length) (Column Technology, Fremont, CA) that directly connected the nano-electrospray ion source to an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific, San Jose, CA). The pump flow was split to achieve a flow rate of 1 μl/min for sample loading and 300 nl/min for MS analysis. The mobile phases consisted of 0.1% formic acid (A) and 0.1% formic acid and 80% ACN (B). A five-step linear gradient of 5% to 30% B in 105 min, 35% to 90% B in 16 min, 90% B in 4 min, 90% to 2% B in 0.5 min, and 2% B for 14.5 min was performed. The spray voltage was set at 2.0 kV, and the temperature of the heated capillary was 240 °C.
For data acquisition, each MS scan was acquired at a resolution of 60,000 (at 400 m/z) with the lock mass option enabled, with acquisition followed by data-dependent top-10 MS/MS scans using collision-induced dissociation. The threshold for precursor ion selection was 500, and the mass window for precursor ion selection was 2.0 Da. The dynamic exclusion duration was 120 s, the repeat count was 1, and the repeat duration was 30 s. The analyzer for the MS scans was Orbitrap, and for the MS/MS scans it was LTQ (37% relative collision energy). Three biological replicates were performed independently from sample collection to the phosphopeptide identification using LC-MS/MS.
The raw files were processed with MaxQuant (version 1.1.1.36) (27) and searched against the NCBI Brachypodium protein database (26,035 entries in total, downloaded on July 16, 2011) concatenated with a decoy of reversed sequences. The following parameters were used for database searches: cysteine carbamidomethylation was selected as a fixed modification; and methionine oxidation, protein N-terminal acetylation, and phosphorylation on serine, threonine, and tyrosine were selected as variable modifications. Up to two missing cleavage points were allowed. The precursor ion mass tolerance was 7 ppm, and the fragment ion mass tolerance was 0.5 Da for MS/MS spectra. The false discovery rate (FDR) was set at <1.0% for both peptide and protein identifications, and the minimum peptide length was set as 6.
Phosphorylation site localization was based on PTM scores that assigned probabilities for each of the possible sites according to their site-determining ions. In this study, MaxQuant (version 1.1.1.36) was used to calculate the PTM scores and PTM localization probabilities. Potential phosphorylation sites were then grouped into three categories depending on their PTM localization probabilities (26, 28), namely, class I (localization probability p ≥ 0.75), class II (0.75 > p ≥ 0.5), and class III (p < 0.5). An FDR of 1% was used for phosphorylation site identification. Spectra without site-determining ions were identified as phosphopeptides with undetermined sites.
Bioinformatic Analysis
Proteins were examined using AgriGO (29) for GO annotation and enrichment analysis. For pathway analysis, the proteins were used to search the KEGG B. distachyon database and mapped to the B. distachyon specific pathways with KEGG Mapper. The significantly enriched phosphorylation motif set was extracted from phosphopeptides with confidently identified phosphorylation sites (class I) using the motif-X algorithm (30). The phosphopeptides were centered at the phosphorylated amino acid residues and aligned, and six positions upstream and downstream of the phosphorylation site were included. For C- and N-terminal peptides, the sequence was completed to 13 amino acids with the required number of X, where X represents any amino acid. Because the upload restriction of motif-X is 10 MB, a FASTA format dataset (nearly 10 MB) containing the protein sequences from the B. distachyon protein database in Phytozome (version 9.0) was used as the background database to normalize the score against a random distribution of amino acids. The occurrence threshold was set at 5% of the input dataset at a minimum of 10 peptides, and the probability threshold was set at p < 10−6. Amino acid sequences around the phosphorylated amino acid based on the alignment of the phosphorylation sites were completed by using the WebLogo program (31). The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database of physical and functional interactions (32) was used to analyze the protein–protein interactions (PPIs), and the PPI network was displayed by Cytoscape software (version 3.0.0) (33). The Phyre2 online server (34) was used to predict the three-dimensional structure of the interested proteins. Then the three-dimensional structures and the phosphorylated site were displayed by SPDBV software (version 4.1) (35).
RESULTS
Physiological Changes of Bd21 Leaf Under Salt Stress
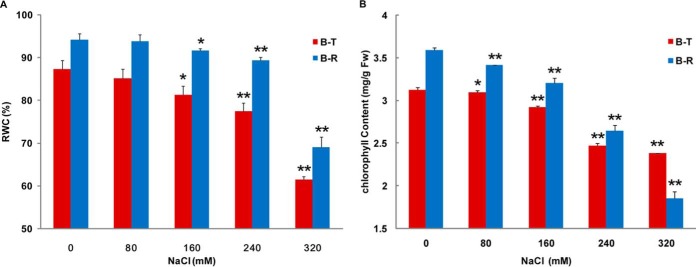
The RWC and chlorophyll content of control, salt-treated, and 48-h recovery Bd21 leaf samples are shown in Fig. 1. For the control sample, both the RWC and the chlorophyll content increased after 48 h of growth. This is normal because of the elongation and stretch of leaves. In the salt-treatment and recovery groups, both physiological indicators decreased to various degrees under all NaCl concentrations relative to the control. As shown in Fig. 1A, the RWC decreased only slightly, by 1.37% to 6.87%, under 80 to 240 mm NaCl treatments relative to the control and were able to recover to the normal level after 48 h of recovery. However, under treatment with 320 mm NaCl, the leaf RWC decreased dramatically by 28.79% and was able to recover to only 72.61% of the control level. For the leaf chlorophyll content, different patterns were shown under different salt concentrations. Increasing NaCl concentration resulted in greater decreases in chlorophyll content (Fig. 1B). Under 80 mm NaCl stress, the chlorophyll content decreased only slightly (4.94%) and was able to quickly recover to the normal level. Under 160 mm NaCl stress, it decreased by 6.47% relative to the control, whereas under 240 and 320 mm NaCl concentrations it decreased by 20.85% and 23.83%, respectively. In the recovery group, the chlorophyll content had varying degrees of recovery relative to the salt-treatment group, except the 320 mm NaCl concentration, which implied irreversible damage to the chloroplast and seriously blocked photosynthesis. The RWC and chlorophyll content both showed a dividing point at 200 mm, indicating that an NaCl concentration of less than 200 mm is a moderate salt concentration, whereas one over 200 mm is a severe salt concentration.
Fig. 1.
Measurement of physiological indices. A, leaf relative water content (RWC) analysis. B, chlorophyll content analysis. “B-T” represents leaves of Bd21 treated with NaCl for 48 h, and “B-R” represents leaves of Bd21 recovered 48 h after treatment with NaCl. Error bars indicate standard error of three biological replicates. Statistically significant differences relative to the control were calculated by independent Student's t tests. *p < 0.05. **p < 0.001.
2-DE Mapping and Identification of DEP Spots
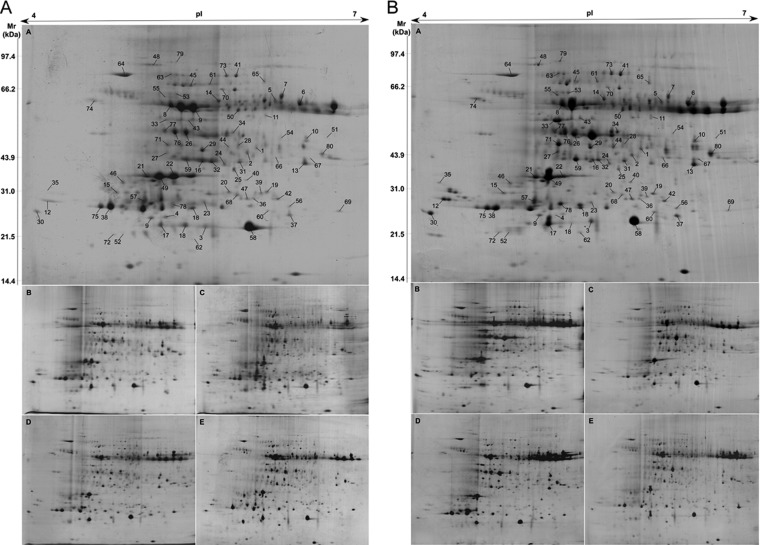
The Bd21 leaf proteome analyses of the salt-treated and recovery groups were conducted via 2-DE with Coomassie Brilliant Blue staining. About 800 protein spots could be reproducibly detected and matched among all gels, of which 108 showed significant changes among the 2-DE maps of NaCl treatment and recovery. Finally, out of the 108 DEP spots, 80 were successfully identified via MALDI-TOF/TOF-MS (Fig. 2 and supplemental Table S1). The mass spectrometry data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (36) with the dataset identifier PXD000470. The identified DEP spots represented 60 unique DEPs. In particular, 16 unique DEPs (Table I) were each identified from two or three DEP spots with different isoelectric points and/or molecular weights. Among them, 13 DEPs were each distributed in two or three DEP spots with different isoelectric points but the same or very similar molecular weights, respectively.
Fig. 2.
A, 2-DE maps of Bd21 leaf proteome in the salt treatment group. Protein spots were visualized using Coomassie Brilliant Blue staining; a–e represent samples treated with 0, 80, 160, 240, and 320 mm NaCl, respectively, for 48 h. B, 2-DE maps of Bd21 leaf proteome in the recovery group. Protein spots were visualized using Coomassie Brilliant Blue staining; a–e represent samples recovered after treatment with 0, 80, 160, 240, and 320 mm NaCl, respectively, for 48 h. The 80 DEP spots are labeled in A-a and B-a.
Table I. The 15 DEPs that were identified from two or three DEP spots on 2-DE maps.
| Protein gi No. | Description | DEP spot IDs | Experimental Mr | Experimental pI | Expression patterns in T | Expression patterns in R |
|---|---|---|---|---|---|---|
| gi 357111658 | Oxygen-evolving enhancer protein 2 | 3/58 | 23.0/23.0 | 5.45/5.83 | c/c | a/b |
| gi 193075554 | ATPase alpha subunit, partial | 5/6/7 | 65.1/63.8/67.8 | 6.06/6.27/6.11 | a/a/b | f/e/c |
| gi 357146802 | Alpha-galactosidase | 10/54 | 46.9/47.1 | 6.32/6.12 | b/b | g/e |
| gi 357149112 | Ferredoxin–NADP reductase | 16/59 | 39.9/40.4 | 5.48/5.34 | c/c | e/g |
| gi 357134997 | Uncharacterized protein At2g37660 | 19/20 | 30.7/30.6 | 5.99/5.66 | f/c | g/b |
| gi 357111487 | Oxygen-evolving enhancer protein 1, isoform 1 | 21/22/49 | 35.0/34.9/34.9 | 4.93/5.19/5.09 | c/a/c | e/a/e |
| gi 357133147 | Phosphoglycerate kinase, isoform 1 | 26/76/77 | 50.3/50.2/51.1 | 5.29/5.25/5.14 | a/e/e | e/b/d |
| gi 357137138 | Phosphoribulokinase | 27/71 | 42.5/45.5 | 5.31/5.18 | f/e | c/b |
| gi 357147655 | Quinone oxidoreductase-like protein At1g23740 | 31/32 | 39.3/39.7 | 5.75/5.52 | b/d | e/c |
| gi 357155664 | Ribulose bisphosphate carboxylase/oxygenase activase A | 33/34/80 | 55.9/50.3/42.4 | 5.13/5.70/6.47 | c/c/f | d/c/g |
| gi 357112766 | l-ascorbate peroxidase 1 | 36/42 | 28.5/26.5 | 5.98/6.07 | c/f | g/g |
| gi 357122389 | Chlorophyll a-b binding protein of LHCII type III | 38/75 | 27.2/28.0 | 4.64/4.56 | c/d | g/g |
| gi 357110873 | Transketolase | 41/73 | 80.4/80.5 | 5.72/5.64 | d/d | e/c |
| gi 357164996 | Elongation factor G | 48/79 | 88.2/90.2 | 5.00/5.16 | c/c | b/c |
| gi 357163385 | 2-Cys peroxiredoxin BAS1 | 52/72 | 21.6/20.2 | 4.77/4.76 | h/c | g/g |
| gi 357111666 | Uncharacterized oxidoreductase At1g06690 | 66/67 | 41.5/41.9 | 6.06/6.37 | f/d | e/g |
T, treatment group; R, recovery group.
