Abstract
Background/Aims
The current study examines the expression of molecular biomarkers in hepatocellular carcinoma (HCC) and whether these findings correlate with the clinicopathologic features of the disease and patient survival.
Methods
We analyzed the immunohistochemical expression of p53, mammalian target of rapamycin (mTOR), c-Met, and insulin-like growth factor 1 receptor (IGF-1R) heat shock protein 70 (HSP70) with the clinicopathologic features of 83 HCCs.
Results
p53 expression was higher in the male patients with undifferentiated histological tumor grades, cirrhosis, and portal vein invasion. High 48 c-Met expression correlated with cirrhosis, and high mTOR expression correlated with the tumor grade and cirrhosis. High IGF-1R expression correlated with the tumor grade and cirrhosis. A multivariate analysis identified a significant relationship between the high expression of p53, tumor grade, and portal vein invasion. In addition, a high expression of mTOR was related to tumor grade and cirrhosis, and a high expression of HSP70 was related to portal vein invasion in a multivariate analysis. The Kaplan-Meier survival curve for patients with high versus low Edmondson grades and p53 expression was statistically significant.
Conclusions
p53, mTOR, and IGF-1R expression correlated with the Edmondson tumor grade in a univariate analysis, while p53 and mTOR correlated with the Edmondson tumor grade in a multivariate analysis. In addition, the tumor grade was found to predict survival. p53 was primarily related to the clinicopathologic features compared to other markers, and it is a poor prognostic factor of survival.
Keywords: p53; mTOR; c-Met; Receptor, IGF type 1; Carcinoma, hepatocellular
INTRODUCTION
Hepatocellular carcinoma (HCC) is the sixth commonest malignancy worldwide and is the third most common cause of cancer-related death. Surgery, including transplantation, remains the only potentially curative modality for HCC, yet recurrence rates are high and long-term survival poor. The ability to predict individual recurrence risk and subsequently prognosis would help to guide surgical and chemotherapeutic treatment.
HCC have extremely heterogeneous nature in the molecular mechanisms responsible for HCC development and progression. HCC can arise anywhere in the liver parenchyma, with or without liver cirrhosis. Various etiologies have been linked to HCC development, the most prominent of which include chronic hepatitis B virus and hepatitis C virus (HCV) infection, chronic alcohol consumption and aflatoxin-B1-contaminated food. Virtually all cirrhosis-inducing conditions can cause HCC, pointing to important interactions with the host microenvironment.1
With advances in understanding of tumor biology, interest in molecular biomarkers of carcinogensis has grown. The biological significance of some proto-oncogenes, tumor suppressor genes, growth factor genes, and virologic factors has been implicated in hepatocarcinogenesis.2 The mechanism of hepatocarcinogenesis is thought to be multifactorial that involves multiple oncogenes.3 Molecular biomarkers not only might predict prognosis for patients with HCC but also may provide targets for potential therapeutic agents.
The individual study such as prognostic significance of p53 overexpression in HCC has been reported,4 but few simultaneous studies on variable molecular biomarkers have been found in the literature in South Korea. We selected molecular biomarkers including mammalian target of rapamycin (mTOR), c-Met, and insulin-like growth factor 1 receptor (IGF-1R) which is related to key signaling pathways in hepatocarcinogenesis and under clinical trial for targeted therapy,5,6 and commonly overexpressed genes in HCC that regulate cell cycle and apoptosis such as p53 and heat shock protein 70 (HSP70). The current study examines the expression of p53, mTOR, c-Met, IGF-1R, and HSP70 in surgically resected HCC and whether the findings correlate with clinicopathologic features and survival.
MATERIALS AND METHODS
1. Patients
From the database of the department of pathology of the Chungnam National University Hospital, 83 cases of radically resected HCCs operated at the department of general surgery from January 1999 to December 2011 were recruited for this study. The case with preoperative portal vein occlusion and transhepatic artery chemoembolization were excluded in this study. We retrospectively reviewed the medical records using electronic medical records. Sixty seven patients were male and 16 were female with ages ranging from 32 to 78. The mean age of the patients was 58.5 years. Regarding the basic liver disease preceding HCC development, 55 cases (66.3%) of liver cirrhosis were recognized. Hepatitis B surface antigen was positive in 59 patients (71.1%) and anti-HCV antibody was positive in eight patients (9.6%) and both were positive in three patients (3.6%). There were 13 patients (15.7%) without an underlying liver disease. The median follow-up period was 39 months (range, 2 to 135 months).
For tissue array production, paraffin blocks from all resected HCC were obtained to conduct a tissue microarray (TMA). Hematoxylin and eosin-stained slides were examined to identify normal and tumor tissue. The tissue arrays were produced by placing single 3-mm cores of tumor tissue and normal from tumor tissue in a recipient block with holes created manually.
The clinicopathologic features of HCCs examined were sex, age, hepatitis infection, tumor size, histological differentiation of tumor cells according to the Edmondson & Steiner's grading system (Fig. 1),7 the presence of liver cirrhosis, portal vein invasion, hepatic artery invasion, microvascular invasion (Table 1).
Fig. 1.
