Summary
We report a long-term follow-up (6.5 years) of a phase I/II clinical trial envisaging the use of autologous genetically modified cultured epidermal stem cells for gene therapy of junctional epidermolysis bullosa, a devastating genetic skin disease. The critical goals of the trial were to evaluate the safety and long-term persistence of genetically modified epidermis. A normal epidermal-dermal junction was restored and the regenerated transgenic epidermis was found to be fully functional and virtually indistinguishable from a normal control. The epidermis was sustained by a discrete number of long-lasting, self-renewing transgenic epidermal stem cells that maintained the memory of the donor site, whereas the vast majority of transduced transit-amplifying progenitors were lost within the first few months after grafting. These data pave the way for the safe use of epidermal stem cells in combined cell and gene therapy for genetic skin diseases.
Highlights
-
•
Long-term restoration of functional epidermis and epidermal-dermal junction
-
•
A defined number of transgenic stem cells sustain the regenerated epidermis
-
•
Human epidermal stem cells maintain the memory of the site of origin
-
•
No adverse events have been reported during a 6.5-year follow-up
De Rosa et al. report a long-term follow up of ex vivo gene therapy of junctional epidermolysis bullosa by means of autologous genetically modified cultured epidermal stem cells. The epidermal-dermal junction and functional epidermis were restored. The transgenic epidermis was sustained by a discrete number of long-lasting, self-renewing epidermal stem cells that maintained the memory of the donor site.
Introduction
The human epidermis is renewed monthly, and daily occurring wounds need timely repair. The processes involved in this regeneration and repair rely on epidermal stem cells, which generate colonies known as holoclones (Barrandon and Green, 1987; Pellegrini et al., 1999a; Rochat et al., 1994). Holoclones produce meroclones and paraclones, which have properties expected of transit-amplifying progenitors (Barrandon and Green, 1987; Pellegrini et al., 1999a). The holoclone-forming cell is the only clonal type that possesses long-term regenerative potential, and is the stem cell of all human squamous epithelia (De Luca et al., 2006). Autologous keratinocyte cultures containing holoclones can permanently restore massive epithelial defects such as skin and ocular burns (Gallico et al., 1984; Pellegrini et al., 1997, 1999b, 2013; Rama et al., 2010; Ronfard et al., 2000).
Inherited epidermolysis bullosa (EB) is a family of rare genetic disorders characterized by structural and mechanical fragility of the integuments, leading to recurrent skin and mucosal blistering and erosions that severely impair the quality of life of EB patients (Fine et al., 2008). Junctional EB (JEB) is marked by blister formation at the level of the lamina lucida of the basement membrane and absence (or severe alteration) of hemidesmosomes. JEB has been divided into three categories: Herlitz (JEB-H), non-Herlitz (JEB-nH), and JEB with pyloric atresia (JEB-PA). JEB-H is an early lethal form and is usually due to deleterious mutations in LAMA3, LAMB3, or LAMC2 genes causing a total absence of laminin 332 (previously known as laminin 5), a heterotrimeric protein that consists of α3, β3, and γ2 chains, and links α6β4 integrins to collagen VII dermal fibrils. Mutations of the same genes cause JEB-nH, which is characterized by reduced expression of laminin 332. JEB-nH can also arise from mutations in COL17A1, the gene encoding collagen XVII, whereas JEB-PA is due to mutations in genes encoding the α6β4 integrin (Fine et al., 2008). There is no cure for EB; treatments are palliative and focused on relieving the devastating clinical manifestations (Carulli et al., 2013).
A phase I/II clinical trial showed that autologous epidermal cultures containing genetically modified holoclones restored a normal epidermis on both upper legs of a patient (Claudio) suffering from a severe form of laminin 332-β3-dependent JEBnH (Mavilio et al., 2006; the phase I/II clinical trial was authorized by the Italian Ministry of Health and approved by the ethics review board of the University of Modena). Epidermal keratinocytes were taken from his palm skin, which, at variance with other affected body sites, contained an appropriate number of holoclones (Mavilio et al., 2006). Cells were transduced ex vivo with a murine leukemia virus (MLV)-based retroviral (RV) vector expressing long terminal repeat (LTR)-driven LAMB3 cDNA and used to prepare transgenic epidermal grafts, which were transplanted onto surgically prepared regions of Claudio’s upper legs. Synthesis of normal levels of functional laminin 332 was observed together with the development of a firmly adherent epidermis that remained stable for 1-year follow-up in the absence of blisters, infections, inflammation, or immune response (Mavilio et al., 2006).