Expression Pattern Analysis of the DEP Spots
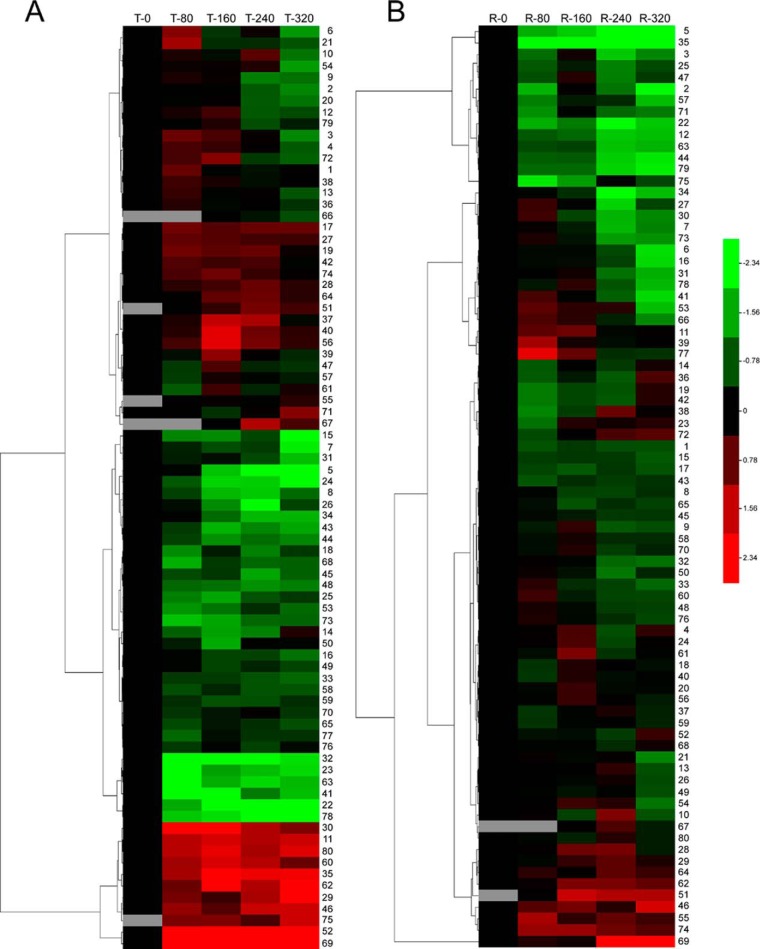
To visualize the coordinately regulated DEP spots, especially for the 16 DEPs identified in two or three spots, hierarchical clustering was used to analyze the proteome dataset based on methods described by Eisen et al. (37). The relative ratios of DEP spots from the proteome dataset listed in supplemental Table S1 were conducted log2 transforming, and then the Euclidean distance similarity metric was used to define the similarity and the hierarchical clusters were assembled using the complete-linkage clustering method. Two hierarchical clusters corresponding to the salt treatment group (Fig. 3A) and the recovery group (Fig. 3B) were constructed. The expression of the DEP spots in the treatment and recovery groups was divided into eight patterns (Fig. 3). Finally, 9 of the 16 DEPs identified from two or three DEP spots showed the same patterns in the treatment group, whereas in the recovery group only 4 of the 16 DEPs displayed the same patterns (Table I).
Fig. 3.
Protein expression clustering analysis of DEP spots. A, hierarchical clustering of DEP spots in the treatment group. B, hierarchical clustering of DEP spots in the recovery group. Each column represents an NaCl concentration. Each row displays the change of a DEP spot using color coding based on the relative ratio listed in supplemental Table S1.
Transcriptional Expression Analysis of the Genes Encoding DEPs
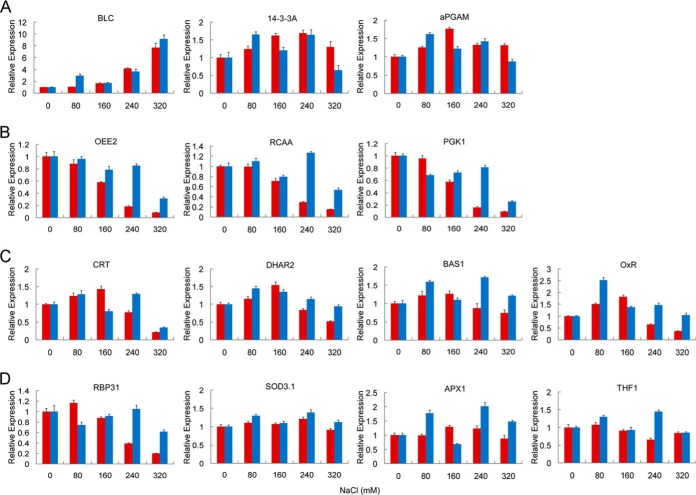
To explore the changes of DEPs at the transcriptional level, 14 representative DEPs (supplemental Table S2) were chosen for transcriptional-level analysis via quantitative RT-PCR. Among the 14 DEPs, 6 were identified in two or three DEP spots on the 2-DE profiles. Finally, the transcriptional expression patterns of these genes in the salt treatment group were divided into four groups as shown in Fig. 4. The first group was up-regulated under salt stress, including genes encoding BLC, 14-3-3A, and aPGAM (Fig. 4A). The second group was down-regulated, with three genes encoding OEE2, RCAA, and PGK1 (Fig. 4B). The gene transcriptional expression in the third group was up-regulated under moderate salt stress but down-regulated under harsh salt stress (Fig. 4C). Four genes belonged to this group, encoding CRT, DHAR2, BAS1, and OxR. The last group displayed an irregular expression trend (Fig. 4D). Compared with the expression at the transcriptional and translational levels of the 14 coding genes, the transcriptional expression trends of 5 genes for BLC, 14-3-3A, RCAA, PGK1, and BAS1 were consistent with their translational expression. The other seven genes displayed similar trends at both transcriptional and translational levels to some extent, but for the two genes for aPGAM and OxR, the consistency of their transcriptional and translational levels was rather poor.
Fig. 4.
Quantitative RT-PCR analysis of the 14 representative DEPs identified from 2-DE. The x-axis is the NaCl concentration. The salt-treatment group is represented by red columns, and the recovery group by blue columns. The y-axis is the relative expression of each gene encoding DEPs after normalization to the reference gene SamDC. A, three genes with transcriptional level up-regulated under salt stress at all NaCl concentrations. B, three genes with transcriptional level down-regulated under salt stress at all NaCl concentrations. C, four genes with transcriptional level up-regulated under moderate salt stress but down-regulated under harsh salt stress. D, four genes with irregular transcriptional trends.
Phosphoprotein Identification and Phosphorylated Site Location
In total, 2389 phosphopeptides with 2839 phosphorylated sites corresponding to 1509 phosphoproteins were identified (supplemental Table S3). The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (36) with the dataset identifier PXD000340. Phosphorylated sites with site location probability belonging to class I, class II, and class III accounted for 73.2%, 18.5%, and 8.3%, respectively. To improve the reliability of the further differential phosphorylation level analysis, only phosphorylated sites belonging to class I were screened. The proportions of pS, pT, and pY sites belonging to class I were estimated as 91.62%, 8.33%, and 0.05%, respectively. Among the 2077 class I phosphorylated sites, 1323 were identified from the control and 1803 from the salt-treated sample. Before the comparison analysis of the phosphorylation levels between the salt stress condition and the control, a range of quality control measures were taken for the three biological replicates of each condition to determine analytical reproducibility (supplemental Fig. S1), and only the phosphopeptides identified from all biological repeats were used for further analysis.
Screening Phosphoproteins with Phosphorylation Level Significantly Changed
The intensity of each phosphopeptide was normalized to the mean of intensities of all phosphopeptides within each biological replicate. Subsequently, the log2 intensity value changes (salt stress/control) in each condition were calculated for each phosphopeptide. Only those phosphopeptides that met the following restrictions were regarded as having undergone a significant change at the phosphorylation level: (1) phosphopeptide detected in all three biological replicates; (2) Student's t test p value of FDR correction < 0.05; (3) |log2 (salt stress/control)| ≥ 1; and (4) phosphorylation site localization probability greater than 0.75 (class I) and phosphorylation site score difference not less than 5. Finally, 496 phosphorylated sites corresponding to 468 phosphoproteins (supplemental Table S4) with the phosphorylation level significantly changed (PLSC) were screened out. Of the 496 phosphorylated sites, 294 were detected only in the salt-treated sample and 81 phosphorylated sites were detected only in the control. Relative to the control, 45 phosphorylated sites were up-regulated and 76 phosphorylated sites were down-regulated at the phosphorylation level in the salt-treated sample (supplemental Table S4).
Identification of Phosphorylated DEPs
Among the 60 DEPs, 14 DEPs were also identified as phosphoproteins (Table II), and interestingly, 6 of them were each identified in two or three DEP spots. For example, oxygen-evolving enhancer protein 2 (GI No. 357111658) was identified from two spots (DEP spots 3 and 58) that showed the same molecular weight (23.0 KDa) but different isoelectric points (5.45 and 5.83) (Table I). It was also a phosphoprotein with two phosphorylated sites (phosphorylated sites 109 and 2896 in supplemental Table S3). One phosphorylated site (phosphorylated site 109) was only identified under salt stress, indicating that this phosphoprotein was also a PLSC phosphoprotein (supplemental Table S4A).