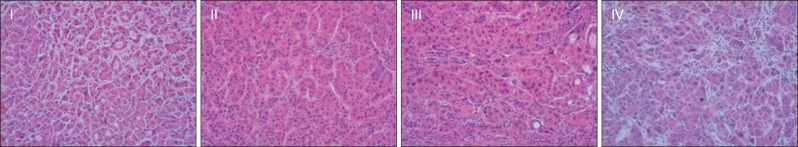
Edmondson-Steiner classifications (grade I to IV) (H&E stain, ×200).
Table 1.
The Clinicopathologic Features of Hepatocellular Carcinoma
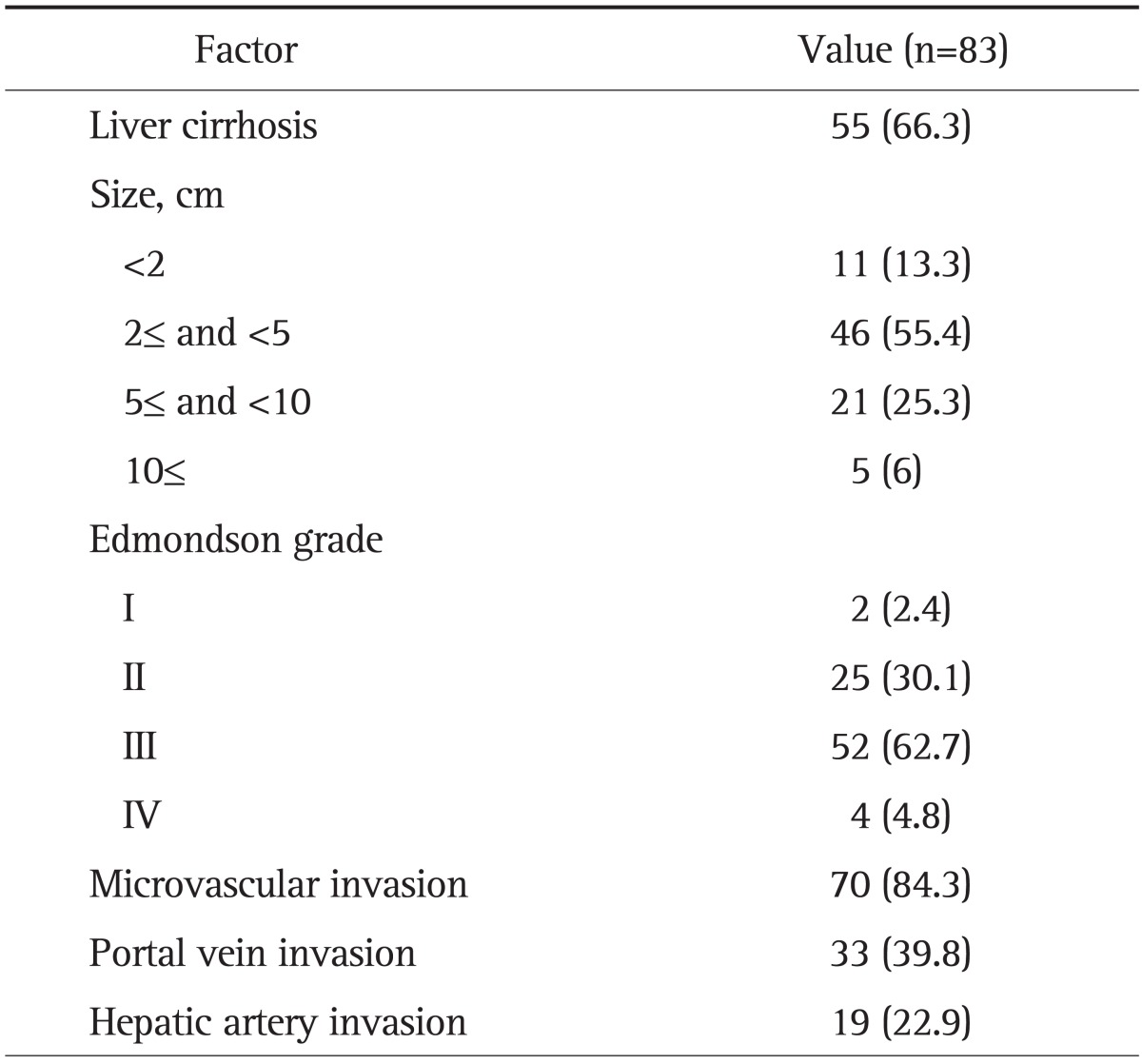
Data are presented as number (%).
2. Immunohistochemical staining
Formalin-fixed, paraffin-embedded sections of tumor tissue obtained from the resected liver specimens of patients with HCC were cut into 4 µm thick sections. The specific antibodies used, sources, dilutions, antigen retrieval, and detection system are listed Table 2. The sections were deparaffinized in xylene and the xylene was subsequently removed with absolute ethanol. Endogenous peroxidase was blocked by treatment with 0.3% hydrogen peroxide in methanol for 20 minutes. Antigen retrieval was performed by autoclave. Sections were then incubated with the primary antibody for 60 minutes at room temperature. DAB (3,3'-diaminobenzidine tetrahydrochloride) was used as a chromogen, and Mayer's hematoxylin counterstain was applied. Omission of primary antibody was used as negative control. We also prepared control groups of normal liver tissue blocks obtained from 10 patients with metastatic colonic carcinoma of the liver.
