Abstract
Background
Aberrant expression of microRNA-146a (miR-146a) has been found in several classes of cancers. However, its expression and clinicopathological contribution in hepatocellular carcinoma (HCC) has not been fully elucidated.
Objective
To explore the clinicopathological significance of the miR-146a level in HCC formalin-fixed paraffin-embedded (FFPE) tissue.
Methods
Eighty-five HCC samples and their para-cancerous normal liver tissues were collected. Total mRNA including miRNA was extracted, and miR-146a expression was determined using real-time RT-PCR. Furthermore, the correlation between the miR-146a expression and clinicopathological parameters was investigated.
Results
MicroRNA-146a expression in HCC tissues was lower compared with that in adjacent non-cancerous hepatic tissues. MicroRNA-146a expression was also related to clinical TNM stage, metastasis, portal vein tumor embolus, and number of tumor nodes.
Conclusions
Down-regulation of miR-146a is related to HCC carcinogenesis and deterioration of HCC. MicroRNA-146a may act as a suppressor miRNA of HCC, and it is therefore a potential prognostic biomarker for HCC patients.
Keywords: Hepatocellular carcinoma, metastasis, miR-146a, oncogenes, paraffin-embedded tissues, pathology, RT-qPCR, tumor biology
Introduction
Primary hepatocellular carcinoma (HCC) is the third leading cause of death from cancers, with an estimated 549,000 deaths per year, which ranks it as the fifth most frequent malignancy worldwide (1-5). The carcinogenesis and progression of HCC are typical of a multistage process. It has been well noted that infection with hepatitis B and C virus (HBV and HCV) is the main etiological factor for the development of HCC (1,4-8). The progression is considered to affect the expression of genes that are critical to cellular processes such as cell cycle control, apoptosis, cell migration and invasion (9-16). Many reports have emphasized the investigation of genes and proteins underlying the progress of HCC; however, their sensitivity and specificity are imperfect (16). Therefore, new biomarkers are urgently required to understand factors causing hepatocarcinogenesis and to link diverse phenotypes in clinical features and prognosis. More vitally, the identification of new biomarkers is needed to predict response possibilities to different therapeutic methods.
MicroRNA (miRNA) has become a global concern for cancer research. These small, non-coding RNAs can inhibit target gene expression by binding the 3’-untranslated region (3’-UTR) of target mRNA, resulting in either mRNA degradation or inhibition of translation to protein. MicroRNAs play critical roles in many normal biological processes involving cell proliferation, differentiation, apoptosis, and stress resistance (17-21). Studies have also revealed that aberrant miRNA expression is correlated with the development and progression of cancers. Thus, miRNAs could be utilized as biomarkers for diagnosis and prognosis prediction of cancers. Furthermore, miRNAs can have oncogenic or tumor suppressor characteristics (20,22-24).
Extensive profiling studies over recent years have shown that a variety of miRNAs are abnormally expressed in HCC (25-29). Nevertheless, there remains a lot to understand as regards the involvement of miRNAs in hepatocarcinogenesis and progression of HCC. Among all the HCC-related miRNAs, miR-146a was reported to be increasingly expressed in serum samples of HCC patients, compared with healthy controls (30). However, an inconsistent expression level of miR-146a in HCC tissue samples was observed by Karakatsanis et al. (31). MicroRNA-146a expression was lower in HCC than that in normal liver tissues. In the present study, we investigated the expression of miRNA-146a in HCC and their matched adjacent non-cancerous liver tissues in formalin-fixed paraffin-embedded (FFPE) surgically resected samples. Moreover, we studied the correlation between miR-146a levels and different clinicopathological parameters of HCC.
Materials and methods
Tissue samples
This retrospective study included 85 cases of HCCs and their paired paraneoplastic liver FFPE tissues. The age of the HCC patients ranged from 29 to 82 years old, with a mean age of 52 years. Clinicopathological information provided from medical records has been summarized in Table I. Adjacent non-cancerous liver tissues were at least 2 cm away from the tumor node. All cases were initial hepatectomies without treatment and were randomly selected from hepatectomies performed over a 1–2 year span in the First Affiliated Hospital of Guangxi Medical University, P. R. China between March 2010 and June 2011. The study protocol was approved by the Ethical Committee of the First Affiliated Hospital of Guangxi Medical University. Written informed consent was obtained from the patients and clinicians for the usage of the samples for research. All samples were reviewed and diagnosed by two independent pathologists.
