Abstract
Background/Aims
A single gene mutation alone cannot explain the poor prognosis of colorectal cancer. This study aimed to establish a correlation between the expression of six proteins and the prognosis of colorectal cancer patients.
Methods
Tissue samples were collected from 266 patients who underwent surgery for colorectal cancer at our institution from January 2006 to December 2007. The expression of six proteins were determined using immunohistochemical staining of specimens.
Results
Cathepsin D, p53, COX-2, epidermal growth factor receptor, c-erbB-2, and Ki-67 expression were detected in 38.7%, 60.9%, 37.6%, 35.7%, 30.1%, and 74.4% of the samples, respectively. The expression of cathepsin D was significantly correlated with reduced cancer-free survival (p=0.036) and colorectal cancer-specific survival (p=0.003), but the other expression levels were not. In a multivariate analysis, cathepsin D expression was found to be an independent prognostic factor for poorer colorectal cancer-specific survival (hazard ratio, 8.55; 95% confidence interval, 1.07 to 68.49). Furthermore, patients with tumors expressing four or more of the proteins had a significantly decreased cancer-free survival rate (p=0.006) and colorectal cancer-specific survival rate (p=0.002).
Conclusions
Patients with cathepsin D positivity had a poorer outcome than patients who were cathepsin D-negative. Thus, cathepsin D may provide an indicator for appropriate intensive follow-up and adjuvant chemotherapy.
Keywords: Cathepsin D, Prognostic factors, Colorectal neoplasms
INTRODUCTION
Traditional prognostic factors of colorectal cancer (CRCA) include the tumor node metastasis (TNM) stage and 'potential' residual disease after initial surgery.1 Other features known to be related to survival include vascular and perineural invasion, tumor necrosis, character of invasive margin, and differentiation.2 Unfortunately, these factors are clearly not sufficient to accurately assess individual risk and to possibly avoid adjuvant systemic therapy. Additional, more refined methods of predicting the prognosis of CRCA patients are required. Assessment of molecular prognostic factors associated with a distinct prognostic outcome would, therefore, be a great help for identification of patients who are likely to benefit from adjuvant therapies, leading to an improvement in prognosis.
Carcinogenesis and development of CRCA are multistep and multistage processes involving cumulative effects of many genes.3,4 For this reason, much effort has been placed on the identification of novel molecular prognostic factors that alone or in combination with clinicopathologic factors may improve the prediction of clinical outcome and determine the appropriate therapeutic approach.
In this study, we analyzed the expression of cathepsin D (CD), an aspartic protease; p53, a tumor suppressor gene; COX-2, a gene involved in carcinogenesis; epidermal growth factor receptor (EGFR), a receptor tyrosine kinase; C-erbB-2, an oncogene; and the Ki-67 protein, a cellular marker for proliferation, that have recently been cited as prognostic factors in patients with CRCA, and determined the correlation between the expression of these proteins and clinicopathologic factors.
MATERIALS AND METHODS
1. Study setting
Tissue was collected from 266 patients with colorectal adenocarcinoma who were candidates for surgical resection. All patients were seen at the Department of Surgery, Yeouido St. Mary's Hospital, The Catholic University of Korea College of Medicine from January 2006 to December 2007. The patient's clinical records were examined, and surgically resected, paraffin-embedded tissue specimens were examined for CD, p53, COX-2, EGFR, C-erbB-2, and Ki-67 expression using direct immunohistochemistry. The tissues were classified as positive or negative for each of the markers, and the clinicopathologic factors of the groups were examined and analyzed. The clinicopathologic factors examined included the patient's age and gender; tumor location, histologic type, degree of differentiation, and stage; degree of bowel wall infiltration; and presence of lymph node (LN) and/or distant metastases. Approval was obtained from the Institutional Review Boards of The Catholic University of Korea College of Medicine. Informed consent was provided according to the Declaration of Helsinki.
2. Study methods
1) Immunohistochemical staining
Immunohistochemical staining was performed using the direct method with the following primary antibodies: anti-CD (Novocastra, Newcastle, UK), anti-p53 (DAKO, Glostrup, Denmark), anti-COX-2 (DAKO), anti-EGFR (DAKO), anti-C-erbB-2 (DAKO), and anti-Ki-67 (DAKO).
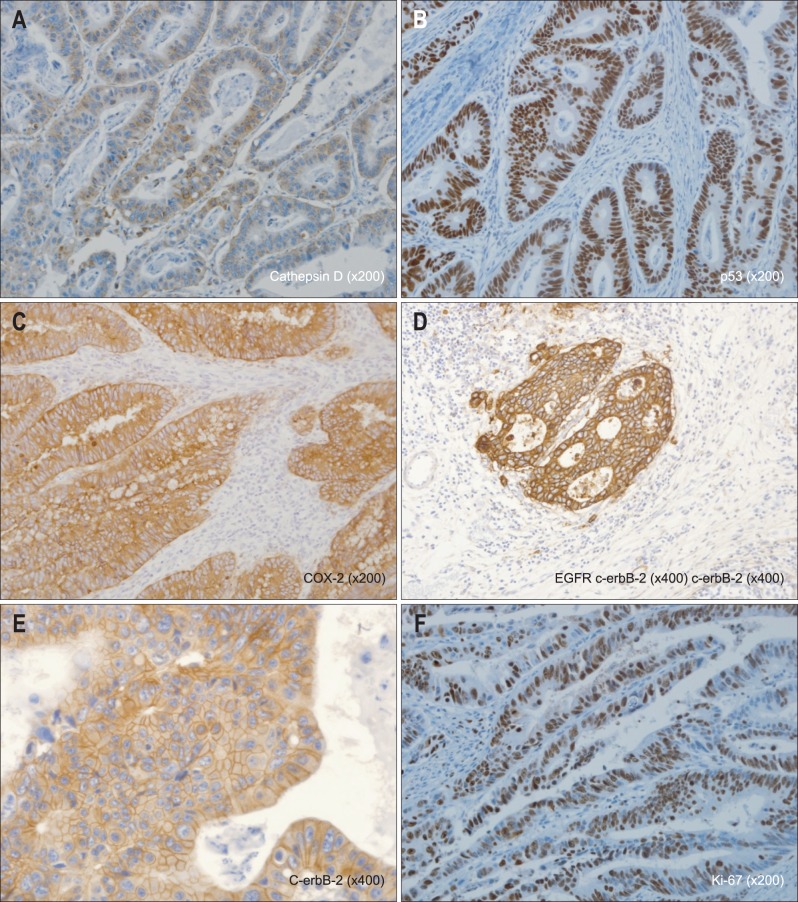
2) Immunohistochemical evaluation (Fig. 1)
Fig. 1.