Table 2.
Primary Antibodies
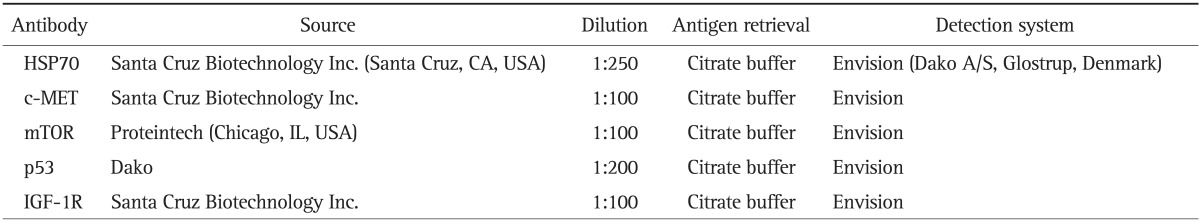
HSP70, heat shock protein 70; mTOR, mammalian target of rapamycin; IGF-1R, insulin-like growth factor 1 receptor.
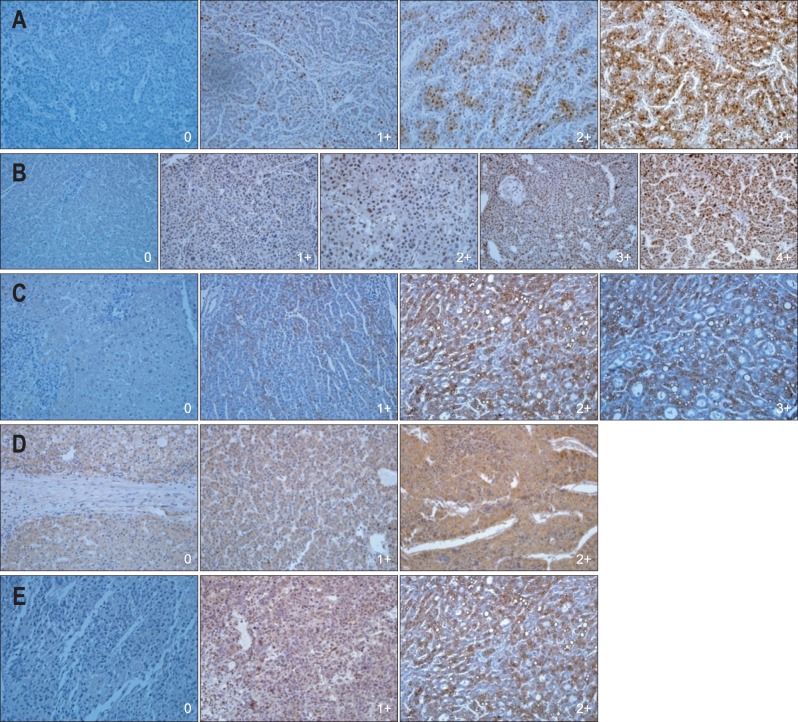
The distribution of positive staining for p53, mTOR, c-Met, IGF-1R, and HSP70 were semiquatitatively assessed by the percentage of positive cells. Three degree of percentage of staining for mTOR, c-Met, IGF-1R, and HSP70 were identified: negative (less than 5%), 1+ (between 5% and 25%), 2+ (between 25% and 50%), 3+ (over 50%); four degree of percentage of staining for p53: negative (less than 5%), 1+ (between 5% and 25%), 2+ (between 25% and 50%), 3+ (between 50% and 75%), 4+ (over 75%) (Fig. 2). Two observers evaluated staining results independently and interpretation differences were resolved by consensus.
Fig. 2.
Immunohistochemical stain. (A) The tumor cells were positive (0, 1+, 2+, 3+) for heat shock protein 70 (×200). (B) The tumor cells were positive (0, 1+, 2+, 3+, 4+) for p53 (×200). (C) The tumor cells were positive (0, 1+, 2+, 3+) for c-Met (×200). (D) The tumor cells were weakly positive (0, 1+, 2+) for mammalian target of rapamycin (×200). (E) The tumor cells were weakly positive (0, 1+, 2+) for insulin-like growth factor 1 receptor (×200).
After the completion of the immunohistochemistry tests, immunohistochemical expression of each markers were correlated with clinicopathologic data and survival in order to ascertain the real prognostic value of these variables. For this purpose the patients were divided into two groups in relation with value of certain parameters, i.e., high (3+, 4+) versus low (negative, 1+, 2+) for p53, high (2+, 3+) versus low (negative, 1+) for mTOR, c-Met, IGF-1R, and HSP70 (Table 3).
Table 3.