Table I.
Relationship between the expression of miR-146a and clinicopathological features in HCC 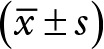 .
.
| Clinicopathological features | n | MiR-146a relevant expression |
|||
|---|---|---|---|---|---|
| 2-Δcq | t | p | |||
| Tissue | HCC | 85 | 0.76 ± 0.53 | –5.532a | < 0.001 |
| Adjacent non-cancerous liver | 85 | 1.30 ± 0.71 | |||
| Age | ≥50 y | 40 | 0.72 ± 0.50 | –0.729 | 0.468 |
| <50 y | 45 | 0.80 ± 0.55 | |||
| Gender | Male | 69 | 0.68 ± 0.47 | –2.447 | 0.024 |
| Female | 16 | 1.11 ± 0.65 | |||
| Differentiation | Well | 5 | 0.47 ± 0.35 | F = 1.01b | 0.369 |
| Moderately | 55 | 0.81 ± 0.57 | |||
| Poorly | 25 | 0.72 ± 0.45 | |||
| Clinical TNM stage | I–II | 22 | 1.24 ± 0.56 | 4.918 | < 0.001 |
| III–IV | 63 | 0.60 ± 0.40 | |||
| Metastasis | Yes | 36 | 0.49 ± 0.36 | –4.498 | < 0.001 |
| No | 49 | 0.96 ± 0.55 | |||
| With cirrhosis | Yes | 43 | 0.72 ± 0.54 | –0.815 | 0.417 |
| No | 42 | 0.81 ± 0.52 | |||
| AFP (μg/L) | ≥400 | 32 | 0.73 ± 0.57 | –0.62 | 0.537 |
| <400 | 36 | 0.81 ± 0.57 | |||
| Portal vein tumor embolus | Yes | 23 | 0.47 ± 0.37 | –3.338 | 0.001 |
| No | 62 | 0.87 ± 0.54 | |||
| Tumor capsular infiltration | No capsular or capsular infiltration | 42 | 0.66 ± 0.44 | –1.763 | 0.081 |
| No capsular infiltration | 43 | 0.86 ± 0.59 | |||
| Tumor nodes | Multi | 35 | 0.56 ± 0.37 | –3.173 | 0.002 |
| Single | 50 | 0.91 ± 0.58 | |||
| Tumor diameter (cm) | ≥5 | 69 | 0.73 ± 0.53 | –1.122 | 0.265 |
| <5 | 16 | 0.90 ± 0.57 | |||
Paired t student's test was performed.
One-way analysis of variance (ANOVA) test was performed.
RT-qPCR
Total RNA including miRNA was extracted from tumor sections using the miRNeasy FFPE Kit (QIAGEN, KJ Venlo, the Netherlands) according to our previous reports (32-34). RNA concentrations were determined by NanoDrop 2000 (Wilmington, DE, USA). A combination of RUN6B and RUN48 was the housekeeping genes for detection of miR-146a expression (33,34). The primers for miR-146a, RNU6B and RNU48, were included in TaqMan® MicroRNA Assays (4427975, Applied Biosystems, Life Technologies, Grand Island, NY, USA). Sequence of miRNA and references used in the paper are as follows: miR-146a (Applied Biosystems Cat. No. 4427975-000468): UGAGAACUGAAUUCCAUGGGUU; RNU6B (Applied Biosystems Cat. No. 4427975-001093): CGCAAGGAUGACACGCAAAUUCGUGAAGCGUUCCAUAUUUUU; RNU48 (Applied Biosystems Cat. No. 4427975-001006): GAUGACCCCAGGUAACUCUGAGUGUGUCGCUGAUGCCAUCACCGCAGCGCUCUGACC. The reverse primers were also used for reverse transcription with TaqMan® MicroRNA Reverse Transcription Kit (4366596, Applied Biosystems, Life Technologies, Grand Island, NY, USA) in a total volume of 10 μL. Real-time qPCR for miRNA was performed with Applied Biosystems PCR7900. The miR-146a abundance in each sample was normalized to its references. The expression of miR-146a in the FFPE experiments was calculated with the formula 2-Δcq (32-35).