Table II. DEPs that were also identified as phosphoproteins with reliable phosphorylated sites.
| DEP spot ID | Phosphoprotein ID | Protein gi number | Description | Phosphorylated peptide and site |
|---|---|---|---|---|
| 1 | 71 | gi 357111159 | Uncharacterized protein At3g63140 | AMpSFDLDDK |
| 3, 58 | 89 | gi 357111658 | Oxygen-evolving enhancer protein 2 | pSTEFVAYSGDGFK |
| pTDSEGGFESDAVATANVLESAAPVVGGK | ||||
| 5, 6, 7 | 18 | gi 193075554 | ATPase alpha subunit, partial | GEIIApSESR |
| 8 | 19 | gi 193075564 | ATPase beta subunit | pSAPAFIELDTK |
| 18 | 849 | gi 357134081 | Triosephosphate isomerase | IIYGGSVTGApSCR |
| 21, 22, 49 | 85 | gi 357111487 | Oxygen-evolving enhancer protein 1, isoform 1 | QLVATGKPEpSFSGPFLVPSYR |
| 26, 76, 77 | 810 | gi 357133147 | Phosphoglycerate kinase, isoform 1 | RPFAAIVGGpSK |
| 30 | 1006 | gi 357138100 | ATP synthase delta chain | DSpSNLIDMSVR |
| 33, 34, 80 | 1418 | gi 357155664 | Ribulose bisphosphate carboxylase/oxygenase activase A |
GLAYDIpSDDQQDITR pSFQCELVFAK |
| 41, 73 | 51 | gi 357110873 | Transketolase | VVPGLLGGSADLApSSNM(ox)TLLK |
| 45 | 486 | gi 357123383 | ATP-dependent zinc metalloprotease FTSH 2 | QVpSVDVPDVR |
| 55 | 1217 | gi 357146493 | Chaperonin CPN60–2 | GIpSM(ox)AVDDVVTNLK |
| 70 | 601 | gi 357125604 | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase | AHGTAVGLPSDDDMGNpSEVGHNALGAGR |
| 74 | 467 | gi 357122930 | Calreticulin | DDpSDDEKPQHANK |
GO Annotation and Enrichment Analysis of DEPs and PLSC Phosphoproteins
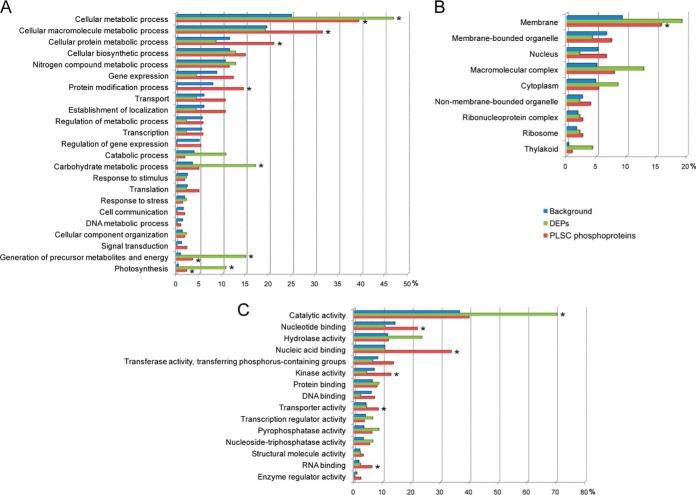
All the identified DEPs and PLSC phosphoproteins were used for GO annotation and enrichment analysis with AgriGO, and the GO annotation of total B. distachyon genome proteins was set as the background dataset. The distribution bar charts for biological process, cellular component, and molecular function are shown in Fig. 5. From the biological process perspective, for DEPs, the generation of precursor metabolites and energy (GO: 0006091, FDR: 1.1E-5), photosynthesis (GO: 0015979, FDR: 7.5E-5), carbohydrate metabolic process (GO: 0005975, FDR: 6.4E-3), and cellular metabolic process (GO: 0044237, FDR: 6.4E-2) were significantly overrepresented (Fig. 5A). For PLSC phosphoproteins, generation of precursor metabolites and energy (GO: 0006091, FDR: 1.9E-2), photosynthesis (GO: 0015979, FDR: 4.8E-2), and cellular protein metabolic process (GO: 0044267, FDR: 1.2E-3) were also significantly enriched. Cellular metabolic process (GO: 0044237, FDR: 8.5E-5), cellular macromolecule metabolic process (GO: 0044260, FDR: 6.8E-4), and protein modification process (GO: 0006464, FDR: 1.2E-2) were only found to be significantly overrepresented in the PLSC phosphoprotein dataset (Fig. 5A). From the cellular component perspective, only membrane (GO: 0016020, FDR: 4.3E-2) was significantly enriched from the PLSC phosphoprotein dataset, and for the DEP dataset, no significantly enriched cellular component was found (Fig. 5B). From the molecular function perspective, for DEPs, only catalytic activity (GO: 0003824, FDR: 1.7E-4) was significantly overrepresented (Fig. 5C), and for PLSC phosphoproteins, nucleic acid binding (GO: 0003676, FDR: 6.8E-19) and RNA binding (GO: 0003723, FDR: 6.1E-4) were highly significantly enriched relative to background. Nucleotide binding (GO: 0000166, FDR: 2.5E-2), kinase activity (GO: 0016301, FDR: 2.8E-2), and transporter activity (GO: 0005215, FDR: 4.2E-2) were also significantly overrepresented (Fig. 5C).
Fig. 5.
GO analysis of the DEPs and PLSC phosphoproteins. A, biological process. B, cellular component. C, molecular function. Significantly overrepresented terms (FDR adjusted p value < 0.05) are marked with an asterisk.
Conservation Analysis of the PLSC Phosphoproteins
Sequences of the PLSC phosphoproteins were used as queries to blast phosphoprotein databases constructed based on datasets from P3DB (38), MORE (39), and PhosPhAt 4.0 (40). Oryza sativa, Arabidopsis thaliana, and Medicago truncatula were compared with B. distachyon to determine the degree of conservation of phosphoproteins between species. The thresholds were set as score ≥ 80, E-value < 1E-10, and identity ≥ 30%. In all, 294 (62.8%) of the 468 phosphoproteins had phosphorylated orthologs in all three species, 122 (26.1%) had phosphorylated orthologs in two of the three species, and 44 (9.4%) had phosphorylated orthologs in only one species (supplemental Table S5). Only eight phosphoproteins had no phosphorylated orthologs in any of the three species. Further searches also revealed that these eight phosphoproteins had no phosphorylated orthologs in other organisms (supplemental Table S5). Further GO enrichment analysis of the 294 highly conserved phosphoproteins showed that protein modification process (GO: 0006464, FDR: 4.6E-3), signal transduction (GO: 0003723, FDR: 6.1E-5), and kinase activity (GO: 0016301, FDR: 1.4E-3) were significantly overrepresented.
KEGG Pathway Analysis of the DEPs and PLSC Phosphoproteins
To reveal the DEP and PLSC phosphoprotein pathways, KEGG pathway analysis was conducted and the relevant proteins were used to map the pathways specific for B. distachyon. A total of 25 maps (supplemental Fig. S2) of B. distachyon KEGG pathways with no less than three hits were highlighted by all of these proteins. For DEPs, carbon metabolism (bdi01200, supplemental Fig. S2B), carbon fixation in photosynthetic organisms (bdi00710, supplemental Fig. S2C), biosynthesis of amino acids (bdi01230, supplemental Fig. S2D), glycolysis/gluconeogenesis (bdi00010, supplemental Fig. S2G), and photosynthesis (bdi00195, supplemental Fig. S2J) were the main pathways. For PLSC phosphoproteins, oxidative phosphorylation (bdi00190, supplemental Fig. S2E), RNA degradation (bdi03018, supplemental Fig. S2F), and RNA transport (bdi03013, supplemental Fig. S2K) were the highlighted pathways. In addition, some pathways that occurred in spliceosomes (bdi03040, supplemental Fig. S2A) and ribosomes (bdi03010, supplemental Fig. S2H) were also highlighted in relation to PLSC phosphoproteins. It is worth noting that the plant hormone signal transduction (bdi03040, supplemental Fig. S2U) pathway was highlighted, indicating that salt stress, an adverse environmental factor, can stimulate various hormone signals such as abscisic acid (ABA) and ethylene-related response and defense pathways in B. distachyon.
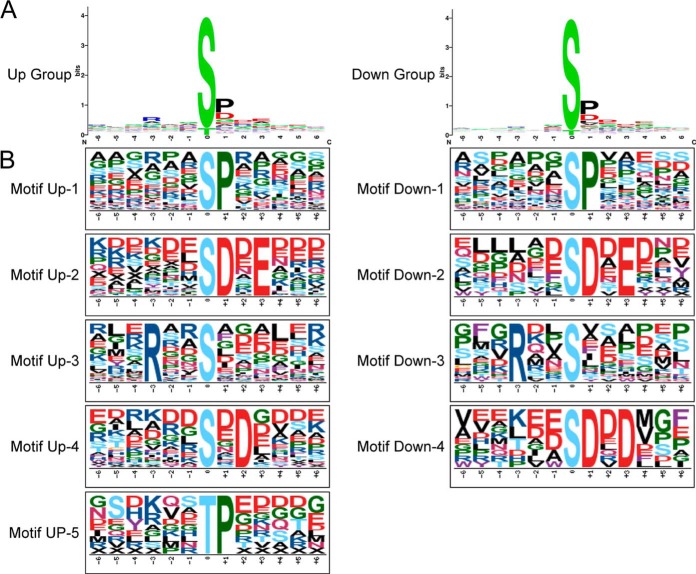
Phosphorylation Motif Analysis for the PLSC Phosphopeptides
WebLogo and motif-X were used to identify the kinase-associated phosphorylation motifs of the PLSC phosphopeptides. The PLSC phosphopeptides were classified into two groups. One of them was the Up group, including the phosphopeptides with phosphorylation specially identified in the salt stress condition and up-regulated phosphorylation levels under the stress condition relative to the control. The other one was the Down group, which represented the phosphopeptides with phosphorylation specially identified in the control and down-regulated phosphorylation levels after salt treatment. Then the phosphopeptides centered at the phosphorylated amino acid residues of each group were used for WebLogo analysis and phosphorylation motif extraction. Results showed Ser was the main phosphorylated amino acid residue in both groups, and the amino acid closely neighboring the Ser/Thr phosphorylated site was mainly proline or aspartate (Fig. 6A). motif-X analysis showed that five phosphorylation motifs were enriched from the Up group and four motifs were enriched from the Down group (Fig. 6B and supplemental Table S6). Three phosphorylation motifs ([sP], [sDxE], and [Rxxs]) were enriched from both Up and Down groups. Two motifs ([tP] and [sxD]) were only found in the Up group, and one motif ([sDxD]) was only found in the Down group. Those phosphorylation motifs were then searched in relevant databases (41–45) to find the specific protein kinases. [sP] and [tP] motifs were the proline-directed motifs, which were potential substrates of mitogen-activated protein kinase (MAPK), cyclin-dependent kinase, and cyclin-dependent kinase–like. [sDxE], [sxD], and [sDxD] motifs were all acidic motifs and were recognized by casein kinase-II (CK-II). The [Rxxs] motif was a basic motif and was recognized by CaMK-II, which was also similar to the 14-3-3 binding motif [Rxxs/txP] (46).
Fig. 6.
Sequence alignment of the amino acids surrounding the identified phosphorylated sites and significantly enriched phosphorylation motifs. A, logos were constructed based on the alignment of all phosphopeptides from the Up group and the Down group using WebLogo. B, phosphorylation motifs extracted from the phosphopeptides in the Up group and the Down group by motif-X.