Immunohistochemical staining for (A) cathepsin D, (B) p53, (C) COX-2, (D) epidermal growth factor receptor (EGFR), (E) C-erbB-2, and (F) Ki-67. Representative images of cytoplasmic staining for cathepsin D (×200) and COX-2 (×200), membrane staining for EGFR (×200) and C-erbB-2 (×400), and nuclear staining for p53 (×200) and Ki-67 (×200).
The stained slides were read by pathologists and categorized as positive or negative. For CD expression,5 any evidence of cytoplasmic staining was considered positive. Additionally, if any of the tumor cells were stained, or if more than 5% of the stromal cell were stained, the sample was considered positive. For p53 expression,1 the nuclear staining in tumor cells was considered positive. Additionally, if more than 10% of tumor cell was stained it was evaluated as positive. For COX-2 expression,6 cytoplasmic staining in at least 10% of the tumor cells was considered positive. For EGFR,7 only the cases showing membrane staining were recognized as positive. If at least one of the tumor cells was stained, it was recognized as positive. For C-erbB-2,7 the American Society of Clinical Oncology/College of American Pathologists guidelines were followed: no staining of tumor cells was scored as 0; partial staining in at least one tumor cell was scored as +; complete staining of the cytoplasm showing moderate staining in more than 10% of the tumor cell was categorized as ++; and complete staining of cytoplasm showing strong staining in more than 10% was categorized as +++. Scores of 0 and + were considered negative, and scores of ++ and +++ were considered positive. Using the Ki-67 proliferation index,1 the percentage of positive cells out of 1,000 cells were calculated at the location where most positive cells expressed in tumor cell nuclei are distributed. Greater than 50% was determined as positive.
3. Statistical analysis
Recurrence was defined on the basis of clinical, radiological, and histopathological results. The survival period was defined as the period between the date of surgery and the date of death. The cancer-free survival period was defined as the date of surgery to the date when any recurrence was discovered. For the analysis of the clinicopathologic factors, a Fisher's exact test or a chi-square test was used. The correlation in the expression of the different proteins, which is dichotomous data, was analyzed using the phi coefficient. The point-biserial correlation (phi) was used. The significance of the univariate prognosis of the variables was evaluated using a univariate COX proportional hazard analysis, and the Kaplan-Meier analysis and log-rank test were used. Multivariate survival analysis was performed for each tumor marker after the data were adjusted for age, gender, and TNM stage. A p-value less than 0.05 was considered significant. SPSS version 14.0 (SPSS Inc., Chicago, IL, USA) was used for all statistical analyses.
RESULTS
1. Patient characteristics
Of the 266 study participants, 160 patients (60.2%) were male, and 106 patients (39.8%) were female, resulting in a male: female ratio of 1.5:1. The average age was 63.0±11.2 years (range, 30 to 87 years). Eighty-one patients (30.5%) had right-side CRCA (defined as cancer located in the ascending and/or transverse colon), and 185 patients (69.5%) had left-side CRCA (defined as cancer located between the descending colon and the rectum). Of the 266 patients, 253 (95.1%) had a nonmucinous adenocarcinoma, and 13 (4.9%) had a mucinous adenocarcinoma. A total of 69 patients had well-differentiated tumors, 177 patients had moderately differentiated tumors, and 12 patients had poorly differentiated tumors. In total, 62 patients were classified as stage I, 78 patients as stage II, 94 patients as stage III, and 32 patients as stage IV (Table 1).
Table 1.
Clinicopathologic Characteristics of the Patients (n=266)
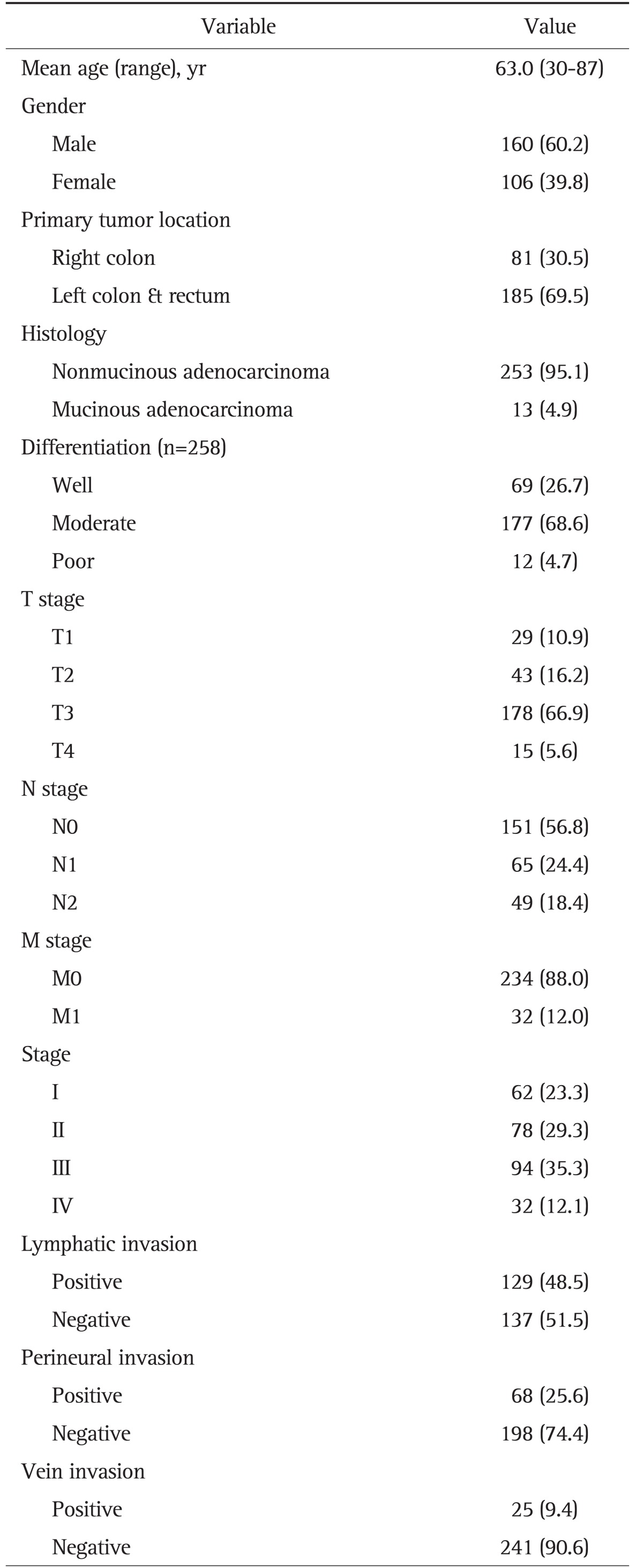
Data are presented as number (%).