Statistical analysis
SPSS 19.0 (Munich, Germany) was used for statistical analysis. Results were representative of three independent experiments unless otherwise stated. Values were presented as the mean ± standard deviation (SD). Student's paired or unpaired t test was used to analyze significance between paired or unpaired groups. One-way analysis of variance (ANOVA) test was used to analyze significance between groups of various differentiation. Statistical significance was determined at a p < 0.05 level.
Results
Expression of miR-146a in the HCC tissues was lower than that in the adjacent non-cancerous hepatic tissues (Table I). Female patients had a higher miR-146a expression than male patients. With regard to clinical TNM stages, miR-146a expression in early stages (I and II) was remarkably higher than that in advanced stages (III and IV). Lower levels of miR-146a were found in HCC patients with metastasis, portal vein tumor embolus, and multiple tumor nodes, compared with patients without corresponding traits. In addition, miR-146a expression was higher in patients with a single tumor node than in those with multiple tumor nodes. However, there was no correlation between miR-146a expression and other clinicopathological features, for instance, age, histological differentiation grades, cirrhosis, plasma AFP concentrations, tumor capsular infiltration, or tumor size.
Discussion
The distinction of miR-146a expression between HCC tissues and non-cancerous liver tissues has only been reported once (31). Using real-time RT-qPCR, miR-146a expression was found to be down-regulated in HCC tissues (60 cases) compared with that in healthy controls (98 cases). The average fold change of miR-146a expression between HCC and normal liver tissues was 0.13 (HCC/normal) (31). In the present study, consistent underexpression of miR-146a in HCC tissues was also observed, as compared with the corresponding adjacent liver tissues in the same patients. The results of our study, together with those of Karakatsanis et al. (31), indicate that miR-146a plays a critical role in the hepatocarcinogenesis, as a tumor suppressor miRNA. However, the average fold change of miR-146a level varied (0.13 versus 0.58). The various controls selected by Karakatsanis et al. (31) and our group (healthy liver tissues versus corresponding non-cancerous liver tissues) may partially explain the disparity. It might be that the miR-146a expression is lower in non-cancerous liver tissue of HCC patients than in liver tissue of healthy controls. It might also be of interest to investigate the dynamic change of miR-146a expression in the hepatocarcinogenesis and progression of HCC. For instance, a comparison of the miR-146a levels in normal liver, cirrhotic tissue, adjacent non-cancerous liver, and HCC tissues would be worthwhile to pursue. Zhu et al. (36) reported that the expression of miR-146a was up-regulated in HUVECs co-cultured with a HCC cell line HCCLM3 compared with HUVECs alone. The miR-146a expression of endothelial cells (ECs) in HCC tissues might also be different. However, this remains to be investigated. We compared the miR-146a level of pure HCC cells from various cell lines (HepG2, HepB3, SNU449, and SMMC7211) with adjacent non-cancerous hepatic tissues without micrangiums by means of CD34 immunostaining (data not shown). Indeed, we found lower miR-146a expression in the HCC cells. This further confirmed the idea that there is down-regulation of miR-146a in HCC tissues.
MicroRNAs can also be identified in serum and plasma in a remarkably stable form, which makes it possible to determine the expression of miRNAs in blood samples (37,38). Gui et al. (30) profiled the circulating miRNAs in seven serum pools (two normal controls, two liver cirrhosis, and three HCC pools) using TaqMan Human MicroRNA Array. MicroRNA-146a was present in a relatively high abundance in serum samples of HCC and cirrhosis patients when compared with normal controls. Gui et al. (30) failed to validate their elevation in a set of serum samples of HCC and liver cirrhosis patients using real-time RT-qPCR. A probable reason for this discrepancy is the variation between the two approaches (microRNA array versus real-time RT-qPCR). However, the cause of the divergence between the miR-146a level in HCC tissues and serum has not been clarified. Further studies will be needed to analyze the relationship of miR-146a expression between HCC tissue and serum samples and to figure out potential mechanisms.