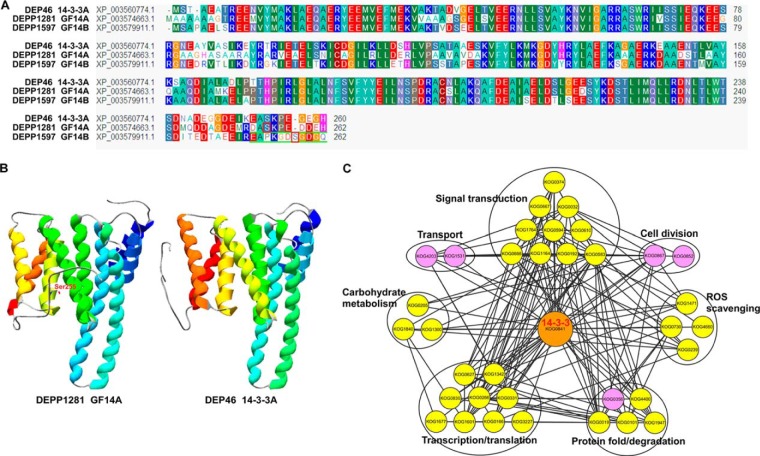
PPI Analysis of DEPs and PLSC Phosphoproteins
The PPI network of the DEPs and PLSC phosphoproteins identified in the current study were analyzed by STRING. A total of 288 eukaryotic orthologous groups (KOGs) representing 48 DEPs and 343 PLSC phosphoproteins (supplemental Tables S7A and S7B) were used to construct the PPI network. To improve the reliability of the PPI analysis, the confidence score was set at the highest level (≥0.900). Finally, a complex PPI network that contained 247 nodes and 1322 edges was displayed through Cytoscape (supplemental Fig. S3). Three 14-3-3 (KOG0841) proteins, 14-3-3-like protein A (14-3-3A) (DEP spot 46), 14-3-3-like protein GF14-A (GF14-A) (PLSC phosphoprotein 1281), and 14-3-3-like protein GF14-B (GF14-B) (PLSC phosphoprotein 1597), were identified. Sequence alignment analysis (Fig. 7A) showed that compared with 14-3-3A, GF14-B possessed an additional phosphorylated Ser. GF14-A contained a Ser phosphorylation site at the C terminus that belonged to the [RxxsxP] motif. 14-3-3A also contains the Ser residue at the same position, but the amino acid residual in position -3 of the Ser residual is lysine (K) instead of arginine (R), so there is no [RxxsxP] motif in 14-3-3A. Comparison of the three-dimensional structures of 14-3-3A and GF14-A showed that their tertiary structures were highly similar (Fig. 7B). Based on these, KOG0841, which represents the three 14-3-3 proteins, was used to extract the potential interacting proteins from the whole PPI network, and a 14-3-3-centered subnetwork was constructed (Fig. 7C), which showed that 14-3-3 proteins interact with 52 proteins (supplemental Table S7C) involved in signal transduction, transcription/translation, protein folding/degradation, cell division, transport, reactive oxygen species (ROS) scavenging, and carbohydrate metabolism.
Fig. 7.
The three 14-3-3 proteins and associated proteins analysis. A, protein sequences comparing 14-3-3A, GF14-A, and GF14-B. The sequences with green underlining represent the phosphopeptides of GF14-A and GF14-B, and the phosphorylated Ser site is marked with a red box. B, three-dimensional structures of 14-3-3A and GF14-A. The phosphorylated Ser site is labeled in the three-dimensional structure of GF14-A. C, protein–protein interaction network of 14-3-3 proteins and related proteins. Nodes with pink background color represent the KOGs of DEPs, nodes with yellow background color represent the KOGs of PLSC phosphoproteins, and nodes with orange background color represent the KOGs of DEPs that were also identified as phosphoproteins with reliable phosphorylated sites (class I).
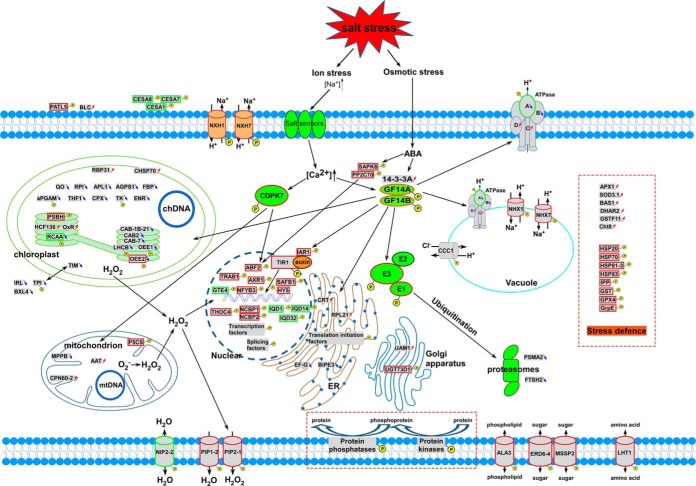
An Overview of Response and Defense Mechanisms of B. distachyon Under Salt Stress
Based on the above results, a systematic salt response and defense pathway in B. distachyon was proposed (Fig. 8). The ion stress signal was transferred by salt sensors on plasma membrane into cells and led to increased Ca2+. Then, signal transduction pathways involved in salt stress response and defense in B. distachyon such as the 14-3-3 signal pathway and the ABA signal pathway were activated through phosphorylation modification mediated by the increased Ca2+. The 14-3-3 signal pathway could also be triggered by ABA, which was generated in response to stimulation by osmotic stress. The signals were then transferred to the nucleus, resulting in changes of the phosphorylation levels of various transcription and translation regulation factors that regulated the expression of response- and defense-related genes. Under salt stress, photosynthesis and carbohydrate metabolism were depressed and superoxide radicals were produced. This caused the proteins associated with ROS scavenging to be up-regulated at the expression level. Some superoxide radicals were transferred into H2O2. As a molecular signal, H2O2 regulated the expressions of some defense-related genes, although excessive H2O2 can lead to cell damage. H2O2 also could diffuse to other cells with PIP2–1 on the membrane. Channels and transporters on the membrane that were associated with water, ion, and nutrient transport were also regulated through phosphorylation or dephosphorylation. Excessive radicals and high Na+ would lead to the misfolding of proteins. Plant cells need various protein-folding-associated proteins to refold the misfolded proteins through changes in the expression level or phosphorylation level. Another strategy for coping with the misfolded proteins was degradation; a ubiquitination degradation pathway that might be centered by E3s was shown in the schematic representation.
Fig. 8.
Schematic representation of a possible comprehensive salt response and defense model in B. distachyon. The DEPs and PLSC phosphoproteins identified in our study were used to construct the schematic representation. Proteins with red arrows represent DEPs up-regulated under salt stress relative to the control, and those with blue arrows represent down-regulated DEPs. PLSC phosphoproteins are represented by a yellow ball. PLSC phosphoproteins with red outlines represent phosphopeptides only detected in salt stress samples or with significantly up-regulated phosphorylation levels under salt stress relative to the control, whereas those with green outlines represent those phosphopeptides only detected in controls or with significantly down-regulated phosphorylation levels under salt stress.
DISCUSSION
In the current study, through a combination of proteomic and phosphoproteomic approaches, we performed a comprehensive analysis of salt stress response and defense in B. distachyon leaves for the first time. Plants respond to salt stress through the transcription and translation of response-related genes, which is a complex mechanism that involves various cross-talk pathways. In addition, post-translational phosphorylation modification can regulate protein functions to respond to abiotic stress (47). Comparison between the DEPs and PLSC phosphoproteins revealed that the DEPs mainly acted as functional proteins, whereas most of the PLSC phosphoproteins were found to be regulatory proteins, suggesting that plants can respond to and defend against salt stress in two ways: (1) through phosphorylation modification changes, mainly involving signal transduction, transcription/translation, and transport; and (2) through protein expression changes, which mainly occur in photosynthesis and energy production. The proteins related to ROS scavenging and protein folding/degradation can respond to and defend against salt stress in both ways.
Perception of Salinity and Signal Transduction
Salinity is communicated by sodium ionic and osmotic stress signals in plants, which can trigger the corresponding signal transduction pathways. Protein kinases and protein phosphatases play opposite functions in signal transduction. Many protein kinases and protein phosphatases were found to be PLSC phosphoproteins in our study. Salinity stress can lead to a transient increase in cytosolic Ca2+, derived from either influx from the apoplastic space or release from internal stores (48). Two calcium-dependent protein kinase 7 phosphoproteins (241 and 435) were identified as PLSC phosphoproteins in our study. Calcium-dependent protein kinase 7 can be activated by Ca2+ to lead to autophosphorylation that plays an important role in the regulation of kinase activity (49). In tobacco, Ser40 and Thr65 of NtCDPK2 are phosphorylated after hyperosmotic stress. Ser40 is a target for a regulatory upstream protein kinase, and correct NtCDPK2 membrane localization is required for Ser40 phosphorylation (50). In our study, PLSC phosphoproteins 241 and 435 were phosphorylated at Ser532 and Ser463 of the C terminus, respectively, only under salt stress, indicating a relationship with osmotic stress response. Protein phosphatase 2C 70 (PLSC phosphoprotein 557), which is encoded by the ABI1 gene, can lead to the dephosphorylation of phosphoproteins; is involved in ABA signal transduction; and interacts with RLK5, SERK1, and CDC48A (51–53). The current study revealed that protein phosphatase 2C 70 was phosphorylated only under salt stress, which might indicate that protein phosphatase 2C 70 functions in the phosphorylation status in response to salt stress. Polyphosphoinositides, as important signaling molecules, are involved in membrane and vesicular trafficking, regulation of the cytoskeleton, and stress response in plants (54). Polyphosphoinositide phosphatases regulate the synthesis and turnover of polyphosphoinositides (54). In our research, two polyphosphoinositide phosphatases, PLSC phosphoproteins 195 and 1016, were identified, with the phosphorylation of the latter occurring only after salt stress.
In parallel, osmotic stress signals induced by salinity activate the synthesis of ABA, of which the signal pathway acts as a vital approach for salt stress defense (55). Roberts et al. (56) reported that 14-3-3 proteins regulate the activities of many proteins involved in signal transduction and play important roles in stress responses in higher plants. In the ABA-mediated osmotic stress response, some 14-3-3 proteins are constituents of transcription factor complexes for ABA-induced gene expression (57). In the current study, differential expression and phosphorylation modification of 14-3-3 proteins were detected in both leaf proteome and phosphoproteome. In particular, 14-3-3A (DEP spot 46) showed dramatically up-regulated expression at both transcriptional (up-regulated 1.68-fold) and translational levels (up-regulated 3.07-fold) under all salt treatments and maintained a higher expression level even after the stimulus was removed. Previous reports indicated that a 14-3-3 protein was also up-regulated in Physcomitrella patens under cold stress (58), and the transcription of another 14-3-3 protein in rice showed an increasing pattern under salt stress (59), which is consistent with our results. In addition, some 14-3-3 proteins could be phosphorylated by several protein kinases in mammals and plants (60). Phosphorylation of 14-3-3 proteins as well as target proteins is important for PPIs (61). Previous studies have shown that 14-3-3 proteins can interact with many proteins, such as calcium-dependent protein kinase and H+-ATPase (56). The phosphorylation of some 14-3-3 proteins may inhibit their interaction with target proteins (61), but other 14-3-3 proteins interact with target proteins in a phosphorylation-dependent manner (62). In our research, GF14-A and GF14-B (PLSC phosphoproteins 1281 and 1597) were both phosphorylated only under salt stress. GF14-A contained a phosphopeptide that belonged to the [Rxxs] motif and might be regulated by CaMK-II. In addition, the Arg (R) residue in GF14-A was replaced by a Lys (K) residue in 14-3-3A at the same position ahead of the phosphorylated Ser residue, which might have led to the difference in function. Different from the [Rxxs] motif of GF14-A, the phosphopeptide of GF14-B belonged to the [sxD] motif, which might be regulated by CK-II. The PPI network in our study showed that 14-3-3 proteins can interact with many proteins in various functional groups, most of which have been demonstrated previously (63).
Auxin plays a pivotal role in many aspects of plant growth and development (64). Recent studies have shown that auxin is involved in many biotic and abiotic stresses (65, 66). Transport inhibitor response 1 (PLSC phosphoprotein 1002), as an auxin receptor, can be induced by auxin and mediate Aux/IAA degradation and auxin-regulated transcription (64, 67). IAA-alanine resistance protein 1 (PLSC phosphoprotein 1278) is required for auxin conjugate sensitivity and is involved in auxin homeostasis (68). In the current study, both transport inhibitor response 1 and IAA-alanine resistance protein 1 were found to be phosphorylated only in salt-treated samples, which suggested that they could participate in the auxin signal pathway and function in phosphorylation status under salt stress. Auxin homeostasis under stress involves interactions with salicylic acid and ABA signals (69). In our study, phosphorylation of ABA-insensitive 5-like protein 5 (PLSC phosphoprotein 565) was identified only under the salt stress condition. ABA-insensitive 5-like protein 5 can interact with the ABA-responsive elements and be involved in ABA or stress responses, and its overexpression affects multiple stress tolerance (70). Serine/threonine protein kinase SAPK8 (PLSC phosphoprotein 231) was also phosphorylated under salt stress in our study. SAPK8 is involved in the ABA signal pathway, which can be activated by hyperosmotic stress and ABA (71) and directly phosphorylate TRAB1 (an ABA-insensitive 5-like protein) in rice (72). Interestingly, bZIP transcription factor TRAB1 (PLSC phosphoprotein 1280) was identified with a significantly higher phosphorylation level under salt stress, and might be phosphorylated by SAPK8.