T, tumor; N, node; M, metastasis.
2. The correlation between the clinicopathologic factors and expression of molecular markers
Of 266 cases, 103 (38.7%) demonstrated positive results for CD, 162 (60.9%) for p53, and 100 (37.6%) for COX-2. And 95 (35.7%) showed positive results for EGFR, 80 (30.1%) for C-erbB-2, and 198 (74.4%) for Ki-67.
Of the six proteins, CD and COX-2 expression were detected more frequently in tumors of the right colon (p=0.037 and p=0.038, respectively). The proportion of tumors expressing CD, p53, and Ki-67 increased as the T stage increased (p=0.035, p=0.004, and p=0.032, respectively). The expression of CD and COX-2 were also correlated with an increase in the N stage (p=0.021 and p=0.005, respectively). The expression of p53 was correlated with the presence of distant metastases (p=0.012), and while increased CD expression was also associated with the presence of distant metastases, this association was not significant (p=0.075). The cancer stage was correlated with CD, p53, and COX-2 expression (p=0.015, p=0.008, and p=0.019, respectively). The expression of all genes, except C-erbB-2, was correlated with the presence of LN metastases. However, vascular invasion and perineural invasion were not correlated with the expression of any of the examined proteins (Table 2).
Table 2.
Relationship between the Clinicopathologic Factors and Molecular Marker Expression
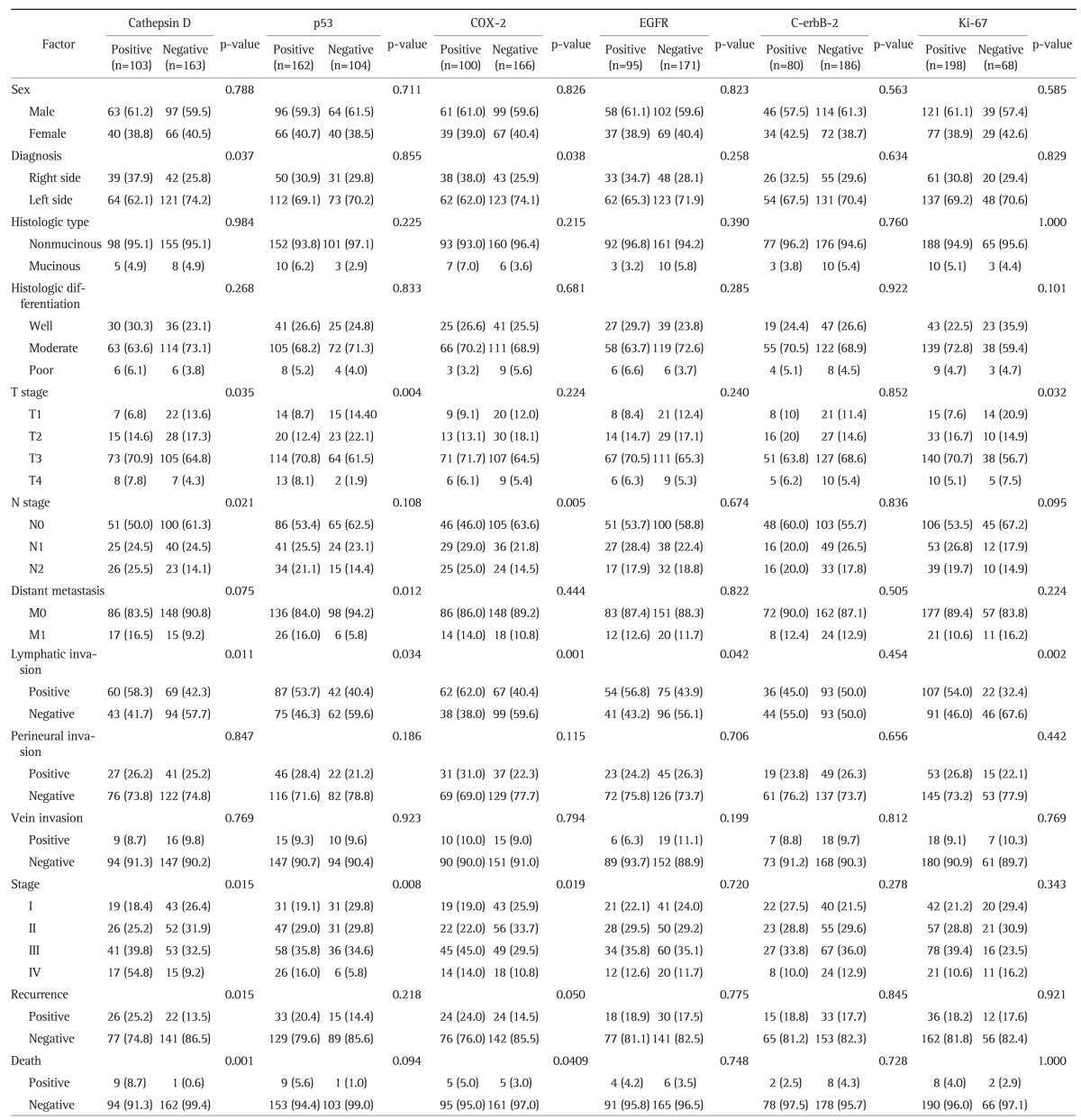
Data are presented as number (%).