It has previously been shown that SNPs rs2910164 in miR-146a were associated with an increased susceptibility to HCC in an Asian population (39,40). The location of rs2910164 is the stem region opposite to the mature miR-146a sequence, which results in a change from G:U pair to C:U mismatch in the stem structure of the miR-146a precursor. Xu et al. reported that the G allele miR-146a precursor displayed increased production of mature miR-146a compared with the C-allele (41). We also performed a PCR-based restriction fragment length polymorphism assay to investigate the relationship between genetic polymorphism in miRNA precursors and miRNA expression in a larger cohort including the 85 cases (data not shown). Among the 85 cases, 29 cases (34.1%) were CC genotype, 42 (49.4%) were GC genotype, and 14 (16.5%) were GG genotype. No relationship was found between different genotypes and the miR-146a expression level. It would be of great interest to explore how polymorphism influences the mature miR-146a expression in HCC.
Concerning the relationship between miR-146a expression and clinicopathological parameters, Karakatsanis et al. (31) found no association between miR-146a level and any clinicopathological feature. In the present study, miR-146a expression in stage III and IV was found to be lower than that in stage I and II. Moreover, miR-146a expression was down-regulated in the metastatic group compared with that in the non-metastatic group. Additionally, miR-146a expression was correlated with the status of portal vein tumor embolus and the number of tumor nodes. The status of portal vein tumor embolus and tumor nodes generally reflects tumor invasion and metastasis and disease deterioration. The results of our study reveal an obvious relation between miR-146a and the infiltration of tumor cells, migration, invasion, and metastasis of HCC. Hence, it may be valuable to clinically examine miR-146a expression for the prediction of metastasis and deterioration of HCC. Interestingly, we found that the miR-146a levels were lower in males than in females, which has not been reported before. Karakatsanis et al. (31) studied the expression of miR-146a of 60 cases, with 16 females included, and no relationship was observed between miR-146a expression and gender. In the present study, there were only 16 female cases. Further studies in a larger cohort will be warranted to investigate the relationship between miR-146a level and gender. The potential mechanism could be related to the correlation between miR-146a and hormones, because gender-specific-miRNAs do exist (42). However, this also needs further investigations.
The mechanisms of miR-146a being down-regulated in advanced stage of HCC could be related to different target genes. Different target genes of miR-146a have been demonstrated in several cancers. Rho-associated protein kinase (ROCK1) has been reported to be a target of miR-146a in androgen-independent PC3 prostate cancer cells. Transfection of miR-146a suppressed more than 82% of the expression of ROCK1, thereby markedly reducing cell proliferation, invasion, and metastasis in human bone-marrow endothelial cell monolayers (43). The L1 cell adhesion molecule and SMAD family member 4 were both identified as novel targets of miR-146a in gastric cancer (44,45). In gliomas, Notch1 was proved to be a direct target of miR-146a (46). The NF-kappaB regulatory kinase interleukin 1 receptor-associated kinase 1 was verified as a target of miR-146a in pancreatic and breast cancer (47,48). EGFR was also documented as a target of miR-146a in various cancers. Re-expression of miR-146a suppressed tumor growth or inhibited the invasive capacity of prostate, breast, pancreatic, and lung cancer cells (32,46-49). Future work will hopefully discover possible target genes of miR-146a in HCC.
Conclusions
Together with previous reports, the current observations strongly suggest that miR-146a acts as a tumor suppressor miRNA, which plays a vital role in the tumorigenesis and deterioration of human HCC. MicroRNA-146a expression in HCC FFPE samples could be a prognostic biomarker for HCC. Further in vitro and in vivo studies are planned to identify the role and mechanism of miR-146a in the malignant phenotype of HCC cells.