Water, Ion, and Nutrient Transport
The physiological maintenance of living cells relies on the homeostasis of intracellular ion concentrations. Under salt stress it is necessary for cells to keep the concentration of toxic ions low and to accumulate essential ions. In our study, both ATP synthase subunit γ chain (DEP 28) and ATP synthase δ chain (DEP 30), involved in ion transport, were up-regulated under salt stress. This was consistent with the result obtained from a study on the salt-tolerant wheat variety Shanrong No. 3, in which vacuolar proton ATPase subunit E displayed specifically up-regulated expression under salt stress (73). In our phosphoproteome analysis, many ion transporters such as potassium ion, calcium ion, chloride ion, zinc ion, copper ion transporter, and sodium/hydrogen exchanger were found. However, most of these ion transporters showed no significant changes at the phosphorylation level between salt stress and control conditions, indicating their fundamental roles in the maintenance of cell homeostasis. Only five of them were identified as PLSC phosphoproteins. Two sodium/hydrogen exchangers, NHX1 and NHX7 (PLSC phosphoproteins 1425 and 1385), were found in phosphorylation status only under salt stress. Overexpression of NHX1 was shown to be related to salt tolerance in Arabidopsis (74). Conservation analysis confirmed that NHX7 in our study had a phosphorylated ortholog that was SOS1 in Arabidopsis. SOS1 is involved in Na+ and K+ homeostasis and required for cytoplasmic Na+ detoxification, which is regulated by SOS2 and SOS3 through phosphorylation modification (75). Cation-chloride cotransporter 1 (PLSC phosphoproteins 53 and 845) can mediate potassium chloride and sodium chloride cotransports and is related to plant development and Cl− homeostasis (76). Interestingly, we found that PLSC phosphoprotein 53 was phosphorylated only in the control, whereas PLSC phosphoprotein 845 was phosphorylated only in the salt stress condition.
Aquaporin can facilitate the transport of water and small neutral solutes across cell membranes (77). Phosphorylation of plasma membrane aquaporin can activate the water channel, and it can be inactivated through dephosphorylation of the phosphorylated plasma membrane aquaporin (78). In vivo, aquaporins are always phosphorylated on Ser residue (79). Three aquaporins were found in our study; two of them (PIP2–1 and PIP1–2; PLSC phosphoproteins 468 and 1627) were only identified under salt stress conditions, and one of them (NIP2–2; PLSC phosphoprotein 560) was down-regulated at the phosphorylation level under salt stress. This demonstrated that they might play different roles under salt stress. PIP2 is related to H2O2 diffusion in phosphorylation status in the response to cold stress in maize (80), suggesting a potentially similar function of the phosphorylation of PIP2–1 in our study.
In this study, several nutrient transporters involved in sugar, amino acid, peptide, and phospholipid transport were found as PLSC phosphoproteins, such as aluminum-activated malate transporter 9 (PLSC phosphoprotein 548), sugar transporter ERD6-like 4-like isoform 1 (PLSC phosphoprotein 764), monosaccharide-sensing protein 2 (PLSC phosphoprotein 1082), lysine histidine transporter 1 (PLSC phosphoprotein 1167), probable peptide/nitrate transporter At5g62680-like (PLSC phosphoprotein 321), and phospholipid-transporting ATPase 3 (PLSC phosphoprotein 1212). Among them, aluminum-activated malate transporter 9, ERD6-like 4-like isoform 1, monosaccharide-sensing protein 2, lysine histidine transporter 1, and phospholipid-transporting ATPase 3 were phosphorylated only under the salt stress condition.
Transcription and Translation
Under salt stress, many response- and defense-related genes are stimulated by upstream transcription regulatory factors. Meanwhile, the genes involved in normal plant growth and development are inhibited. Our results revealed that most of the DEPs associated with translation showed down-regulation under salt stress, except 31-kDa ribonucleoprotein (DEP spot 35), which was up-regulated by up to 6.66-fold under 160 mm NaCl stress. 31-kDa ribonucleoprotein is involved in the 3′-end processing of chloroplast mRNAs (81), and the up-regulation of this protein may be related to the translation of defense-related genes in chloroplast.
The phosphorylation or dephosphorylation of some transcription- and translation-associated proteins such as transcription factors, splicing factors, translation initiation factors, and nucleolar proteins may play important roles. Transcription factor HY5 (PLSC phosphoprotein 902) is a bZIP transcription factor that promotes photomorphogenesis and can be phosphorylated by CK-II (82). In our results, HY5 was phosphorylated at a Ser residue only under salt stress, with its phosphopeptide belonging to the [sDxE] motif, which was a substrate of CK-II. HY5 can interact with COP1 when unphosphorylated, which negatively regulates HY5 activity (83). Transcription factor GTE4 (PLSC phosphoprotein 1094) plays a role in cell cycle regulation and specifically in the maintenance of the mitotic cell cycle (84). Different from HY5, GTE4 was phosphorylated only under normal conditions. Scaffold attachment factor B1 (PLSC phosphoprotein 1099) functions in different cellular processes involved in RNA processing (85) and stress response (86). Scaffold attachment factor B1 was phosphorylated only under salt stress in our study, suggesting that it might play a crucial role in salt stress defense. Nuclear cap-binding protein subunit 2 (PLSC phosphoprotein 1347) can combine with nuclear cap-binding protein subunit 1 to form the nuclear cap-binding complex, which mediates the recognition of capped RNA and is involved in spliceosome assembly (87). THO complex subunit 4 (THOC4) (PLSC phosphoprotein 344) is a component of the THO subcomplex of the TREX complex. The TREX complex is recruited to splice mRNAs by a splicing-coupled mechanism (88, 89). The recruitment occurs via an interaction between ALYREF/THOC4 and nuclear cap-binding protein subunit 1 (90). The nuclear export of HSP70 mRNA depends on ALYREF/THOC4 in conjunction with THOC5 (91). In our research, nuclear cap-binding protein subunit 2 and THOC4 were both phosphorylated only under salt stress. IQ-domain 1 (IQD1) (PLSC phosphoprotein 875), IQD14 (PLSC phosphoprotein 735), and IQD32 (PLSC phosphoprotein 1664) were all IQD proteins that linked calcium signaling pathways to the regulation of gene expression. IQD1 can stimulate glucosinolate accumulation and plant defense (92).
Photosynthesis and Carbohydrate Metabolism
Photosynthesis is usually depressed under salt stress in glycophytes. Our results indicated that Bd21, as a glycophyte, could tolerate NaCl treatment up to 200 mm based on the measurement of RWC and chlorophyll content (Fig. 1). When the NaCl concentration exceeded this boundary, the photosynthesis process was disturbed. At the translational level, the expression of most photosynthesis-associated proteins decreased. Fructose-1,6-bisphosphatase (DEP spot 44) is involved in gluconeogenesis and the Calvin cycle. The down-regulation of fructose-1,6-bisphosphatase in both stress-treatment and recovery groups reflected the depression of photosynthesis. Chlorophyll a/b binding proteins (LHCB; DEP spots 17, 38, 57, and 75) displayed an up-regulated expression pattern at different NaCl concentration levels (Fig. 3A), which was generally consistent with the results obtained for Arabidopsis treated with 150 mm NaCl (9). LHCB can be significantly phosphorylated under NaCl treatment in the halotolerant green alga Dunaliella salina (93). In our phosphoproteomic results, two LHCB proteins (PLSC phosphoproteins 1505 and 1595) were identified as PLSC phosphoproteins. One of them (PLSC phosphoprotein 1505) was significantly up-regulated at the phosphorylation level under salt stress, whereas the other one (PLSC phosphoprotein 1595) was down-regulated (supplemental Table S4).
Under salt treatment, energy production is also inhibited. With increasing NaCl concentration, the depression of energy is aggravated, which is caused by ROS damage. At the protein expression level, most of the DEPs were down-regulated under salt stress. For example, transketolase (DEP spots 41 and 73), the key enzyme of the reductive and oxidative pentose phosphate pathway, was down-regulated to one-fifth in expression under salt stress relative to the control. In the recovery group, transketolase displayed slight up-regulation under an 80 mm NaCl concentration. But with increasing NaCl concentration, the expression of transketolase decreased (Fig. 3A). Bernacchia et al. (94) found that transketolases played roles in the conversion of sugars and were involved in the rehydration process of desiccated plants. Our results suggested that the pentose phosphate pathway could be seriously disturbed by salt stress, and whether this pathway could be recovered depended on the NaCl concentration level.
In our study, 10 DEPs associated with photosynthesis or carbohydrate metabolism were also identified as PLSC phosphoproteins (Table II), which means these proteins are regulated at both translational and phosphorylation levels in response to salt stress. For example, oxygen-evolving enhancer protein 2 (DEP spots 3 and 58) was also identified as a PLSC phosphoprotein. The intensity of DEP spot 3 in 2-DE maps was far less than that of DEP spot 58. Results revealed that DEP spot 3 should represent the phosphorylation status of DEP spot 58. This demonstrated that some DEPs with more than one spot on 2-DE profiles might have different phosphorylation statuses at various phosphorylated sites.
ROS Scavenging
Under treatment with different NaCl concentrations, proteins related to ROS scavenging were up-regulated to different extents at the translational level. Mitochondrion is the organelle for producing energy that yields a lot of peroxides and superoxides. The plant cells need various antioxidants to detoxify those oxides in mitochondria. Salt stress generally induces a reduction of molecular oxygen by membrane-bound NADPH oxidase, giving rise to superoxide radicals. These radicals are, to some degree, detoxified by discrimination into H2O2, which is also toxic but can serve as a signal for gene expression that leads to programmed cell death or the production of radical scavenging proteins. Thus the balancing of production and detoxification of ROS is vital for maintaining cellular functions (95). Upon exposure to abiotic stress, the transcription and translation of enzymes involved in ROS scavenging are induced or up-regulated (96, 97). Superoxide dismutase [Mn] 3.1 (DEP spot 37) can transform superoxide into H2O2 in mitochondria. 2-Cys peroxiredoxin BAS1 (DEP spots 52 and 72) is involved in the detoxification of hydroperoxides and can be induced by oxidative stress in Arabidopsis (98). Both glutathione S-transferase F11 (DEP spot 56) and glutathione S-transferase DHAR2 (DEP spot 60) are involved in redox homeostasis (99). In our research, 2-cys peroxiredoxin BAS1, glutathione S-transferase F11, glutathione S-transferase DHAR2, and superoxide dismutase [Mn] 3.1 were all up-regulated dramatically under salt stress and returned to normal in the recovery period, which reflects the crucial roles of these proteins in ROS scavenging. The expression of ascorbate peroxidase 1 (DEP spots 36 and 42) increased sharply to a higher level under salt treatment in the 80 to 160 mm NaCl range (Fig. 3A), suggesting that ascorbate peroxidase 1 is a main scavenging enzyme for defense against salt stress. Under higher salt stress, its accumulation returned to a normal level, suggesting that the extent of oxidative damage exceeded the regulatory capacity of Bd21 leaf cells. For the purpose of signaling, small fluctuations of ROS are adequate to activate transduction pathways. ROS production and scavenging enzymes are likely to be regulated by PTMs (47). In our phosphoproteomic analysis, glutathione S-transferase (PLSC phosphoprotein 1522) and glutathione peroxidase 4 (PLSC phosphoprotein 118) were both only phosphorylated under salt stress. All of these DEPs and PLSC phosphoproteins are components of the oxidative stress defense system and can be used as potential salt stress markers in further research.