The critical goals of this trial were to evaluate the safety and long-term persistence of the transgenic epidermis. Assessment of these parameters was crucial for continuing the trial, which has been halted since 2007 to allow our cell-culture facility to conform to the 2007 EU directive 1394 imposing Good Manufacturing Practices for any advanced therapy, and to develop ex vivo gene therapy for other forms of EB. We therefore analyzed the epidermis that regenerated on Claudio’s upper legs after a very long (6.5 years) follow-up, during which time the transgenic epidermis underwent a minimum of 80 complete renewing cycles.
Results and Discussion
Clinical Evaluation of the Transplanted Upper Legs
During the 6.5-year follow-up, the epidermis of both of Claudio’s upper legs was normal looking, normally pigmented, and robust, and did not itch or form blisters, either spontaneously or after induced mechanical stress (such as biopsy withdrawal). Tactile and pain sensitivity was present in both legs. In contrast, blisters were consistently observed around the transplanted area. Approximately 3 years after grafting, the patient received a strong contusion on his right upper leg, which would have caused severe blistering in the diseased skin. Blisters did not appear on the bruised area. Three punch biopsies, representative of the whole transplanted area, were taken from Claudio’s upper legs and used for further analyses.
Long-Term Restoration of a Normal Functional Epidermis and Dermal-Epidermal Junction
In situ hybridization using vector-specific laminin 332-β3 probes showed that the regenerated epidermis consisted only of transgenic keratinocytes (Figures 1A and 1B, arrowheads). Of note, the stratum corneum of the transgenic epidermis was thicker than that observed in control leg skin (Figures 1A and 1B, asterisks).
Figure 1.
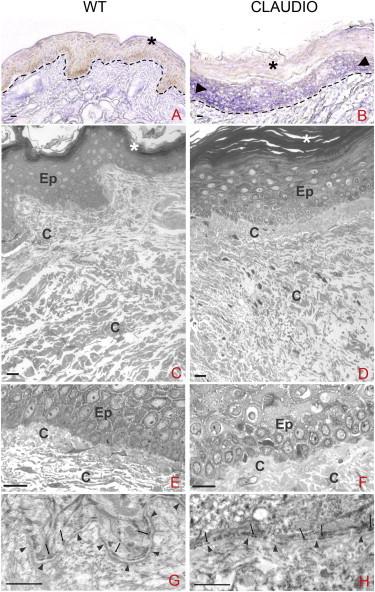
Regeneration of a Functional Transgenic Epidermis
(A and B) In situ hybridization with a vector-specific probe on 20-μm-thick skin sections shows the homogeneous expression of laminin 332-β3 transcripts in all epidermal layers (B, arrowheads). Sections from normal skin were used as a control (A). Dotted lines indicate the basal lamina. Asterisks mark the stratum corneum. Scale bars, 10 μm.
(C–F) Light microscopy of 0.5 μm sections from a skin biopsy of the upper leg of a healthy donor (C and E) and Claudio (D and F) were stained with toluidine blue. In both cases, normal-looking epidermis (Ep) and dermis with well-organized collagen bundles (c) are evident. Asterisks mark the stratum corneum, which is thicker in the regenerated epidermis. Scale bars, 10 μm.
(G and H) Transmission electron microscopy of 70 nm skin sections shows that basement membranes (arrowheads) and hemidesmosomes (arrows) are clearly evident in both control (G) and transgenic (H) skin. Scale bars, 1 μm.