Acknowledgements
The study was supported by the Fund of Guangxi Natural Scientific Research (No. 2010GXNSFB013059, No. 2013GXNSFBA01919), Guangxi Provincial Health Bureau Scientific Research Project (Z2013201), and the Fund of National Natural Science Foundation of China (NSFC 81360327). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. It is a pleasure to thank Jurgen Huylebroeck from Belgium who has given useful suggestions in the manuscript preparation. Minhua Rong and Rongquan He contributed equally to the writing of this paper.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- 1.Yamazaki K, Masugi Y, Sakamoto M. Molecular pathogenesis of hepatocellular carcinoma: altering transforming growth factor-beta signaling in hepatocarcinogenesis . Dig Dis. 2011;29:284–8. doi: 10.1159/000327560. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000 . Int J Cancer. 2001;94:153–6. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 3.Liu B, Mao A, Liu D. The hypothesis of an effective strategy for resistance of hepatocellular carcinoma to therapy-autophagy . West Indian Med J. 2011;60:666–8. [PubMed] [Google Scholar]
- 4.Kudo M. Hepatocellular carcinoma in 2011 and beyond: from the pathogenesis to molecular targeted therapy . Oncology. 2011;81:1–10. doi: 10.1159/000333252. [DOI] [PubMed] [Google Scholar]
- 5.Meguro M, Mizuguchi T, Kawamoto M, Hirata K. The molecular pathogenesis and clinical implications of hepatocellular carcinoma . Int J Hepatol. 2011;2011:818672. doi: 10.4061/2011/818672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zemel R, Issachar A, Tur-Kaspa R. The role of oncogenic viruses in the pathogenesis of hepatocellular carcinoma . Clin Liver Dis. 2011;15:261–79. doi: 10.1016/j.cld.2011.03.001. vii–x. [DOI] [PubMed] [Google Scholar]
- 7.Vertemati M, Moscheni C, Petrella D, Lamperti L, Cossa M, Gambacorta M, et al. Morphometric analysis of hepatocellular nodular lesions in HCV cirrhosis . Pathol Res Pract. 2012;208:240–4. doi: 10.1016/j.prp.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Abdo AA, Al-Ahdal MN, Khalid SS, Helmy A, Sanai FM, Alswat K, et al. IL28B polymorphisms predict the virological response to standard therapy in patients with chronic hepatitis C virus genotype 4 infection . Hepatol Int. 2013;7:533–8. doi: 10.1007/s12072-013-9421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu L, Chen G, Yu H, Qiu X. Clinicopathological significance of RASSF1A reduced expression and hypermethylation in hepatocellular carcinoma . Hepatol Int. 2010;4:423–32. doi: 10.1007/s12072-010-9164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen G, Luo D. Expression of decoy receptor 3 in liver tissue microarrays . Natl Med J India. 2008;21:275–8. [PubMed] [Google Scholar]
- 11.Chen G, Dang YW, Luo DZ, Feng ZB, Tang XL. Expression of heparanase in hepatocellular carcinoma has prognostic significance: a tissue microarray study . Oncol Res. 2008;17:183–9. doi: 10.3727/096504008785114138. [DOI] [PubMed] [Google Scholar]
- 12.Chen G, Luo DZ, Liu L, Feng ZB, Guo F, Li P. Hepatic local micro-environmental immune status in hepatocellular carcinoma and cirrhotic tissues . West Indian Med J. 2006;55:403–8. doi: 10.1590/s0043-31442006000600007. [DOI] [PubMed] [Google Scholar]
- 13.Chen G, Luo D. Over-expression of decoy receptor 3 in gastric precancerous lesions and carcinoma . Ups J Med Sci. 2008;113:297–304. doi: 10.3109/2000-1967-240. [DOI] [PubMed] [Google Scholar]
- 14.Yang M, Chen G, Dang Y, Luo D. Significance of decoy receptor 3 in sera of hepatocellular carcinoma patients . Ups J Med Sci. 2010;115:232–7. doi: 10.3109/03009734.2010.516410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruan ZP, Xu R, Lv Y, Tian T, Wang WJ, Guo H, et al. PTEN enhances the sensitivity of human hepatocellular carcinoma cells to sorafenib . Oncol Res. 2012;20:113–21. doi: 10.3727/096504012x13477145152995. [DOI] [PubMed] [Google Scholar]