Protein Folding and Degradation
Under salt stress, many proteins are misfolded, and these proteins cannot be assembled and function as normal. CPN60–2 (DEP spots 53 and 55) is involved in protein import and macromolecular assembly in mitochondria (100), which may prevent misfolding and promote the refolding and proper assembly of unfolded polypeptides generated under salt stress. Stromal 70-kDa heat shock-related protein (CHSP70) (DEP spot 64) can conjunct with CPN60, which is involved in the assembly of RuBisCO in plastids (101, 102). Under salt stress and recovery, the up-regulation of CPN60–2 and CHSP70 might function to maintain the protein conformation. The protein GrpE (PLSC phosphoprotein 1467) can function as a nucleotide exchange factor for HSP70 and participates in the response to hyperosmosis and heat shock through preventing the aggregation of stress-denatured proteins (103, 104). In our study, GrpE was only phosphorylated in the salt-treated sample, which indicated that it played roles in phosphorylation status under salt stress. Two HSP80 proteins (PLSC phosphoproteins 1290 and 1655), one HSC70 (PLSC phosphoprotein 178), and one HSP20 (PLSC phosphoprotein 1004) were only phosphorylated in the stress-treated sample (supplemental Table S4A). Our result provided more insights than a previous study by Cole and Meyers (105) that revealed an increase of HSP70 at the phosphorylation level but no change at the expression level under osmotic stress.
Another mechanism involving misfolded proteins is degradation. Our results showed that three DEP spots were identified as degradation-associated proteins. Among them was mitochondrial-processing peptidase subunit β (DEP spot 14), a heterodimeric enzyme playing an essential role in mitochondrial protein import. It cleaves off the N-terminal targeting signals of nuclear encoded mitochondrial proteins upon their transport into the organelle (106). The proteasome is a very large protein complex (26S) containing a 20S core particle that is a multicatalytic protease that degrades proteins using an ATP-dependent mechanism (107). The 20S core particle is a barrel-shaped structure composed of four stacked rings, each containing seven subunits (α7β7β7α7). The two outer rings are composed of seven different α subunits that regulate the opening of the proteasome to misfolded proteins. In our research, proteasome subunit α type-2 (DEP spot 23) was down-regulated in the salt-treated group but up-regulated in the recovery group (Fig. 3), suggesting that it mainly functioned in the recovery from the abnormal condition instead of in the defense against salt stress. There are some proteasome interacting proteins, including 14-3-3-like protein, glyceraldehyde-3-phosphate dehydrogenase, transaldolase, actin, translation elongation factor, enolase, ATPase, and eukaryotic initiation factor (108). The ATP-dependent zinc metalloprotease FTSH2 (DEP spot 45) is an ATP- and Zn2+-dependent metalloprotease. Besides being a protease, FTSH also acts as a molecular chaperone influencing protein assembly in and through the cytoplasmic membrane and associates with denatured alkaline phosphatase without degrading it (109). Therefore, FTSH may serve to maintain quality control of some cytoplasmic and membrane proteins. Ubiquitin protein ligases (E3s), along with the ubiquitin-activating enzymes (E1s) and ubiquitin-conjugating enzymes (E2s), catalyze the attachment of ubiquitin to target proteins. The specificity of the ubiquitination pathway is controlled mainly by the E3s, and most stress-related E3s facilitate responses to environmental stress by modulating the abundance of key downstream stress-responsive transcription factors (110). Stone et al. (111) found a novel E3 containing 12 HERC2-like repeats that was required for ABA signaling of Arabidopsis seedlings. Here, two E3s (PLSC phosphoproteins 608 and 1428) were only phosphorylated under salt stress. NEDD8-activating enzyme E1 catalytic subunit (PLSC phosphoprotein 723) was found with a significantly higher phosphorylation level under salt stress relative to the control (supplemental Table S4C). NEDD8-activating enzyme E1 catalytic subunit is highly diverged from characterized E1 enzymes and lacks a key cysteine residue that is essential for E1 activity. NEDD8-activating enzyme E1 catalytic subunit may have a novel function in cellular regulation that is unrelated to ubiquitin conjugation (102). del Pozo et al. (112) found that mutation of the AXR1 gene could lead to a reduction of auxin response and diverse defects in auxin-regulated growth and development. The degradation-associated proteins were down-regulated at the translational level under salt stress and then restored to normal expression levels or up-regulated after recovery. At the same time, the phosphorylation levels of proteins associated with the ubiquitination degradation pathway were raised under salt stress. Degradation is an energy-consuming process, and energy production is depressed under salt stress. Thus, we speculated that plants handle misfolded proteins mainly through refolding under stress treatment and through degradation in recovery. Changes at the phosphorylation level instead of in protein abundance may be the main strategy for the ubiquitination degradation pathway under salt stress.
Other Functions
Under harsh salt stress, cell structure might be damaged, and several DEPs or PLSC phosphoproteins associated with dynamic changes of cell membrane and cell wall were found in the current study. Outer membrane lipoprotein BLC is involved in the storage or transport of lipids necessary for membrane maintenance and is up-regulated during cold acclimation and heat-shock treatment (113). Our results also showed that BLC (DEP spot 62) was up-regulated dramatically at the translational level with increasing NaCl concentration in the stress group (up to 6.80) and the recovery group (up to 1.98). Quantitative RT-PCR analysis of the gene encoding BLC also showed a consistent result. With increasing NaCl concentration, the transcription of this gene increased sharply to 7.69-fold under 320 mm NaCl stress and maintained a high level (5.41-fold) in the recovery group (Fig. 4A). Five cellulose synthase A catalytic subunits (CESA2, CESA4, CESA7, CESA8, and CESA9) were found with down-regulated phosphorylation levels under salt stress or only found in the normal condition. CESA2 and CESA9 (PLSC phosphoproteins 242 and 1478) are mainly responsible for cellulose production during the formation of the primary cell wall (114), whereas CESA4, CESA7, and CESA8 (PLSC phosphoproteins 935, 1219, and 73) are required for secondary cell wall formation (115–117). Phosphorylation of the catalytic subunit of CESA plays a key role in the regulation of cellulose synthesis in the primary cell wall (118). Phosphorylation of CESA7 has been linked to its degradation via a 26S proteasome-dependent pathway (119). The down-regulated phosphorylation level of CESAs might lead to a hard-to-stretch cell wall and thus hindered leaf growth and development. Two patellin-5 proteins (PLSC phosphoproteins 823 and 824) were only phosphorylated under salt stress. Patellin-5 is similar to patellin-1, which may be involved in membrane-trafficking events associated with cell-plate expansion or maturation and points to the involvement of phosphoinositides in cell-plate biogenesis (120). Chitinase belongs to the pathogenesis-related proteins and has mostly been detected in plants exposed to phytopathogenic viruses, bacteria, or fungi. Some studies revealed that chitinase is also involved in plants' defense against abiotic stress, such as that caused by salt (26), wounds (121), or heavy metals (122). In our study, a chitinase protein (Cht8; DEP spot 69) was up-regulated up to 8.58- and 18.04-fold in the stress and recovery groups, respectively (supplemental Table S1), which reflected its important roles in salt stress defense.
CONCLUSION
In summary, for the first time, the model plant Bd21 was studied in a comprehensive analysis at both translational and post-translational levels under salt stress. Proteome analysis of salt stress treatment and recovery from moderate to harsh salt concentrations displayed the protein expression characters for salt stress response and defense in Brachypodium. Phosphoproteome analysis of nontreated and salt-treated samples revealed the difference at the phosphorylation level and a range of phosphoproteins and phosphorylation sites involved in the stress signaling cascade. In total, 60 DEPs and 468 PLSC phosphoproteins were identified through proteomic and phosphoproteomic analyses. GO enrichment and KEGG pathway analyses revealed the function and pathway distribution of the DEPs and PLSC phosphoproteins. For the 496 PLSC phosphopeptides, 9 phosphorylation motifs were extracted, which were mainly regulated by MAPK, cyclin-dependent kinase, CaMK-II, and CK-II. A complex PPI network of all DEPs and PLSC phosphoproteins and a 14-3-3 subnetwork were constructed and revealed the potential crucial roles of the three 14-3-3 proteins identified in this work. Among the 60 DEPs, 14 were also identified as phosphoproteins. Many proteins and phosphoproteins have been screened out and can be potential salt stress markers for further research. Our results broaden the understanding of salt stress response and defense in plants.
Supplementary Material
Footnotes
Author contributions: D.L., W.M., and Y.Y. designed research; D.L. performed research; S.S., M.C., and X.Y. contributed new reagents or analytic tools; D.L., S.S., M.C., X.Y., X.L., R.A., and D.S. analyzed data; D.L., W.M., and Y.Y. wrote the paper.
* This research was financially supported in part by grants from the Chinese Ministry of Science and Technology (2009CB118300), the National Natural Science Foundation of China (31271703), the China-Australia Cooperation Project from the Chinese Ministry of Science and Technology (2013DFG30530), and the National Key Projects for Transgenic Crops of China (2011ZX08009-003-004). The data deposition to the ProteomeXchange Consortium was supported by PRIDE Team, EBI.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- Bd21
- Brachypodium distachyon 21
- PTM
- post-translational modification
- GO
- gene ontology
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- RWC
- relative water content
- DEP
- differentially expressed protein
- ACN
- acetonitrile
- FDR
- false discovery rate
- PPI
- protein–protein interaction
- 2-DE
- two-dimensional gel electrophoresis
- PLSC
- phosphorylation level significantly changed
- ABA
- abscisic acid
- CK-II
- casein kinase-II
- KOG
- eukaryotic orthologous group
- 14-3-3A
- 14-3-3-like protein A
- GF14
- 14-3-3-like protein GF14
- ROS
- reactive oxygen species
- THOC
- THO complex
- IQD
- IQ-domain.