3. The correlation between expression of molecular markers and cancer-free survival and colorectal cancer specific survival
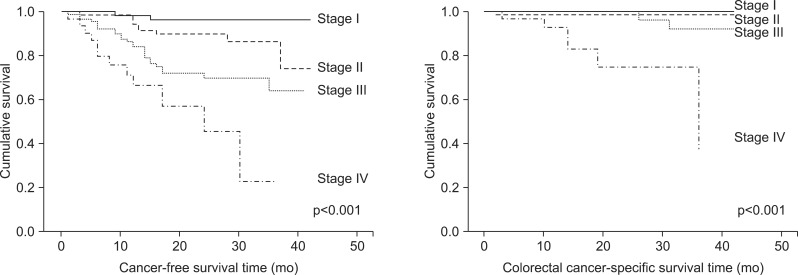
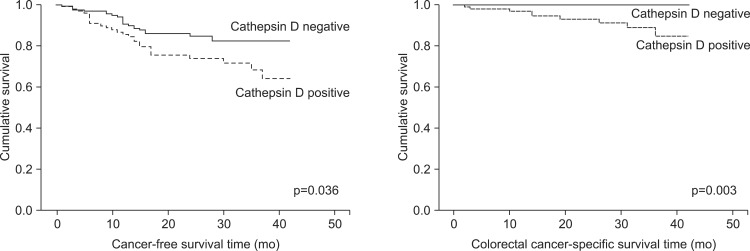
The cancer-free survival rate and colorectal cancer specific survival rate of the patients varied depending on the TNM stage (p<0.001) (Fig. 2). Patients with CD-expressing tumors had a lower cancer-free survival rate (p=0.036) and a lower colorectal cancer specific survival rate (p=0.003) (Fig. 3). Furthermore, in patients with a CD-positive tumors, the risk of recurrence was high (hazard ratio [HR], 1.82; 95% confidence interval [CI], 1.02 to 3.22; p=0.04), and the risk of death was very high (HR, 12.12; 95% CI, 1.52 to 96.52; p=0.02) (Table 3). The multivariate survival analysis, when adjusted for age, gender, and TNM stage, demonstrated that tumoral CD expression did not affect cancer-free survival, but had a significant effect on colorectal cancer specific survival (HR, 8.55; 95% CI, 1.07 to 68.49; p=0.04) (Table 3). However, the expression of the other proteins did not significantly impact cancer-free survival or colorectal cancer specific survival.
Fig. 2.
Cancer-free survival and colorectal cancer-specific survival of patients grouped based on TNM stage.
Fig. 3.
Cancer-free survival and colorectal cancer-specific survival of patients grouped based on cathepsin D expression.
Table 3.
A COX Proportional-Hazard Regression Analysis for Cancer-Free Survival and Colorectal Cancer-Specific Survival
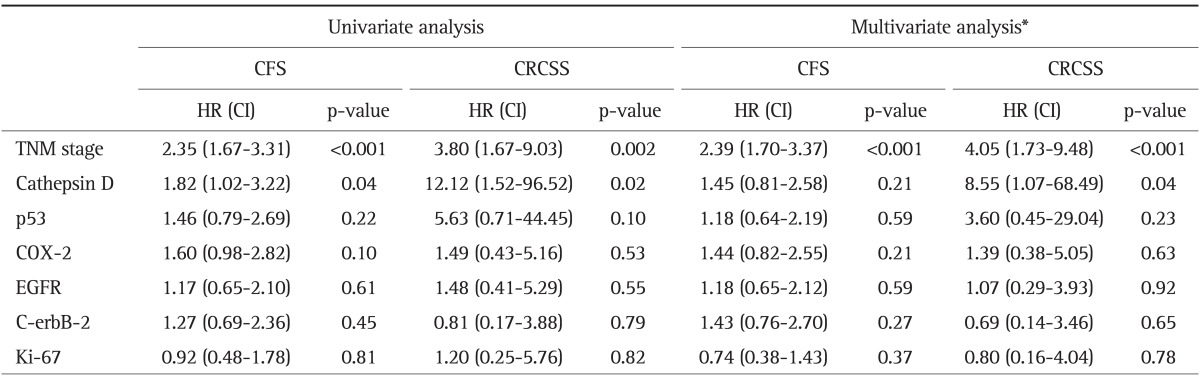
CFS, cancer-free survival; CRCSS, colorectal cancer-specific survival; HR, hazard ratio; CI, confidence interval; TNM, tumor node metastasis; EGFR, epidermal growth factor receptor.
*Adjusted for age, gender, TNM stage, and each tumor marker.
4. The correlations in protein expression and their association with survival
Ki-67 expression was correlated with CD (φ=0.200, p<0.001), C-erbB-2, EGFR, and p53 (p<0.05) expression, but there were no other correlations in the expression of the other proteins (Table 4).
Table 4.