The Correlation between Biomarker Overexpression and the Clinicopathologic Parameters of Hepatocellular Carcinoma
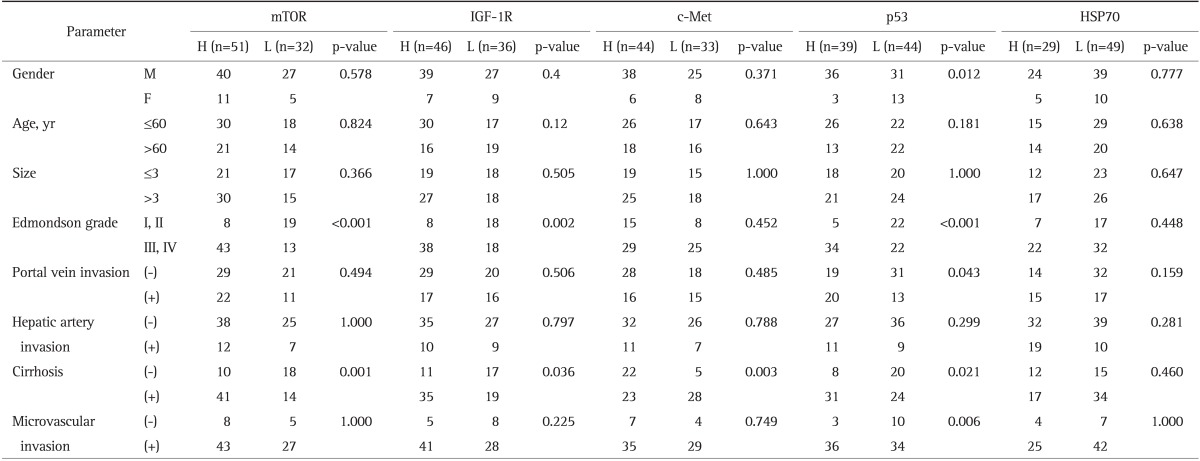
mTOR, mammalian target of rapamycin; IGF-1R, insulin-like growth factor 1 receptor; HSP70, heat shock protein 70; H, high expression; L, low expression; (-) , absence; (+), presence.
3. Statistics
Fisher exact test was used for categorical variables, and logistic regression model for multivariate analysis. Cumulative overall and disease-free survival curve were constructed using the Kaplan-Meier method and survival curve were calculated using log rank test. Factors identified as statistically significant were included by using the Cox multiple regression models. A p<0.05 was considered statistically significant.
4. Ethics statement
The study protocol was approved by the Institutional Review Board of Chungnam National University Hospital (IRB no. 2012-07-002). When research was embarked, Informed consents for human derived material were waived based on traditional version of Regulations of Bioethics in South Korea.
RESULTS
1. Immunohistochemical expression of mTOR
The mTOR expression was positive in 80 of 83 cases (96.4%): mTOR high expression was observed in 51 of the 83 HCCs and low expression was in 32 of the 83 HCCs. The correlation between biomarker overexpression and the clinicopathologic parameters of HCC are shown in Table 3. Univariate analysis identified significant correlation between mTOR overexpression and poor histological differentiation (Edmondson grade) (p<0.001) and liver cirrhosis (p=0.001). Multivariate analysis identified significant correlations between mTOR overexpression and Edmondson grade (p=0.029) and liver cirrhosis (p=0.031) (Table 4).
Table 4.
A Multiple Regression Analysis of the Clinicopathologic Features Demonstrating p53, and mTOR as Independent Prognostic Factors
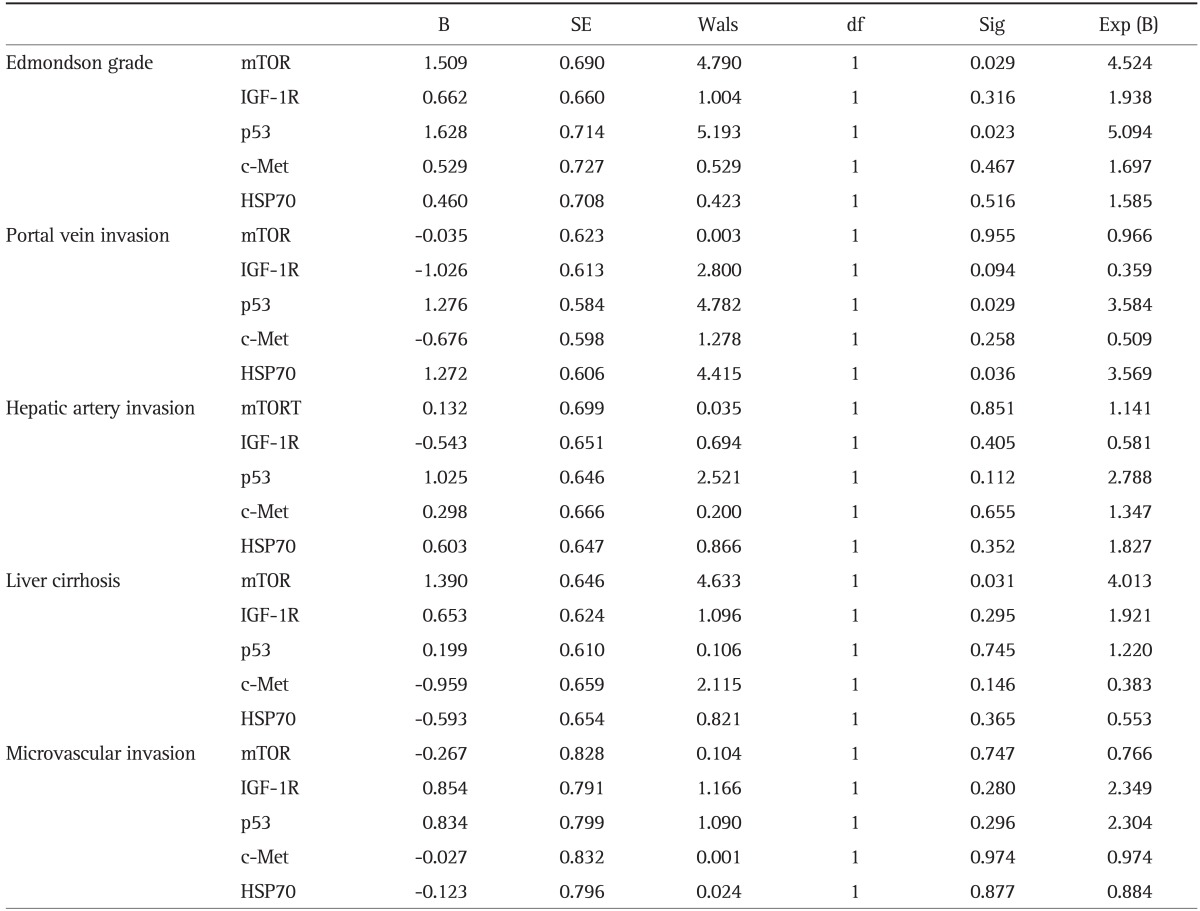
B, regression coefficient; SE, standard error; Wals, Wald statistic; df, degree of freedom; Sig, significance probability; Exp (B), odds ratio; mTOR, mammalian target of rapamycin; IGF-1R, insulin-like growth factor 1 receptor; HSP70, heat shock protein 70.
2. Immunohistochemical expression of IGF-1R
From 82 HCCs (unsatisfactory data, one case), IGF-1R expression was positive in all cases: IGF-1R high expression was observed in 46 of the 82 HCCs and low expression was in 36 of the 82 HCCs. Univariate analysis identified significant correlation between IGF-1R overexpression and Edmondson grade (p=0.002) and liver cirrhosis (p=0.036) (Table 3).
3. Immunohistochemical expression of c-Met
From 77 HCCs (unsatisfactory data, six cases), c-Met expression was positive in 66 of 77 cases (92.8%): c-Met high expression was observed in 44 of the 77 HCCs and low expression was in 33 of the 77 HCCs. Univariate analysis identified significant correlation between c-Met overexpression and liver cirrhosis (p=0.003) (Table 3).