As shown in Figures 1C–1F, the morphology and stratification of the transgenic epidermis (Figures 1D and F) were virtually indistinguishable from those of a normal control (Figures 1C and E), with the exception of the stratum corneum, which was thicker in the transgenic epidermis (Figure 1D, asterisks) as compared with a normal upper leg (Figure 1C, asterisks). The density and organization of collagen bundles in the papillary dermis were consistent with restoration of mechanical strength. Although the transgenic skin had a reduced number of dermal papillae, no blisters, ruptures, and/or detachment of the epidermis from the underlying dermis were ever observed. Transmission electron microscopy (Figures 1G and 1H) showed that the thickness and continuity of the basement membrane (arrowheads) and the number and morphology of hemidesmosomes (arrows) were virtually indistinguishable between control (Figure 1G) and transgenic (Figure 1H) skin, clearly demonstrating that a functional epidermal-dermal junction had been restored.
Laminin 332-β3 was undetectable in the affected skin of Claudio, including the palm keratinocytes that were used to establish cell cultures. A tiny amount of the protein was detected only after immunoprecipitation on cultured cells (Mavilio et al., 2006). In contrast, control and transgenic epidermis expressed virtually identical amounts of laminin 332-β3, which was properly located at the epidermal-dermal junction (Figures 2A and 2B). The absence of laminin 332-β3 is associated with a decrease of α3 and γ2 chains in the protein (and its α6β4 integrin receptor) due to both reduced transcription and increased protein degradation (Matsui et al., 1998; McMillan et al., 1997; Ryan et al., 1999). As shown in Figure 2, the expression of laminin 332-γ2 (Figures 2C and D), 332-α3 (Figures 2E and 2F), α6 integrin (Figures 2G and 2H), and β4 integrin (Figures 2I and 2J) was identical in transgenic (Figures 2B, 2D, 2F, 2H, and 2J) and normal (Figures 2A, 2C, 2E, 2G, and 2I) epidermis.
Figure 2.
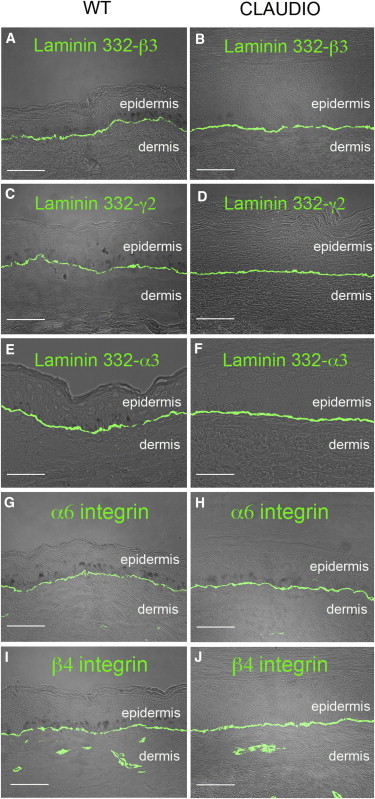
Expression of LAM332 and α6β4 Integrins
(A–J) IF analysis of laminin 332-β3 (A and B), 332-γ2 (C and D) 332-α3 (E and F), α6 integrin (G and H), and β4 integrin (I and J) in control (WT) and transgenic (Claudio) skin sections. The transgenic epidermis expresses normal amounts of laminin 332 and α6β4 integrins properly located at the epidermal-dermal junction. Scale bars, 40 μm.
As shown in Figures 3A–3D, the transgenic epidermis contained normal amounts of keratin 14 (K14, a marker of the epidermal proliferative compartment) and involucrin (INV, a keratinocyte differentiation marker), properly located in basal and suprabasal cells, respectively, suggesting that the balance between proliferation and differentiation had been restored. Elastin (ELN) was equally expressed in normal dermis (Figure 3E) and in the reticular dermis underlying the transgenic epidermis (Figure 3F). However, whereas the control dermis showed peculiar vertical fine fibers (elaunin [arrowheads] and oxytalan [asterisks]; Uitto et al., 2013), Claudio’s dermis contained a diffuse, not well organized elastin fiber network. Previous chronic inflammation and/or continuous wound healing may have caused this alteration of the fine morphology of the elastic fibers.