- 16.Aravalli RN, Steer CJ, Cressman EN. Molecular mechanisms of hepatocellular carcinoma . Hepatology. 2008;48:2047–63. doi: 10.1002/hep.22580. [DOI] [PubMed] [Google Scholar]
- 17.Winter J, Diederichs S. MicroRNA biogenesis and cancer . Methods Mol Biol. 2011;676:3–22. doi: 10.1007/978-1-60761-863-8_1. [DOI] [PubMed] [Google Scholar]
- 18.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas . Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Omelia EJ, Uchimoto ML, Williams G. Quantitative PCR analysis of blood- and saliva-specific microRNA markers following solid-phase DNA extraction . Anal Biochem. 2013;435:120–2. doi: 10.1016/j.ab.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 20.Hui A, How C, Ito E, Liu FF. Micro-RNAs as diagnostic or prognostic markers in human epithelial malignancies . BMC Cancer. 2011;11:500. doi: 10.1186/1471-2407-11-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Leva G, Briskin D, Croce CM. MicroRNA in cancer: new hopes for antineoplastic chemotherapy . Ups J Med Sci. 2012;117:202–16. doi: 10.3109/03009734.2012.660551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types . Eur J Cancer. 2010;46:298–311. doi: 10.1016/j.ejca.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 23.Siegrist F, Singer T, Certa U. MicroRNA expression profiling by bead array technology in human tumor cell lines treated with interferon-alpha-2a . Biol Proced Online. 2009;11:113–29. doi: 10.1007/s12575-009-9012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papagiannakopoulos T, Kosik KS. MicroRNAs: regulators of oncogenesis and stemness . BMC Med. 2008;6:15. doi: 10.1186/1741-7015-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Augello C, Vaira V, Caruso L, Destro A, Maggioni M, Park YN, et al. MicroRNA profiling of hepatocarcinogenesis identifies C19MC cluster as a novel prognostic biomarker in hepatocellular carcinoma . Liver Int. 2012;32:772–82. doi: 10.1111/j.1478-3231.2012.02795.x. [DOI] [PubMed] [Google Scholar]
- 26.Negrini M, Gramantieri L, Sabbioni S, Croce CM. MicroRNA involvement in hepatocellular carcinoma . Anticancer Agents Med Chem. 2011;11:500–21. doi: 10.2174/187152011796011037. [DOI] [PubMed] [Google Scholar]
- 27.Sato F, Hatano E, Kitamura K, Myomoto A, Fujiwara T, Takizawa S, et al. MicroRNA profile predicts recurrence after resection in patients with hepatocellular carcinoma within the Milan Criteria . PLoS ONE. 2011;6:e16435. doi: 10.1371/journal.pone.0016435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diao S, Zhang JF, Wang H, He ML, Lin MC, Chen Y, et al. Proteomic identification of microRNA-122a target proteins in hepatocellular carcinoma . Proteomics. 2010;10:3723–31. doi: 10.1002/pmic.201000050. [DOI] [PubMed] [Google Scholar]
- 29.Burchard J, Zhang C, Liu AM, Poon RT, Lee NP, Wong KF, et al. MicroRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma . Mol Syst Biol. 2010;6:402. doi: 10.1038/msb.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gui J, Tian Y, Wen X, Zhang W, Zhang P, Gao J, et al. Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies . Clin Sci (Lond) 2011;120:183–93. doi: 10.1042/CS20100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karakatsanis A, Papaconstantinou I, Gazouli M, Lyberopoulou A, Polymeneas G, Voros D. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance . Mol Carcinog. 2013;52:297–303. doi: 10.1002/mc.21864. [DOI] [PubMed] [Google Scholar]