4. Immunohistochemical expression of p53
The p53 expression was positive in 80 of 83 cases (96.4%): p53 high expression was observed in 39 of the 83 HCCs and low expression was in 44 of the 83 HCCs. Univariate analysis identified significant correlation between p53 overexpression and male (p=0.012), Edmondson grade (p<0.001), portal vein invasion (p=0.043), liver cirrhosis (p=0.021) (Table 3). Multivariate analysis identified significant correlations between p53 overexpression and Edmondson grade (p=0.023), portal vein invasion (p=0.029) (Table 4).
5. Immunohistochemical expression of HSP70
From 78 HCCs (unsatisfactory data, five cases), HSP70 expression was positive in 41 of 78 cases (52.7%): HSP70 high expression was observed in 29 of the 78 HCCs and low expression was in 49 of the 78 HCCs. Multivariate analysis identified significant correlation between HSP70 overexpression and portal vein invasion (p=0.036) (Table 4).
6. Survival rates and molecular biomarkers and clinicopathologic data
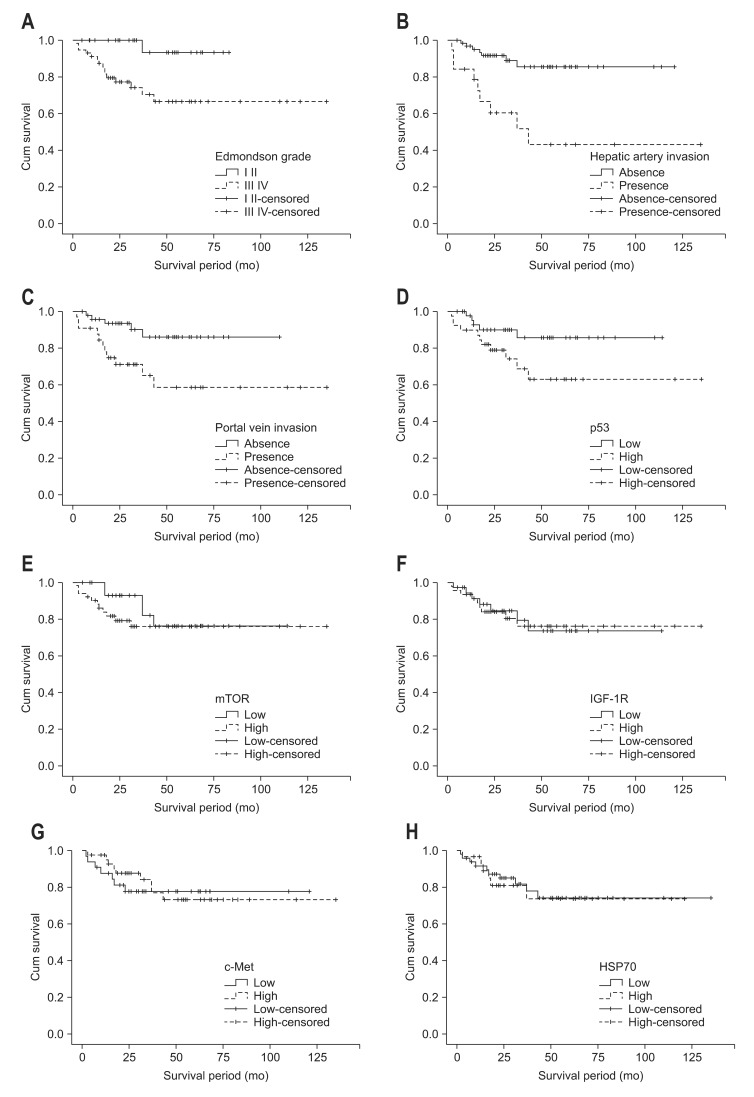
The different survival rate of the patient with mild or over expression of biomarkers as well as clinicopathologic data was evaluated. The Kaplan-Mier survival curves until 60 months of follow-up are shown in Fig. 3. The Kaplan Meier survival for patient with grade III, IV versus grade I, II of Edmondson grade which correlated with biomarkers in this study was statistically significant (66.5% vs 93.3%; p=0.012) (Fig. 3A). In addition portal vein invasion and hepatic artery invasion have prognostic value (Fig. 3B and C). The Kaplan Meier survival for patients with high versus low expression of p53 was statistically significant (63.0% vs 85.6%; p=0.05) (Fig. 3D). No statistic correlations were found in other biomarkers for the overall survival rates (Fig. 3E-H). Cox regression showed tumor grade and hepatic artery invasion was associated with an increased risk of death.
Fig. 3.
The survival curves after surgical resection. (A) Subdivided by the Edmondson grade (Edmondson grades I and II vs Edmondson grades III and IV). (B) Subdivided by hepatic artery invasion (presence vs absence). (C) Subdivided by portal vein invasion (presence vs absence). (D) Subdivided by p53 expression (low p53 expression vs high p53 expression). (E) Subdivided by mammalian target of rapamycin (mTOR) expression (low mTOR expression vs high mTOR expression). (F) Subdivided by insulin-like growth factor 1 receptor (IGF-1R) expression (low IGF-1R expression vs high IGF-1R expression). (G) Subdivided by c-Met expression (low c-Met expression vs high c-Met expression). (H) Subdivided by heat shock protein 70 (HSP70) expression (low HSP70 expression vs high HSP70 expression).
DISCUSSION