Figure 3.
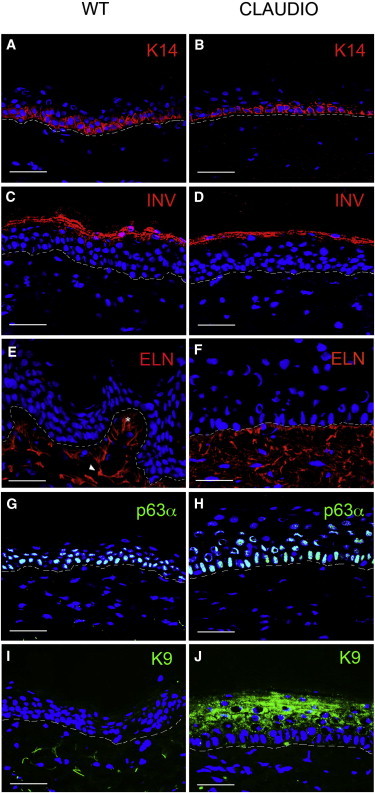
Expression of Epidermal Markers
(A–D) IF analysis of K14 (A and B) and involucrin (C and D) in control (WT) and transgenic (Claudio’s) epidermis.
(E and F) IF analysis of elastin fibers. Network fibers (oxytalan [asterisk] and elaunin [arrowhead]) are expressed at comparable levels, but are differently organized in WT skin (E) and Claudio’s skin (F).
(G and H) IF analysis shows that the ΔNp63α transcription factor is expressed at similar levels and in a comparable number of cells in control (G) and transgenic (H) epidermis.
(I and J) IF analysis shows that K9 is expressed in the upper layers of the transgenic epidermis (J), but is not detected in normal body epidermis (I).
Scale bars, 40 μm. Dotted lines indicate the basal lamina.
The ΔNp63α transcription factor (Mills et al., 1999; Yang et al., 1998, 1999), which is an essential regulator of epithelial stem cell maintenance (Senoo et al., 2007), is highly expressed in holoclones (Di Iorio et al., 2005; Pellegrini et al., 2001), and is instrumental in the clinical performance and long-term persistence of epithelial cultures (Pellegrini et al., 2013; Rama et al., 2010), was expressed at similar levels and in a comparable number of cells in control (Figure 3G) and transgenic (Figure 3H) epidermis.
A Defined Number of Transduced Stem Cells Sustain the Regenerated Epidermis
The human epidermis is renewed monthly; hence, Claudio’s epidermis underwent ∼80 complete renewing cycles in 6.5 years. The long-term maintenance of the regenerated epidermis must be determined by the engraftment of self-renewing transduced epidermal stem cells.
A genome-wide analysis of RV integration sites was performed on DNA extracted from ∼10 mm2 of transgenic epidermis. Libraries of vector-genome junctions, generated by linker-mediated (LM) nested PCR and sequenced to saturation, retrieved six independent integrations unambiguously mapped on the human genome (Table 1). Proviruses were classified as intergenic when they occurred at a distance of >50 kb from any “known gene” (UCSC definition), perigenic when they occurred ≤50 kb upstream or downstream of the transcription start site (TSS) of a known gene, and intragenic when they occurred within the transcribed portion of at least one known gene (Table 1). Three out six integrations were intergenic. One of the three intragenic integrations landed in a gene-dense region, since it was surrounded by five genes in a less than ±40 kb window. None of these integrations belong to a comprehensive compilation of proto-oncogenes and genes associated with common insertion sites (CIS) in mouse tumors (http://microb230.med.upenn.edu/protocols/cancergenes.html). Three of the integrations, whose TSS is more proximal to the MLV integration site, are expressed (according to Affymetrix GeneChip analysis) on keratinocytes cultured under the same conditions used for transduction. The two intragenic integrations landed in the first and second introns of expressed genes, confirming the known integration preferences of γ-RV vectors in human cells (Cattoglio et al., 2010a; Cattoglio et al., 2010b).