- 32.Chen G, Umelo IA, Lv S, Teugels E, Fostier K, Kronenberger P, et al. MiR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells . PLoS ONE. 2013;8:e60317. doi: 10.1371/journal.pone.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dang Y, Luo D, Rong M, Chen G. Underexpression of miR-34a in hepatocellular carcinoma and its contribution towards enhancement of proliferating inhibitory effects of agents targeting c-MET . PLoS ONE. 2013;8:e61054. doi: 10.1371/journal.pone.0061054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rong M, Chen G, Dang Y. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro . BMC Cancer. 2013;13:21. doi: 10.1186/1471-2407-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen G, Kronenberger P, Teugels E, Umelo IA, De Greve J. Targeting the epidermal growth factor receptor in non-small cell lung cancer cells: the effect of combining RNA interference with tyrosine kinase inhibitors or cetuximab . BMC Med. 2012;10:28. doi: 10.1186/1741-7015-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu K, Pan Q, Zhang X, Kong LQ, Fan J, Dai Z, et al. MiR-146a enhances angiogenic activity of endothelial cells in hepatocellular carcinoma by promoting PDGFRA expression . Carcinogenesis. 2013;34:2071–9. doi: 10.1093/carcin/bgt160. [DOI] [PubMed] [Google Scholar]
- 37.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases . Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 38.Brase JC, Wuttig D, Kuner R, Sultmann H. Serum microRNAs as non-invasive biomarkers for cancer . Mol Cancer. 2010;9:306. doi: 10.1186/1476-4598-9-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu M, Zhao L, Hu S, Yang J. The association between two common polymorphisms in MicroRNAs and hepatocellular carcinoma risk in Asian population . PLoS ONE. 2013;8:e57012. doi: 10.1371/journal.pone.0057012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z, Cao Y, Jiang C, Yang G, Wu J, Ding Y. Lack of association of two common polymorphisms rs2910164 and rs11614913 with susceptibility to hepatocellular carcinoma: a meta-analysis . PLoS ONE. 2012;7:e40039. doi: 10.1371/journal.pone.0040039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu T, Zhu Y, Wei QK, Yuan Y, Zhou F, Ge YY, et al. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma . Carcinogenesis. 2008;29:2126–31. doi: 10.1093/carcin/bgn195. [DOI] [PubMed] [Google Scholar]
- 42.Morgan CP, Bale TL. Sex differences in microRNA regulation of gene expression: no smoke, just miRs . Biol Sex Differ. 2012;3:22. doi: 10.1186/2042-6410-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin SL, Chiang A, Chang D, Ying SY. Loss of mir-146a function in hormone-refractory prostate cancer . RNA. 2008;14:417–24. doi: 10.1261/rna.874808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hou Z, Yin H, Chen C, Dai X, Li X, Liu B, et al. MicroRNA-146a targets the L1 cell adhesion molecule and suppresses the metastatic potential of gastric cancer . Mol Med Report. 2012;6:501–6. doi: 10.3892/mmr.2012.946. [DOI] [PubMed] [Google Scholar]
- 45.Xiao B, Zhu ED, Li N, Lu DS, Li W, Li BS, et al. Increased miR-146a in gastric cancer directly targets SMAD4 and is involved in modulating cell proliferation and apoptosis . Oncol Rep. 2012;27:559–66. doi: 10.3892/or.2011.1514. [DOI] [PubMed] [Google Scholar]
- 46.Mei J, Bachoo R, Zhang CL. MicroRNA-146a inhibits glioma development by targeting Notch1 . Mol Cell Biol. 2011;31:3584–92. doi: 10.1128/MCB.05821-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells . Oncogene. 2008;27:5643–7. doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Vandenboom TG., II Wang Z, Kong D, Ali S, Philip PA, et al. MiR-146a suppresses invasion of pancreatic cancer cells . Cancer Res. 2010;70:1486–95. doi: 10.1158/0008-5472.CAN-09-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu B, Wang N, Wang X, Tong N, Shao N, Tao J, et al. MiR-146a suppresses tumor growth and progression by targeting EGFR pathway and in a p-ERK-dependent manner in castration-resistant prostate cancer . Prostate. 2012;72:1171–8. doi: 10.1002/pros.22466. [DOI] [PubMed] [Google Scholar]