As understanding of hepatocarcinogenesis has increased, the myriad of genetic and molecular events that drive the hepatocarcinogenic disease process have been identified. At the molecular level, the mechanisms responsible for the etiopathogenesis of HCC can be summarized into two major groups. First is the activation of specific pathways triggering cancer development and subsequent proliferation, such as those of the epidermal growth factor receptor/mitogen activated protein-kinase, Wnt, IGF, or mTOR. The second group includes the activation of more generic mechanisms/pathways, shared by nearly all types of cancer, which are responsible for the activation of angiogenesis (e.g., vascular endothelial growth factor, platelet-derived growth factor, and relative receptors), insensitivity to apoptosis (Bcl-2, p53, and PI3K/Akt), the inactivation of specific cell cycle check-points (e.g., p53, Rb, TP53, p21, and cycline D1), or for preserving unlimited replicative potential.5,6
The aim of current study was to assess prognostic value of p53, mTOR, c-Met, IGF-1R, and HSP70 and their correlation to clinicopathologic features. Additionally, we investigate whether p53, mTOR, c-Met, IGF-1R, and HSP70 could equally predict the malignant behavior of HCCs. Our result showed that p53 was mostly related to clinicopathologic features compared to other markers and is poor prognostic factor of survival.
TMAs are useful in gene/protein expression profiling of large number of tumors.8 One of the limitations of TMAs is that they are not representative of the entire tumor, and may not show the heterogeneity seen whole sections. Microarray could reveal limitation in some tumors such as ovary or breast carcinoma, composed of heterogeneous tumor cells or prominent fibrous stroma. However, tumor heterogeneity in the HCC tissue section is very rare compared to the other tumors. In present study, TMA technology allowed high-throughput immunohistochemical profiling of cancer specimen.
p53 mutations have been identified in more than a half of all human tumors, including HCC, and they represent the most common genetic abnormality in human cancers.9 Mutations in the p53 gene result in stabilisation of the protein, permitting nuclear accumulation and immunohistochemical detection. Using differing methodologies, p53 has been found to be mutated in 24% to 69% of HCC.10 p53 positive expression was observed in 96.4% in present study. Previous study reported p53 point mutation is common in populations in which hepatitis B viral infection and aflatoxin B1 are prevalent.11 Other study reported p53 mutation in HCC has racial and regional differences.12 Majority of studies provided evidence that the tumor suppressor gene p53 play major role in hepatocarcinogenisis and the prognosis of HCC.13-17 In current study, high expression of p53 correlated to majority of clinicopathologic data (male, Edmondson grade, portal vein invasion, liver cirrhosis) compared to other molecular markers and level of p53 expression affected survival difference, proving most potent prognostic value.