Table 1.
List of Retroviral Integration Sites in Skin Biopsies 6.5 Years after Grafting
| ID # | Chromosome | Position | Target Gene | Gene ID | RefSeq | Location | Distance from TSS (kb) | Orientation | Expression |
|---|---|---|---|---|---|---|---|---|---|
| 1sx | 17q21.31 | 42286063 | UBTF | 7343 | NM014233.3 | intron 17 | 11.0 | rev | intermediate |
|
ATXN7L3 TMUB2 ASB16-AS1 ASB16 |
56970 79089 339201 92591 |
NM020218.1 NM024107.2 NR049729.1 NM080863.4 |
upstream downstream upstream downstream |
−10.6 21.7 −22.0 38.0 |
rev for rev for |
intermediate intermediate absent absent |
|||
| 2sx | 7q21.11 | 80704124 | intergenic | ||||||
| 1dx | 1p21.1 | 103642610 | intergenic | ||||||
| 2dx | 15q22.2 | 59637265 | MYO1E | 4643 | NM004998.3 | intron 1 | 27.8 | for | high |
| 3dx | 9p22.3 | 16062571 | intergenic | ||||||
| 4dx | 10q22.2 | 75313406 | USP54 | 159195 | NM152586.3 | intron 2 | 22.0 | rev | intermediate |
For each integration, columns indicate (from left to right) the identification number and biopsy of origin, chromosomal location, nucleotide position, target gene symbol, target gene identification, RefSeq identifier, position with respect to the hit gene, distance from the TSS, provirus orientation (for, forward; rev, reverse), and expression level in cultured keratinocytes as determined by Affymetrix microarray analysis. Expression values are classified as absent, low, intermediate, or high (Mavilio et al., 2006).
Considering an average of two proviral copies per genome and an overall cloning efficiency of ∼30%, we estimate the presence of approximately five to ten independently transduced stem cells in 10 mm2 of epidermis. Since virtually all keratinocytes contain LAM332-β3 transcripts (Figure 1B), it is clear that the entire regenerated epidermis is sustained only by those few engrafted stem cells.
A 10 mm2 sample of cultured epidermis contains ∼15,000 keratinocytes, ∼3,000 of which are clonogenic and the vast majority of which (>95%) are transit-amplifying progenitors. Thus, <150 stem cells are usually contained in 10 mm2 of a cultured graft. Despite years of clinical applications of epidermal cultures, we have no sense of the number of stem cells that can engraft on the wound bed. It is possible that many stem cells are lost during engraftment owing to a hostile in vivo microenvironment. A slightly higher, though comparable, number of stem cells (i.e., 36 and 26 per 10 mm2) was identified at 1- and 4-month follow-up, respectively (Mavilio et al., 2006). However, the initial genome-wide analysis performed after such a short-term follow-up cannot rule out the presence of residual transit-amplifying cells that are still endowed with a significant proliferative potential and/or stem cells at the end of their natural lifespan. That said, the presence of approximately five to ten stem cells per 10 mm2 of epidermis is not far from the estimated stem cell content of a normal epidermis (Pellegrini et al., 1999b; Rochat et al., 1994) and is compatible with the presence of an almost normal repertoire of genetically corrected epidermal stem cells in the regenerated skin. The remarkable proliferative and self-renewal potential of epithelial stem cells (Barbaro et al., 2007; Rochat et al., 1994) is likely to sustain the regenerated transgenic epidermis for the lifetime of the patient.
MLV-RV vectors raised some concerns about the genotoxic risk associated with their uncontrolled insertion into the genome. Insertional activation of a T cell proto-oncogene has been correlated with the occurrence of lymphoproliferative disorders in gene therapy trials of X-linked severe combined immunodeficiency (X-SCID) (Hacein-Bey-Abina et al., 2003, 2008) and Wiscott-Aldrich syndrome (WAS) (Aiuti et al., 2012; Boztug et al., 2010). Such adverse events were not reported when MLV-RV-transduced hematopoietic stem cells were used to treat adenosine deaminase (ADA)-SCID (Aiuti et al., 2009). Thus, specific risk factors may have contributed to the malignant progression observed in X-SCID or WAS. Although MLV-RV integrates preferentially into active regions of the genome (Bushman et al., 2005; Maruggi et al., 2009), insertional mutagenesis might require other oncogenetic factors, which may be related to the cell type, patient’s genetic background, disease, and transgene or other mutations, to determine the onset of a tumor (Cavazza et al., 2013; Howe et al., 2008).