Correlation Coefficient (Phi) for the Tumor Markers
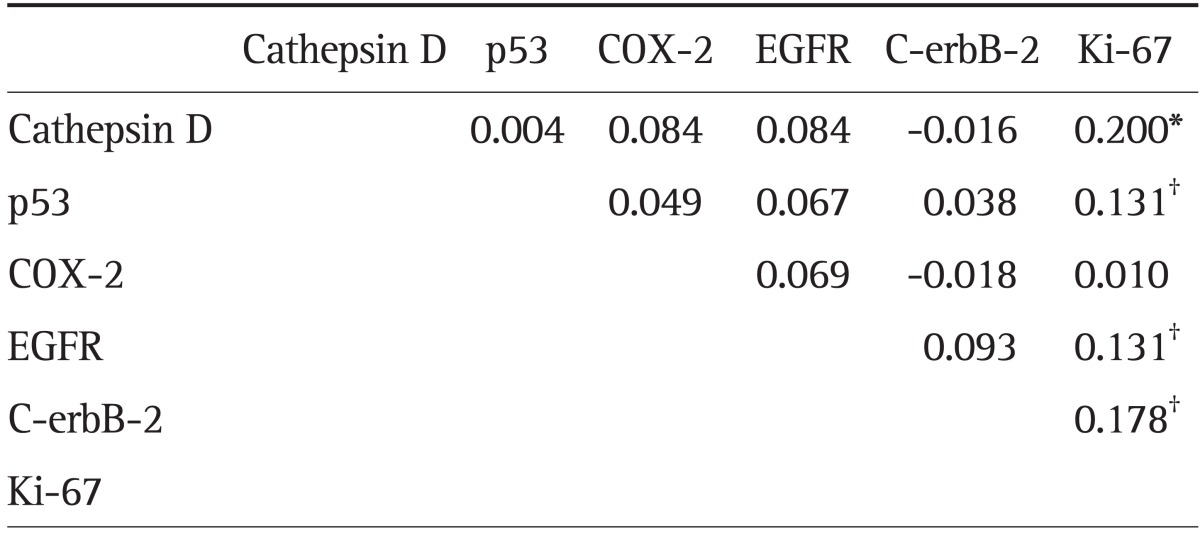
Methodology: the correlations between the tumor markers were evaluated using the Phi (φ) test.
EGFR, epidermal growth factor receptor.
*p<0.001; †p<0.05.
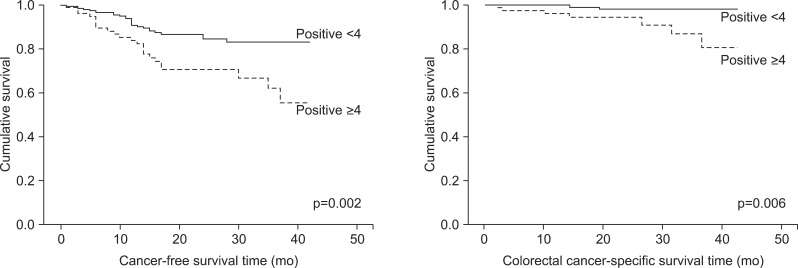
Additionally, we examined the relationship between both the cancer-free survival rate and the colorectal cancer specific survival rate and the number of expressed proteins. There was a significant difference in both the colorectal cancer specific survival rate (p=0.006) and the cancer-free survival rate (p=0.002) of patients with tumors expressing four or more of the proteins when compared to patients expressing fewer than four proteins (Fig. 4). When the number of tumor-expressed proteins was four or more, the risk of recurrence (univariate HR, 2.34; 95% CI, 1.32 to 4.12; p=0.03; multivariate HR, 2.17; 95% CI, 1.22 to 3.86; p=0.008) and the risk of death (univariate HR, 5.48; 95% CI, 1.41 to 21.34; p=0.014; multivariate HR, 4.53; 95% CI, 1.12 to 18.32; p=0.034) were significantly higher than the risks seen in patients with tumors expressing fewer than four of the examined proteins.
Fig. 4.
Cancer-free survival and colorectal cancer-specific survival of patients grouped based on the number of expressed proteins (adjusted for TNM stage).
DISCUSSION
Theodoropoulos et al.5 used immunohistochemistry to examine the expression of CD in tumor cells and reported that CD expression could be detected in 41.6% (25/60) of the tumors based on tumor cell. We observed a lower frequency of CD-positive tumors in our study (38.7%), despite the fact that similar criteria were used to define positive tumors. In contrast, Mayer et al.8 reported a high positivity rate of 87.7% (93/106) based on immunohistochemical detection of CD. The frequency of p53 overexpression also varies significantly in the literature, with rates ranging from 27% to 76%.9 In the current study, the p53-positivity rate was 60.9%. While the COX-2-positivity rate of the tumors was low in this study (37.6%), several studies have reported a COX-2-positivity rate of approximately 80%.2,10,11 Previous studies have shown EGFR expression in 8% to 97% of tumors and C-erbB-2 expression in 0% to 87% of tumors.7 In our study, we detected EGFR expression in 35.7% of the samples and C-erbB-2 expression in 30.1% of the samples. Using a Ki-67 labeling index (LI) greater than 50% as a cutoff, 74.4% of the tumors were Ki-67 positive. A previous study by Huh et al.1 reported the detection of Ki-67 in 52.0% of tumors. Explanations for the wide variation in the expression include differences in methodology, such as using a different antibody; differences in the immunohistochemical staining method; and differences in the evaluation criteria for analysis.