REFERENCES
- 1. Munns R. (2005) Genes and salt tolerance: bringing them together. New Phytol. 167, 645–663 [DOI] [PubMed] [Google Scholar]
- 2. Wang W., Vinocur B., Altman A. (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218, 1–14 [DOI] [PubMed] [Google Scholar]
- 3. Levitt J. (1980) Responses of Plants to Environmental Stresses. Volume II: Water, Radiation, Salt, and Other Stresses, Academic Press, New York [Google Scholar]
- 4. Khan P., Hoffmann L., Renaut J., Hausman J. (2007) Current initiatives in proteomics for the analysis of plant salt tolerance. Curr. Sci. 93, 807–817 [Google Scholar]
- 5. Ouerghi Z., Rémy R., Ouelhazi L., Ayadi A., Brulfert J. (2000) Two-dimensional electrophoresis of soluble leaf proteins, isolated from two wheat species (Triticum durum and Triticum aestivum) differing in sensitivity towards NaCl. Electrophoresis 21, 2487–2491 [DOI] [PubMed] [Google Scholar]
- 6. Salekdeh G., Siopongco J., Wade L. J., Ghareyazie B., Bennett J. (2002) Proteomic analysis of rice leaves during drought stress and recovery. Proteomics 2, 1131–1145 [DOI] [PubMed] [Google Scholar]
- 7. Yan S., Tang Z., Su W., Sun W. (2005) Proteomic analysis of salt stress-responsive proteins in rice root. Proteomics 5, 235–244 [DOI] [PubMed] [Google Scholar]
- 8. Peng Z., Wang M., Li F., Lv H., Li C., Xia G. (2009) A proteomic study of the response to salinity and drought stress in an introgression strain of bread wheat. Mol. Cell. Proteomics 8, 2676–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pang Q., Chen S., Dai S., Chen Y., Wang Y., Yan X. (2010) Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila. J. Proteome Res. 9, 2584–2599 [DOI] [PubMed] [Google Scholar]
- 10. Vogel J. P., Garvin D. F., Mockler T. C., Schmutz J., Rokhsar D., Bevan M. W., Barry K., Lucas S., Harmon-Smith M., Lail K. (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768 [DOI] [PubMed] [Google Scholar]
- 11. Draper J., Mur L. A., Jenkins G., Ghosh-Biswas G. C., Bablak P., Hasterok R., Routledge A. P. (2001) Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 127, 1539–1555 [PMC free article] [PubMed] [Google Scholar]
- 12. Vogel J., Hill T. (2008) High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21–3. Plant Cell Rep. 27, 471–478 [DOI] [PubMed] [Google Scholar]
- 13. Thingholm T. E., Jensen O. N., Larsen M. R. (2009) Analytical strategies for phosphoproteomics. Proteomics 9, 1451–1468 [DOI] [PubMed] [Google Scholar]
- 14. Andersson L., Porath J. (1986) Isolation of phosphoproteins by immobilized metal (Fe3+) affinity chromatography. Anal. Biochem. 154, 250–254 [DOI] [PubMed] [Google Scholar]
- 15. Thingholm T. E., Jensen O. N. (2009) Enrichment and characterization of phosphopeptides by immobilized metal affinity chromatography (IMAC) and mass spectrometry. Methods Mol. Biol. 527, 47–56 [DOI] [PubMed] [Google Scholar]
- 16. Pinkse M. W., Uitto P. M., Hilhorst M. J., Ooms B., Heck A. J. (2004) Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Biochem. 76, 3935–3943 [DOI] [PubMed] [Google Scholar]
- 17. Wu J., Shakey Q., Liu W., Schuller A., Follettie M. T. (2007) Global profiling of phosphopeptides by titania affinity enrichment. J. Proteome Res. 6, 4684–4689 [DOI] [PubMed] [Google Scholar]
- 18. Larsen M. R., Thingholm T. E., Jensen O. N., Roepstorff P., Jørgensen T. J. (2005) Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 4, 873–886 [DOI] [PubMed] [Google Scholar]
- 19. Sugiyama N., Masuda T., Shinoda K., Nakamura A., Tomita M., Ishihama Y. (2007) Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol. Cell. Proteomics 6, 1103–1109 [DOI] [PubMed] [Google Scholar]
- 20. Jensen S. S., Larsen M. R. (2007) Evaluation of the impact of some experimental procedures on different phosphopeptide enrichment techniques. Rapid Commun. Mass Spectrom. 21, 3635–3645 [DOI] [PubMed] [Google Scholar]
- 21. Gao L., Yan X., Li X., Guo G., Hu Y., Ma W., Yan Y. (2011) Proteome analysis of wheat leaf under salt stress by two-dimensional difference gel electrophoresis (2D-DIGE). Phytochemistry 72, 1180–1191 [DOI] [PubMed] [Google Scholar]
- 22. Arnon D. I. (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang W., Scali M., Vignani R., Spadafora A., Sensi E., Mazzuca S., Cresti M. (2003) Protein extraction for two-dimensional electrophoresis from olive leaf, a plant tissue containing high levels of interfering compounds. Electrophoresis 24, 2369–2375 [DOI] [PubMed] [Google Scholar]
- 24. Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J. A. (2007) Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35, W71–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hong S.-Y., Seo P. J., Yang M.-S., Xiang F., Park C.-M. (2008) Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol. 8, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 27. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 28. Nakagami H., Sugiyama N., Mochida K., Daudi A., Yoshida Y., Toyoda T., Tomita M., Ishihama Y., Shirasu K. (2010) Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 153, 1161–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schwartz D., Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 31. Crooks G. E., Hon G., Chandonia J.-M., Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P. (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 35. Guex N., Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 36. Vizcaíno J. A., Côté R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J. (2013) The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yao Q., Bollinger C., Gao J., Xu D., Thelen J. J. (2012) P3DB: an integrated database for plant protein phosphorylation. Front. Plant Sci. 3, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rose C. M., Venkateshwaran M., Volkening J. D., Grimsrud P. A., Maeda J., Bailey D. J., Park K., Howes-Podoll M., den Os D., Yeun L. H. (2012) Rapid phosphoproteomic and transcriptomic changes in the rhizobia-legume symbiosis. Mol. Cell. Proteomics 11, 724–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heazlewood J. L., Durek P., Hummel J., Selbig J., Weckwerth W., Walther D., Schulze W. X. (2008) PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res. 36, D1015–D1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Amanchy R., Periaswamy B., Mathivanan S., Reddy R., Tattikota S. G., Pandey A. (2007) A curated compendium of phosphorylation motifs. Nat. Biotechnol. 25, 285–286 [DOI] [PubMed] [Google Scholar]
- 42. Villén J., Beausoleil S. A., Gerber S. A., Gygi S. P. (2007) Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U.S.A. 104, 1488–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Prasad T. K., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A. (2009) Human protein reference database—2009 update. Nucleic Acids Res. 37, D767–D772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reiland S., Messerli G., Baerenfaller K., Gerrits B., Endler A., Grossmann J., Gruissem W., Baginsky S. (2009) Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol. 150, 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Durek P., Schmidt R., Heazlewood J. L., Jones A., MacLean D., Nagel A., Kersten B., Schulze W. X. (2010) PhosPhAt: the Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 38, D828–D834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Muslin A. J., Tanner J. W., Allen P. M., Shaw A. S. (1996) Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84, 889–897 [DOI] [PubMed] [Google Scholar]
- 47. Barjaktarović Ž., Schütz W., Madlung J., Fladerer C., Nordheim A., Hampp R. (2009) Changes in the effective gravitational field strength affect the state of phosphorylation of stress-related proteins in callus cultures of Arabidopsis thaliana. J. Exp. Bot. 60, 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sanders D., Brownlee C., Harper J. F. (1999) Communicating with calcium. Plant Cell 11, 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dammann C., Ichida A., Hong B., Romanowsky S. M., Hrabak E. M., Harmon A. C., Pickard B. G., Harper J. F. (2003) Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiol. 132, 1840–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Witte C.-P., Keinath N., Dubiella U., Demoulière R., Seal A., Romeis T. (2010) Tobacco calcium-dependent protein kinases are differentially phosphorylated in vivo as part of a kinase cascade that regulates stress response. J. Biol. Chem. 285, 9740–9748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Meyer K., Leube M. P., Grill E. (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264, 1452–1455 [DOI] [PubMed] [Google Scholar]
- 52. Stone J. M., Collinge M. A., Smith R. D., Horn M. A., Walker J. C. (1994) Interaction of a protein phosphatase with an Arabidopsis serine-threonine receptor kinase. Science 266, 793–795 [DOI] [PubMed] [Google Scholar]
- 53. Rienties I. M., Vink J., Borst J. W., Russinova E., de Vries S. C. (2005) The Arabidopsis SERK1 protein interacts with the AAA-ATPase AtCDC48, the 14-3-3 protein GF14lambda and the PP2C phosphatase KAPP. Planta 221, 394–405 [DOI] [PubMed] [Google Scholar]
- 54. Gillaspy G. E. (2010) Signaling and the polyphosphoinositide phosphatases from plants. In Lipid Signaling in Plants (Munnik T., ed.), pp. 117–130, Springer, Berlin [Google Scholar]
- 55. Shi H., Zhu J.-K. (2002) Regulation of expression of the vacuolar Na+/H+ antiporter gene AtNHX1 by salt stress and abscisic acid. Plant Mol. Biol. 50, 543–550 [DOI] [PubMed] [Google Scholar]
- 56. Roberts M. R., Salinas J., Collinge D. B. (2002) 14-3-3 proteins and the response to abiotic and biotic stress. Plant Mol. Biol. 50, 1031–1039 [DOI] [PubMed] [Google Scholar]
- 57. Fulgosi H., Soll J., de Faria Maraschin S., Korthout H. A., Wang M., Testerink C. (2002) 14-3-3 proteins and plant development. Plant Mol. Biol. 50, 1019–1029 [DOI] [PubMed] [Google Scholar]
- 58. Wang X. Q., Yang P. F., Liu Z., Liu W. Z., Hu Y., Chen H., Kuang T. Y., Pei Z. M., Shen S. H., He Y. K. (2009) Exploring the mechanism of Physcomitrella patens desiccation tolerance through a proteomic strategy. Plant Physiol. 149, 1739–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kidou S.-I., Umeda M., Kato A., Uchimiya H. (1993) Isolation and characterization of a rice cDNA similar to the bovine brain-specific 14-3-3 protein gene. Plant Mol. Biol. 21, 191–194 [DOI] [PubMed] [Google Scholar]
- 60. Dubois T., Howell S., Amess B., Kerai P., Learmonth M., Madrazo J., Chaudhri M., Rittinger K., Scarabel M., Soneji Y. (1997) Structure and sites of phosphorylation of 14-3-3 protein: role in coordinating signal transduction pathways. J. Protein Chem. 16, 513–522 [DOI] [PubMed] [Google Scholar]
- 61. Dubois T., Rommel C., Howell S., Steinhussen U., Soneji Y., Morrice N., Moelling K., Aitken A. (1997) 14-3-3 is phosphorylated by casein kinase I on residue 233. Phosphorylation at this site in vivo regulates Raf/14-3-3 interaction. J. Biol. Chem. 272, 28882–28888 [DOI] [PubMed] [Google Scholar]
- 62. Ngok S. P., Geyer R., Kourtidis A., Storz P., Anastasiadis P. Z. (2013) Phosphorylation-mediated 14-3-3 protein binding regulates the function of the Rho-specific guanine nucleotide exchange factor (RhoGEF) Syx. J. Biol. Chem. 288, 6640–6650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Johnson C., Crowther S., Stafford M., Campbell D., Toth R., MacKintosh C. (2010) Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem. J. 427, 69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dharmasiri N., Estelle M. (2004) Auxin signaling and regulated protein degradation. Trends Plant Sci. 9, 302–308 [DOI] [PubMed] [Google Scholar]
- 65. Ghanashyam C., Jain M. (2009) Role of auxin-responsive genes in biotic stress responses. Plant Signal. Behav. 4, 846–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shibasaki K., Uemura M., Tsurumi S., Rahman A. (2009) Auxin response in Arabidopsis under cold stress: underlying molecular mechanisms. Plant Cell 21, 3823–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dharmasiri N., Dharmasiri S., Estelle M. (2005) The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445 [DOI] [PubMed] [Google Scholar]