The Met tyrosine kinase is a cell surface receptor for hepatocyte growth factor (HGF). HGF is secreted by mesenchymal cells have a major role in liver regeneration.18 Met activation by HGF can induce cell scattering, invasion, protection from apoptosis and angiogenesis, thereby acting as a powerful expedient for cancer dissemination.19 In current study high expression of c-Met correlated with liver cirrhosis among clinicopathologic data investigated.
Target of rapamycin (TOR) is an essential protein that is conserved in eukaryotes which directly or indirectly regulates the translation of ribosomal proteins, acting as a gatekeeper for cell-cycle progression from G1 to S phase.20 TOR is regulated by phosphatidylinositol 3-kinase (PI3K) or its downstream effector AKT in PI3K/Akt/mTOR pathway. It has been reported that activation of PI3K/Akt/mTOR pathway predicts poor prognosis in HCC.21 The TOR pathway is also upregulated in many human cancers and oncogenic transformation might sensitize cells to TOR inhibitors, therefore representing a novel therapeutic target. In current study high expression of mTOR correlated with Edmondson grade and liver cirrhosis, proving prognostic value.
The IGF-1 signaling pathway has important roles in regulating cellular proliferation and apoptosis.22 Recent studies have discovered changes in the IGF axis that affect the molecular pathogenesis of HCC, including the autocrine production of IGFs, IGF binding proteins (IGFBPs), IGFBP proteases, and IGF receptor expression. Overexpression of IGF-I receptor observed in 20% in HCC.23 In current study high expression of IGF-1R correlated with Edmondson grade and liver cirrhosis, proving prognostic value.
HSP70 belongs to a class of genes (heat shock proteins) implicated in tumorigenesis, in the regulation of cell-cycle progression and apoptosis.24 Chuma et al.25 reported HSP70 as the most abundantly up-regulated gene, among a set of 12,600 genes, in early HCC and significantly overexpressed in advanced HCC as compared to early HCC. Previous study showed diverse result regarding correlation between HSP70 expression and clinocopathologic parameters. In current study high expression of HSP70 correlated with portal vein invasion among clinicopathologic data investigated. Further study is needed regarding clinicopathologic significance of the expression of HSP70.
Among clinicopathologic data investigated, tumor grade, portal vein invasion, and hepatic artery invasion correlated with survival. A correlation between the tumor grade and prognosis has been reported by some26,27 but disputed by others.28 In current study Edmondson tumor grade was found to predict survival. Among molecular biomarker examined in current study, only p53 overexpression correlated with survival. However, high expression of p53, mTOR, and IGF-1R correlated with tumor grade in univariate analysis and high expression of p53, mTOR correlated with tumor grade multiple regression analysis. Correlation between molecular biomarker and histological grade possibly account for pathogenesis of HCC.
The present study has some limitations. High expression of molecular markers did not correlated with clinicopathologic feature such as tumor size, microvascular invasion, hepatic artery invasion statistically. The results may have been caused by selection bias. To obtain well stored eligible paraffin blocks the case with preoperative portal vein occlusion and transcatheter arterial chemoembolization were excluded in this study. In addition, clinocopathologic parameters did not highly correlated with HSP70 as previously expected, and it would call a further study regarding HSP70 as a prognostic indicator. However we attempt to compare a set of markers that might be of prognostic significance in HCC in a multivariated model.
In summary, molecular markers such as p53, mTOR, and IGF-1R correlated with tumor grade. Among them, multivariate analysis identified relation with p53, mTOR, and tumor grade. In addition, tumor grade was found to predict survival. Our result show p53 is most potent molecular predictor for carcinogenesis and malignancy compared to other markers. This suggests that high expression of p53, mTOR, and IGF-1R as prognostic markers could become further criteria in the selection of adequate therapies for HCC.