CD, a lysosomal aspartyl endopeptidase, is essential for regulating cell growth and tissue homeostasis of colon epithelium,12 may be involved in CRCA development and growth.5 It has also been associated with the invasion and metastasis of tumor cells.5,13 The increased expression of CD is associated with a number of tumors and is also associated with poor prognosis in breast cancer patients. However, the prognostic value of CD overexpression in CRCA remains unclear.13
The correlation between CD expression in CRCA and specific clinicopathologic factors is controversial. Regarding correlation of CD and tumor stage, some authors have described a significant relationship between overexpression of CD and a trend towards advanced tumor stage.8,13 However, in the majority of the investigations, CD expression was not correlated with stage.13,14 Alternatively, CD expression in tumor stromal cells has been reported to be significantly correlated with lymphatic invasion and LN metastasis.5 In this study, CD expression was significantly correlated with the T stage, N stage, and location of the tumor. We also found an association between CD expression and both lymphatic invasion and recurrence. Importantly, unlike in previous studies, in our study, CD expression was associated with decreased cancer-free survival and colorectal cancer specific survival. Similar to our finding, Kirana et al.15 reported that CD expression in cells from the main tumor body was highly elevated in late stage CRC and showed significant correlation with subsequent distant metastasis and shorter cancer-specific survival. In the multivariate analysis, after adjusting for age, sex, and TNM stage, a significant risk (HR, 8.55; 95% CI, 1.07 to 68.49; p=0.004) was detected, even after the results were adjusted for stage, demonstrating that CD has the potential to be a single-gene prognostic factor for CRCA. However, this result was limited in colorectal cancer specific survival, not in cancer-free survival.
The oncosuppressor protein p53 is a stress response protein that mediates growth suppression through cell cycle arrest or induction of apoptosis in response to DNA damage.9 Furthermore, the functional loss of p53 was proposed as a late event in the transition from adenoma to carcinoma.16 Examining the relationship between p53 expression and the various clinicopathologic factors, the expression of p53 was correlated with the T stage, the presence of distant metastasis, and the stage. Huh et al.1 reported a significant correlation between p53 overexpression and tumor grade, the presence of LN metastasis, stage, and lymphatic invasion. However, contrary to our study, this report did not detect an association with the T stage.1 While several studies have reported the value of p53 as a prognostic factor for CRCA, this remains controversial. A number of studies suggest that p53 expression is associated with a poor prognosis;17,18 however, some studies conclude that p53 expression is associated with a good prognosis.19,20 Furthermore, some studies have reported that there is no correlation between p53 expression and survival.21,22 In agreement with these studies, we also failed to find an association between p53 expression and survival in this study.
Recently, COX-2 has been shown to play a role in both tumor cell growth and the inhibition of apoptosis.11,23 After the publication of a study reporting the decreased relative risk of CRCA in individuals who regularly take nonsteroidal anti-inflammatory drugs (e.g., aspirin),24 many studies have focused on the relationship between COX-2 expression and CRCA. Sheehan et al.2 reported that COX-2 expression was correlated with a higher Duke stage, larger tumors, and the presence of LN metastasis. Furthermore, Tomozawa et al.10 reported a correlation between COX-2 expression and cancer recurrence. They reported that COX-2 expression was the only significant prognostic factor while the traditional TNM staging did not reach statistical significance. Yamauchi et al.11 reported that COX-2 was correlated with a more differentiated tumor, greater invasion, a higher stage, and hepatic metastasis. Additionally, they reported that the cancer-free survival rate was lower in patients with COX-2-positive tumors. In our study, we did find that the expression of COX-2 was associated with a higher N stage, stage and location of the tumor, as well as with recurrence. However, unlike previous studies, there was no significant correlation between COX-2 expression and differentiation, invasion, or hepatic metastasis. And univariate and multivariate analysis have shown that COX-2 expression is not a significant prognostic factor for colorectal cancer specific survival. Studies are conflicting regarding prognostic significance of COX-2 in CRCA with some10,25 supporting and others2,26 refuting independent adverse effect of COX-2. This difference is based upon differences in patient cohorts, COX-2 detection methods, criteria for COX-2 overexpression, and multivariate survival analysis.
The Ki-67 protein is present in the nucleus of all cells during mitosis and is involved in the regulation of the cell cycle.1 Generally, Ki-67 expression is used as a marker of cell proliferation, and while it has been recognized as an independent prognostic factor in prostate and breast cancer,1,27,28 its prognostic value in CRCA remains controversial. Chen et al.29 reported that increased Ki-67 expression was associated with a poor prognosis in CRCA patients. In contrast, Allegra et al.30 reported that Ki-67 expression was associated with a good prognosis in CRCA patients. In our study, Ki-67 expression was only correlated with the T stage. As the T stage increased, the number of tumors with a Ki-67 LI greater than 50% also increased (p=0.032), suggesting that the cancer cells were actively dividing and invading the barrier. Additionally, the expression of CD and Ki-67 were correlated (φ=0.200, p<0.001), indicating that protease expression and active cell division are essential to cancer progression.
Carcinogenesis represents a complex process that involves multiple changes in the controlling pathways of cell proliferation, apoptosis, invasiveness and metastatic spread.3 CRCA results from the progressive accumulation of genetic and epigenetic alterations that lead to cellular transformation and tumor progression.4 Currently, the diagnosis, prognosis, and treatment decisions for CRCA are based on the clinicopathologic analysis of the CRCA tissue. The tumor stage, histological classification, presence or absence of LN and/or distant metastasis, and preoperative serum carcinoembryonic antigen levels have all been recognized as prognostic factors. However, these characteristics cannot completely predict the clinical outcome, and as a result, some patients may undergo unnecessary chemotherapy. Therefore, new diagnostic methods are required to better predict the course of CRCA and to assist in personalizing treatments to each patient. The available diagnostic platforms include multigene-based assays and gene microarrays that may provide reliable information on the prognosis of a patient and/or their sensitivity to treatment.31 Gene expression profiling is a genetic microarray analysis of genetic transcriptional variations between normal and malignant cells, has demonstrated the heterogeneity of breast cancer on the genomic level.32 Perou et al.33 used this method to analyze 65 breast cancer samples and reported six different intrinsic subtypes of breast cancer. These intrinsic subtypes have been recognized as prognostic indicator.34,35 Studies using microarrays to analyze CRCA have also been published. Bertucci et al.36 examined the 5-year survival rate of 22 patients who had been divided into low-risk and high-risk groups based on their gene expression signatures. Interestingly, they reported a 5-year survival of 100% and 30% in the low-risk and high-risk groups, respectively. Furthermore, Eschrich et al.37 developed a 43-gene signature based on 78 tissue samples from Stage II and III CRCA patients and claimed that this signature can predict the 3-year survival rate with 90% accuracy. However, in spite of this progress, no single gene that can be used alone as a prognostic factor for CRCA has been identified.8,21,22,38,39 In this study, we examined the correlation between the number of proteins that were expressed and the clinical outcome. After adjusting for the tumor stage, we detected a significant difference in the colorectal cancer specific survival (p=0.006) and cancer-free survival (p=0.002) of the patients with tumors expressing four or more of the examined proteins compared with patients with tumors expressing less than four of the proteins. While estimating the prognosis of a patient using the expression of a single gene or protein may be difficult, the examination of several genes/proteins may increase the accuracy of the prediction.