- 68. Lasswell J., Rogg L. E., Nelson D. C., Rongey C., Bartel B. (2000) Cloning and characterization of IAR1, a gene required for auxin conjugate sensitivity in Arabidopsis. Plant Cell 12, 2395–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Park C. M. (2007) Auxin homeostasis in plant stress adaptation response. Plant Signal. Behav. 2, 306–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kim S., Kang J. Y., Cho D. I., Park J. H., Kim S. Y. (2004) ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 40, 75–87 [DOI] [PubMed] [Google Scholar]
- 71. Kobayashi Y., Yamamoto S., Minami H., Kagaya Y., Hattori T. (2004) Differential activation of the rice sucrose nonfermenting1–related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 16, 1163–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kobayashi Y., Murata M., Minami H., Yamamoto S., Kagaya Y., Hobo T., Yamamoto A., Hattori T. (2005) Abscisic acid−activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 44, 939–949 [DOI] [PubMed] [Google Scholar]
- 73. Wang M. C., Peng Z. Y., Li C. L., Li F., Liu C., Xia G. M. (2008) Proteomic analysis on a high salt tolerance introgression strain of Triticum aestivum/Thinopyrum ponticum. Proteomics 8, 1470–1489 [DOI] [PubMed] [Google Scholar]
- 74. Apse M. P., Aharon G. S., Snedden W. A., Blumwald E. (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285, 1256–1258 [DOI] [PubMed] [Google Scholar]
- 75. Shi H., Ishitani M., Kim C., Zhu J.-K. (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U.S.A. 97, 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Colmenero-Flores J. M., Martínez G., Gamba G., Vázquez N., Iglesias D. J., Brumós J., Talón M. (2007) Identification and functional characterization of cation-chloride cotransporters in plants. Plant J. 50, 278–292 [DOI] [PubMed] [Google Scholar]
- 77. Kaldenhoff R., Bertl A., Otto B., Moshelion M., Uehlein N. (2007) Characterization of plant aquaporins. Methods Enzymol. 428, 505–531 [DOI] [PubMed] [Google Scholar]
- 78. Azad A. K., Sawa Y., Ishikawa T., Shibata H. (2004) Phosphorylation of plasma membrane aquaporin regulates temperature-dependent opening of tulip petals. Plant Cell Physiol. 45, 608–617 [DOI] [PubMed] [Google Scholar]
- 79. Azad A. K., Katsuhara M., Sawa Y., Ishikawa T., Shibata H. (2008) Characterization of four plasma membrane aquaporins in tulip petals: a putative homolog is regulated by phosphorylation. Plant Cell Physiol. 49, 1196–1208 [DOI] [PubMed] [Google Scholar]
- 80. Aroca R., Amodeo G., Fernández-Illescas S., Herman E. M., Chaumont F., Chrispeels M. J. (2005) The role of aquaporins and membrane damage in chilling and hydrogen peroxide induced changes in the hydraulic conductance of maize roots. Plant Physiol. 137, 341–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ohta M., Sugita M., Sugiura M. (1995) Three types of nuclear genes encoding chloroplast RNA-binding proteins (cp29, cp31 and cp33) are present in Arabidopsis thaliana: presence of cp31 in chloroplasts and its homologue in nuclei/cytoplasms. Plant Mol. Biol. 27, 529–539 [DOI] [PubMed] [Google Scholar]
- 82. Hardtke C. S., Gohda K., Osterlund M. T., Oyama T., Okada K., Deng X. W. (2000) HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. Sci. Signal. 19, 4997–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ang L.-H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A., Deng X.-W. (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1, 213–222 [DOI] [PubMed] [Google Scholar]
- 84. Airoldi C. A., Rovere F. D., Falasca G., Marino G., Kooiker M., Altamura M. M., Citterio S., Kater M. M. (2010) The Arabidopsis BET bromodomain factor GTE4 is involved in maintenance of the mitotic cell cycle during plant development. Plant Physiol. 152, 1320–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Nayler O., Stratling W., Bourquin J. P., Stagljar I., Lindemann L., Jasper H., Hartmann A. M., Fackelmayer F. O., Ullrich A., Stamm S. (1998) SAF-B protein couples transcription and pre-mRNA splicing to SAR/MAR elements. Nucleic Acids Res. 26, 3542–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Denegri M., Chiodi I., Corioni M., Cobianchi F., Riva S., Biamonti G. (2001) Stress-induced nuclear bodies are sites of accumulation of pre-mRNA processing factors. Mol. Biol. Cell 12, 3502–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Mazza C., Ohno M., Segref A., Mattaj I. W., Cusack S. (2001) Crystal structure of the human nuclear cap binding complex. Mol. Cell 8, 383–396 [DOI] [PubMed] [Google Scholar]
- 88. Strasser K., Masuda S., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondon A. G., Aguilera A., Struhl K., Reed R., Hurt E. (2002) TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417, 304–308 [DOI] [PubMed] [Google Scholar]
- 89. Masuda S., Das R., Cheng H., Hurt E., Dorman N., Reed R. (2005) Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 19, 1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Cheng H., Dufu K., Lee C. S., Hsu J. L., Dias A., Reed R. (2006) Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 127, 1389–1400 [DOI] [PubMed] [Google Scholar]
- 91. Katahira J., Inoue H., Hurt E., Yoneda Y. (2009) Adaptor Aly and co-adaptor Thoc5 function in the Tap-p15-mediated nuclear export of HSP70 mRNA. EMBO J. 28, 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Levy M., Wang Q., Kaspi R., Parrella M. P., Abel S. (2005) Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J. 43, 79–96 [DOI] [PubMed] [Google Scholar]
- 93. Liu X.-D., Shen Y.-G. (2004) NaCl-induced phosphorylation of light harvesting chlorophyll a/b proteins in thylakoid membranes from the halotolerant green alga, Dunaliella salina. FEBS Lett. 569, 337–340 [DOI] [PubMed] [Google Scholar]
- 94. Bernacchia G., Schwall G., Lottspeich F., Salamini F., Bartels D. (1995) The transketolase gene family of the resurrection plant Craterostigma plantagineum: differential expression during the rehydration phase. EMBO J. 14, 610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Apel K., Hirt H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399 [DOI] [PubMed] [Google Scholar]
- 96. Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004) Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498 [DOI] [PubMed] [Google Scholar]
- 97. Jiang P., Zhang X., Zhu Y., Zhu W., Xie H., Wang X. (2007) Metabolism of reactive oxygen species in cotton cytoplasmic male sterility and its restoration. Plant Cell Rep. 26, 1627–1634 [DOI] [PubMed] [Google Scholar]
- 98. Horling F., Lamkemeyer P., König J., Finkemeier I., Kandlbinder A., Baier M., Dietz K.-J. (2003) Divergent light-, ascorbate-, and oxidative stress-dependent regulation of expression of the peroxiredoxin gene family in Arabidopsis. Plant Physiol. 131, 317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Dixon D. P., Davis B. G., Edwards R. (2002) Functional divergence in the glutathione transferase superfamily in plants. Identification of two classes with putative functions in redox homeostasis in Arabidopsis thaliana. J. Biol. Chem. 277, 30859–30869 [DOI] [PubMed] [Google Scholar]
- 100. Tsugeki R., Mori H., Nishimura M. (1992) Purification, cDNA cloning and Northern-blot analysis of mitochondrial chaperonin 60 from pumpkin cotyledons. Eur. J. Biochem. 209, 453–458 [DOI] [PubMed] [Google Scholar]
- 101. Gething M.-J., Sambrook J. (1992) Protein folding in the cell. Nature 355, 33–45 [DOI] [PubMed] [Google Scholar]
- 102. Langer T., Lu C., Echols H., Flanagan J., Hayer M. K., Hartl F. U. (1992) Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature 356, 683–689 [DOI] [PubMed] [Google Scholar]
- 103. Groemping Y., Reinstein J. (2001) Folding properties of the nucleotide exchange factor GrpE from Thermus thermophilus: GrpE is a thermosensor that mediates heat shock response. J. Mol. Biol. 314, 167–178 [DOI] [PubMed] [Google Scholar]
- 104. Harrison C. (2003) GrpE, a nucleotide exchange factor for DnaK. Cell Stress Chaperones 8, 218–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Cole J. A., Meyers S. A. (2011) Osmotic stress stimulates phosphorylation and cellular expression of heat shock proteins in rhesus macaque sperm. J. Androl. 32, 402–410 [DOI] [PubMed] [Google Scholar]
- 106. Taylor A. B., Smith B. S., Kitada S., Kojima K., Miyaura H., Otwinowski Z., Ito A., Deisenhofer J. (2001) Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences. Structure 9, 615–625 [DOI] [PubMed] [Google Scholar]
- 107. Babbitt S. E., Kiss A., Deffenbaugh A. E., Chang Y.-H., Bailly E., Erdjument-Bromage H., Tempst P., Buranda T., Sklar L. A., Baumler J. (2005) ATP hydrolysis-dependent disassembly of the 26S proteasome is part of the catalytic cycle. Cell 121, 553–565 [DOI] [PubMed] [Google Scholar]
- 108. Dong C.-H., Agarwal M., Zhang Y., Xie Q., Zhu J.-K. (2006) The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. U.S.A. 103, 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Schumann W. (1999) FtsH—a single-chain charonin? FEMS Microbiol. Rev. 23, 1–11 [DOI] [PubMed] [Google Scholar]
- 110. Lyzenga W. J., Stone S. L. (2012) Abiotic stress tolerance mediated by protein ubiquitination. J. Exp. Bot. 63, 599–616 [DOI] [PubMed] [Google Scholar]
- 111. Stone S. L., Williams L. A., Farmer L. M., Vierstra R. D., Callis J. (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18, 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. del Pozo J. C., Dharmasiri S., Hellmann H., Walker L., Gray W. M., Estelle M. (2002) AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell 14, 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Frenette Charron J.-B., Breton G., Badawi M., Sarhan F. (2002) Molecular and structural analyses of a novel temperature stress-induced lipocalin from wheat and Arabidopsis. FEBS Lett. 517, 129–132 [DOI] [PubMed] [Google Scholar]
- 114. Fagard M., Desnos T., Desprez T., Goubet F., Refregier G., Mouille G., McCann M., Rayon C., Vernhettes S., Höfte H. (2000) PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12, 2409–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Taylor N. G., Howells R. M., Huttly A. K., Vickers K., Turner S. R. (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. U.S.A. 100, 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Somerville C., Bauer S., Brininstool G., Facette M., Hamann T., Milne J., Osborne E., Paredez A., Persson S., Raab T. (2004) Toward a systems approach to understanding plant cell walls. Sci. Signal. 306, 2206. [DOI] [PubMed] [Google Scholar]
- 117. Brown D. M., Zeef L. A., Ellis J., Goodacre R., Turner S. R. (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17, 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Chen S., Ehrhardt D. W., Somerville C. R. (2010) Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proc. Natl. Acad. Sci. U.S.A. 107, 17188–17193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Taylor N. G. (2007) Identification of cellulose synthase AtCesA7 (IRX3) in vivo phosphorylation sites—a potential role in regulating protein degradation. Plant Mol. Biol. 64, 161–171 [DOI] [PubMed] [Google Scholar]
- 120. Peterman T. K., Ohol Y. M., McReynolds L. J., Luna E. J. (2004) Patellin1, a novel Sec14-like protein, localizes to the cell plate and binds phosphoinositides. Plant Physiol. 136, 3080–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Takenaka Y., Nakano S., Tamoi M., Sakuda S., Fukamizo T. (2009) Chitinase gene expression in response to environmental stresses in Arabidopsis thaliana: chitinase inhibitor allosamidin enhances stress tolerance. Biosci. Biotechnol. Biochem. 73, 1066–1071 [DOI] [PubMed] [Google Scholar]
- 122. Békésiová B., Hraška Š., Libantová J., Moravčíková J., Matušíková I. (2008) Heavy-metal stress induced accumulation of chitinase isoforms in plants. Mol. Biol. Rep. 35, 579–588 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.