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 2.Tabor E. Tumor suppressor genes, growth factor genes, and oncogenes in hepatitis B virus-associated hepatocellular carcinoma. J Med Virol. 1994;42:357–365. doi: 10.1002/jmv.1890420406. [DOI] [PubMed] [Google Scholar]
- 3.Rogler CE, Chisari FV. Cellular and molecular mechanisms of hepatocarcinogenesis. Semin Liver Dis. 1992;12:265–278. doi: 10.1055/s-2007-1007398. [DOI] [PubMed] [Google Scholar]
- 4.Sung CO, Yoo BC, Koh KC, Cho JW, Park CK. Prognostic significance of p53 overexpression after hepatic resection of hepatocellular carcinoma. Korean J Gastroenterol. 2005;45:425–430. [PubMed] [Google Scholar]
- 5.Pang RW, Poon RT. From molecular biology to targeted therapies for hepatocellular carcinoma: the future is now. Oncology. 2007;72(Suppl 1):30–44. doi: 10.1159/000111705. [DOI] [PubMed] [Google Scholar]
- 6.Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology. 2008;48:1312–1327. doi: 10.1002/hep.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462–503. doi: 10.1002/1097-0142(195405)7:3<462::aid-cncr2820070308>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 8.Hoos A, Cordon-Cardo C. Tissue microarray profiling of cancer specimens and cell lines: opportunities and limitations. Lab Invest. 2001;81:1331–1338. doi: 10.1038/labinvest.3780347. [DOI] [PubMed] [Google Scholar]
- 9.Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 10.Osada S, Saji S, Kuno T. Clinical significance of combination study of apoptotic factors and proliferating cell nuclear antigen in estimating the prognosis of hepatocellular carcinoma. J Surg Oncol. 2004;85:48–54. doi: 10.1002/jso.20006. [DOI] [PubMed] [Google Scholar]
- 11.Fujimoto Y, Hampton LL, Wirth PJ, Wang NJ, Xie JP, Thorgeirsson SS. Alterations of tumor suppressor genes and allelic losses in human hepatocellular carcinomas in China. Cancer Res. 1994;54:281–285. [PubMed] [Google Scholar]
- 12.Song TJ, Fong Y, Cho SJ, et al. Comparison of hepatocellular carcinoma in American and Asian patients by tissue array analysis. J Surg Oncol. 2012;106:84–88. doi: 10.1002/jso.23036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staib F, Hussain SP, Hofseth LJ, Wang XW, Harris CC. TP53 and liver carcinogenesis. Hum Mutat. 2003;21:201–216. doi: 10.1002/humu.10176. [DOI] [PubMed] [Google Scholar]
- 14.Buendia MA. Genetics of hepatocellular carcinoma. Semin Cancer Biol. 2000;10:185–200. doi: 10.1006/scbi.2000.0319. [DOI] [PubMed] [Google Scholar]
- 15.Ng IO, Lai EC, Chan AS, So MK. Overexpression of p53 in hepatocellular carcinomas: a clinicopathological and prognostic correlation. J Gastroenterol Hepatol. 1995;10:250–255. doi: 10.1111/j.1440-1746.1995.tb01089.x. [DOI] [PubMed] [Google Scholar]
- 16.Jeng KS, Sheen IS, Chen BF, Wu JY. Is the p53 gene mutation of prognostic value in hepatocellular carcinoma after resection? Arch Surg. 2000;135:1329–1333. doi: 10.1001/archsurg.135.11.1329. [DOI] [PubMed] [Google Scholar]
- 17.Nishida N, Fukuda Y, Kokuryu H, et al. Role and mutational heterogeneity of the p53 gene in hepatocellular carcinoma. Cancer Res. 1993;53:368–372. [PubMed] [Google Scholar]
- 18.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43(2 Suppl 1):S45–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 19.Comoglio PM, Giordano S, Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nat Rev Drug Discov. 2008;7:504–516. doi: 10.1038/nrd2530. [DOI] [PubMed] [Google Scholar]
- 20.Bjornsti MA, Houghton PJ. The TOR pathway: a target for cancer therapy. Nat Rev Cancer. 2004;4:335–348. doi: 10.1038/nrc1362. [DOI] [PubMed] [Google Scholar]
- 21.Schmitz KJ, Wohlschlaeger J, Lang H, et al. Activation of the ERK and AKT signalling pathway predicts poor prognosis in hepatocellular carcinoma and ERK activation in cancer tissue is associated with hepatitis C virus infection. J Hepatol. 2008;48:83–90. doi: 10.1016/j.jhep.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 22.Pollak MN, Schernhammer ES, Hankinson SE. Insulin-like growth factors and neoplasia. Nat Rev Cancer. 2004;4:505–518. doi: 10.1038/nrc1387. [DOI] [PubMed] [Google Scholar]
- 23.Scharf JG, Braulke T. The role of the IGF axis in hepatocarcinogenesis. Horm Metab Res. 2003;35:685–693. doi: 10.1055/s-2004-814151. [DOI] [PubMed] [Google Scholar]
- 24.Didelot C, Schmitt E, Brunet M, Maingret L, Parcellier A, Garrido C. Heat shock proteins: endogenous modulators of apoptotic cell death. Handb Exp Pharmacol. 2006;(172):171–198. doi: 10.1007/3-540-29717-0_8. [DOI] [PubMed] [Google Scholar]
- 25.Chuma M, Sakamoto M, Yamazaki K, et al. Expression profiling in multistage hepatocarcinogenesis: identification of HSP70 as a molecular marker of early hepatocellular carcinoma. Hepatology. 2003;37:198–207. doi: 10.1053/jhep.2003.50022. [DOI] [PubMed] [Google Scholar]
- 26.Haratake J, Takeda S, Kasai T, Nakano S, Tokui N. Predictable factors for estimating prognosis of patients after resection of hepatocellular carcinoma. Cancer. 1993;72:1178–1183. doi: 10.1002/1097-0142(19930815)72:4<1178::aid-cncr2820720408>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 27.Lauwers GY, Terris B, Balis UJ, et al. Prognostic histologic indicators of curatively resected hepatocellular carcinomas: a multi-institutional analysis of 425 patients with definition of a histologic prognostic index. Am J Surg Pathol. 2002;26:25–34. doi: 10.1097/00000478-200201000-00003. [DOI] [PubMed] [Google Scholar]
- 28.Lai CL, Wu PC, Lam KC, Todd D. Histologic prognostic indicators in hepatocellular carcinoma. Cancer. 1979;44:1677–1683. doi: 10.1002/1097-0142(197911)44:5<1677::aid-cncr2820440522>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]