In this study, the expression of CD was correlated with a poor prognosis in terms of the cancer-free survival and the colorectal cancer specific survival. Importantly, the high HR (HR, 8.55; 95% CI, 1.07 to 68.49; p=0.04) associated with CD expression from the multivariate analysis, even after adjusting for the tumor stage, demonstrates its potential as an independent, single-gene prognostic factor. However, this result was limited in colorectal cancer specific survival, not in cancer-free survival.
The expression of four or more of the examined proteins was significantly correlated with a poor prognosis, even after adjusting for the stage. Currently, the prognostic value of a single gene marker in CRCA is very controversial, but based on these results, we believe that the number of expressed genes/proteins may be helpful in identifying patients with both early-stage cancer and a potentially poor prognosis, which will help to determine if adjuvant chemotherapy is necessary.
ACKNOWLEDGEMENTS
In Kyu Lee wishes to acknowledge the financial supports of Korean Society of Coloproctology, CJ-Aventis New Research Grant and Grant Number #5-2010-D0166-00004 from Yuhan.
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Huh JW, Lee JH, Kim HR. Expression of p16, p53, and Ki-67 in colorectal adenocarcinoma: a study of 356 surgically resected cases. Hepatogastroenterology. 2010;57:734–740. [PubMed] [Google Scholar]
- 2.Sheehan KM, Sheahan K, O'Donoghue DP, et al. The relationship between cyclooxygenase-2 expression and colorectal cancer. JAMA. 1999;282:1254–1257. doi: 10.1001/jama.282.13.1254. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Rajagopalan H, Nowak MA, Vogelstein B, Lengauer C. The significance of unstable chromosomes in colorectal cancer. Nat Rev Cancer. 2003;3:695–701. doi: 10.1038/nrc1165. [DOI] [PubMed] [Google Scholar]
- 5.Theodoropoulos GE, Panoussopoulos D, Lazaris AC, Golematis BC. Evaluation of cathepsin D immunostaining in colorectal adenocarcinoma. J Surg Oncol. 1997;65:242–248. doi: 10.1002/(sici)1096-9098(199708)65:4<242::aid-jso4>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 6.Nasir A, Kaiser HE, Boulware D, et al. Cyclooxygenase-2 expression in right- and left-sided colon cancer: a rationale for optimization of cyclooxygenase-2 inhibitor therapy. Clin Colorectal Cancer. 2004;3:243–247. doi: 10.3816/CCC.2004.n.005. [DOI] [PubMed] [Google Scholar]
- 7.Kountourakis P, Pavlakis K, Psyrri A, et al. Clinicopathologic significance of EGFR and Her-2/neu in colorectal adenocarcinomas. Cancer J. 2006;12:229–236. doi: 10.1097/00130404-200605000-00012. [DOI] [PubMed] [Google Scholar]
- 8.Mayer A, Fritz E, Fortelny R, Kofler K, Ludwig H. Immunohistochemical evaluation of cathepsin D expression in colorectal cancer. Cancer Invest. 1997;15:106–110. doi: 10.3109/07357909709115762. [DOI] [PubMed] [Google Scholar]
- 9.Tejpar S, Bertagnolli M, Bosman F, et al. Prognostic and predictive biomarkers in resected colon cancer: current status and future perspectives for integrating genomics into biomarker discovery. Oncologist. 2010;15:390–404. doi: 10.1634/theoncologist.2009-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomozawa S, Tsuno NH, Sunami E, et al. Cyclooxygenase-2 overexpression correlates with tumour recurrence, especially haematogenous metastasis, of colorectal cancer. Br J Cancer. 2000;83:324–328. doi: 10.1054/bjoc.2000.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamauchi T, Watanabe M, Kubota T, et al. Cyclooxygenase-2 expression as a new marker for patients with colorectal cancer. Dis Colon Rectum. 2002;45:98–103. doi: 10.1007/s10350-004-6120-5. [DOI] [PubMed] [Google Scholar]
- 12.Saftig P, Hetman M, Schmahl W, et al. Mice deficient for the lysosomal proteinase cathepsin D exhibit progressive atrophy of the intestinal mucosa and profound destruction of lymphoid cells. EMBO J. 1995;14:3599–3608. doi: 10.1002/j.1460-2075.1995.tb00029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sis B, Sağol O, Küpelioğlu A, et al. Prognostic significance of matrix metalloproteinase-2, cathepsin D, and tenascin-C expression in colorectal carcinoma. Pathol Res Pract. 2004;200:379–387. doi: 10.1016/j.prp.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 14.Talieri M, Papadopoulou S, Scorilas A, et al. Cathepsin B and cathepsin D expression in the progression of colorectal adenoma to carcinoma. Cancer Lett. 2004;205:97–106. doi: 10.1016/j.canlet.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 15.Kirana C, Shi H, Laing E, et al. Cathepsin D expression in colorectal cancer: from proteomic discovery through validation using Western blotting, immunohistochemistry, and tissue microarrays. Int J Proteomics. 2012;2012:245819. doi: 10.1155/2012/245819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker SJ, Preisinger AC, Jessup JM, et al. p53 gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res. 1990;50:7717–7722. [PubMed] [Google Scholar]
- 17.Belluco C, Guillem JG, Kemeny N, et al. p53 nuclear protein overexpression in colorectal cancer: a dominant predictor of survival in patients with advanced hepatic metastases. J Clin Oncol. 1996;14:2696–2701. doi: 10.1200/JCO.1996.14.10.2696. [DOI] [PubMed] [Google Scholar]
- 18.Yamaguchi A, Kurosaka Y, Fushida S, et al. Expression of p53 protein in colorectal cancer and its relationship to short-term prognosis. Cancer. 1992;70:2778–2784. doi: 10.1002/1097-0142(19921215)70:12<2778::aid-cncr2820701209>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 19.Lan YT, Chang SC, Li AF, et al. p53 protein accumulation as a prognostic marker in sporadic colorectal cancer. Int J Colorectal Dis. 2007;22:499–506. doi: 10.1007/s00384-006-0194-6. [DOI] [PubMed] [Google Scholar]
- 20.Soong R, Grieu F, Robbins P, et al. p53 alterations are associated with improved prognosis in distal colonic carcinomas. Clin Cancer Res. 1997;3:1405–1411. [PubMed] [Google Scholar]
- 21.Nathanson SD, Linden MD, Tender P, Zarbo RJ, Jacobsen G, Nelson LT. Relationship among p53, stage, and prognosis of large bowel cancer. Dis Colon Rectum. 1994;37:527–534. doi: 10.1007/BF02050985. [DOI] [PubMed] [Google Scholar]
- 22.Smith DR, Ji CY, Goh HS. Prognostic significance of p53 overexpression and mutation in colorectal adenocarcinomas. Br J Cancer. 1996;74:216–223. doi: 10.1038/bjc.1996.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsujii M, DuBois RN. Alterations in cellular adhesion and apoptosis in epithelial cells overexpressing prostaglandin endoperoxide synthase 2. Cell. 1995;83:493–501. doi: 10.1016/0092-8674(95)90127-2. [DOI] [PubMed] [Google Scholar]
- 24.Thun MJ, Namboodiri MM, Heath CW., Jr Aspirin use and reduced risk of fatal colon cancer. N Engl J Med. 1991;325:1593–1596. doi: 10.1056/NEJM199112053252301. [DOI] [PubMed] [Google Scholar]
- 25.Soumaoro LT, Uetake H, Higuchi T, Takagi Y, Enomoto M, Sugihara K. Cyclooxygenase-2 expression: a significant prognostic indicator for patients with colorectal cancer. Clin Cancer Res. 2004;10:8465–8471. doi: 10.1158/1078-0432.CCR-04-0653. [DOI] [PubMed] [Google Scholar]
- 26.Fux R, Schwab M, Thon KP, Gleiter CH, Fritz P. Cyclooxygenase-2 expression in human colorectal cancer is unrelated to overall patient survival. Clin Cancer Res. 2005;11:4754–4760. doi: 10.1158/1078-0432.CCR-04-2586. [DOI] [PubMed] [Google Scholar]
- 27.Bubendorf L, Sauter G, Moch H, et al. Ki67 labelling index: an independent predictor of progression in prostate cancer treated by radical prostatectomy. J Pathol. 1996;178:437–441. doi: 10.1002/(SICI)1096-9896(199604)178:4<437::AID-PATH484>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 28.Jansen RL, Hupperets PS, Arends JW, et al. MIB-1 labelling index is an independent prognostic marker in primary breast cancer. Br J Cancer. 1998;78:460–465. doi: 10.1038/bjc.1998.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen YT, Henk MJ, Carney KJ, et al. Prognostic significance of tumor markers in colorectal cancer patients: DNA index, S-phase fraction, p53 expression, and Ki-67 index. J Gastrointest Surg. 1997;1:266–272. doi: 10.1016/s1091-255x(97)80119-9. [DOI] [PubMed] [Google Scholar]
- 30.Allegra CJ, Paik S, Colangelo LH, et al. Prognostic value of thymidylate synthase, Ki-67, and p53 in patients with Dukes' B and C colon cancer: a National Cancer Institute-National Surgical Adjuvant Breast and Bowel Project collaborative study. J Clin Oncol. 2003;21:241–250. doi: 10.1200/JCO.2003.05.044. [DOI] [PubMed] [Google Scholar]
- 31.Tabernero J, Baselga J. Multigene assays to improve assessment of recurrence risk and benefit from chemotherapy in early-stage colon cancer: has the time finally arrived, or are we still stage locked? J Clin Oncol. 2010;28:3904–3907. doi: 10.1200/JCO.2010.30.0806. [DOI] [PubMed] [Google Scholar]
- 32.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 33.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 34.van de Vijver MJ, He YD, van't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 35.van 't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 36.Bertucci F, Salas S, Eysteries S, et al. Gene expression profiling of colon cancer by DNA microarrays and correlation with histoclinical parameters. Oncogene. 2004;23:1377–1391. doi: 10.1038/sj.onc.1207262. [DOI] [PubMed] [Google Scholar]
- 37.Eschrich S, Yang I, Bloom G, et al. Molecular staging for survival prediction of colorectal cancer patients. J Clin Oncol. 2005;23:3526–3535. doi: 10.1200/JCO.2005.00.695. [DOI] [PubMed] [Google Scholar]
- 38.Kavanagh DO, Chambers G, O'Grady L, et al. Is overexpression of HER-2 a predictor of prognosis in colorectal cancer? BMC Cancer. 2009;9:1. doi: 10.1186/1471-2407-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKay JA, Murray LJ, Curran S, et al. Evaluation of the epidermal growth factor receptor (EGFR) in colorectal tumours and lymph node metastases. Eur J Cancer. 2002;38:2258–2264. doi: 10.1016/s0959-8049(02)00234-4. [DOI] [PubMed] [Google Scholar]