We did not observe tumor development or obtain any evidence of clonal expansion in vivo. Although every biopsy has a unique pattern of integration, the notion that the transgenic epidermis is sustained by only a few engrafted stem cells (five to ten per 10 mm2) indeed minimizes the potential (theoretical) risk of insertional oncogenesis, which has never been reported in human epidermal keratinocytes. Furthermore, transforming human keratinocytes in vitro is quite an awkward task. In evaluating the risk/benefit ratio, one should also consider that severely affected EB patients usually develop aggressive skin cancer as a consequence of the progression of the disease (Fine et al., 2008), and the epidermis can be easily removed if necessary.
Epidermal Stem Cell Plasticity
Epidermal grafts were prepared from palm-derived keratinocytes (Mavilio et al., 2006). Keratin 9 (K9) is expressed in palm and sole keratinocytes, but not in the epidermis covering all other body sites (Langbein et al., 1993). Semiquantitative PCR analysis using K9-specific primers showed that K9 transcripts were equally expressed in control palm keratinocytes and in the transgenic epidermis at 4-month follow-up (not shown). As shown in Figure 3, K9 was still expressed in the upper layers of the transgenic epidermis (Figure 3J) after 6.5 years, whereas it was undetectable in normal body skin (Figure 3I). These findings are consistent with the presence of a thick stratum corneum, which is another hallmark of palm and sole epidermis, and demonstrate that epidermal stem cells maintain the memory of their origin even after 80 complete renewing cycles in vivo, even if they have been transplanted onto a virtually undamaged dermis.
This observation is relevant to all somatic human stem cells. It has been suggested that some somatic stem cells might be capable of differentiating across tissue lineage boundaries and hence might represent versatile effectors of therapeutic tissue regeneration. However, studies proposing such “plasticity” remain very controversial, and existing evidence suggests that such transformations are exceedingly rare (if they occur at all) in vivo and can be accounted for by alternative explanations (Bianco et al., 2013). The notion that palm-derived epidermal stem cells do not possess sufficient plasticity to generate a body epidermis makes one reconsider the supposed plasticity of any somatic stem cell. It formally confirms that the in vivo potential of a stem cell is system restricted and cell autonomous, and strengthens the concept that a stem cell’s function should be verified by its ability to reconstitute a tissue in vivo (Bianco et al., 2013).
Conclusions
In summary, these data demonstrate that (1) the regenerated transgenic epidermis is fully functional and virtually indistinguishable from a normal epidermis, (2) the vast majority of transduced keratinocytes are transit-amplifying progenitors that are lost within a few months after grafting, and (3) the regenerated epidermis is sustained by a discrete number of engrafted, long-lasting, self-renewing transgenic stem cells. These data pave the way for the safe use of epidermal stem cells in combined cell and gene therapy for genetic skin diseases.
Experimental Procedures
Light Microscopy, Transmission Electron Microscopy, and Immunofluorescence
Skin biopsies were fixed in 2.5% glutaraldehyde in Tyrode’s saline pH 7.2 (24 hr at 4°C), postfixed in 1% osmium tetroxide (Electron Microscopy Sciences) for 2 hr at room temperature, dehydrated in ethanol and propylene oxide, and embedded in Spurr resin (Polysciences) as previously described (Quaglino et al., 1991). Semithin sections were stained with toluidine blue and observed with a Zeiss Axiophot light microscope. Ultrathin sections were collected on copper grids, stained with uranyl acetate and lead citrate, and observed with a Jeol 1200 EXII (Jeol) electron microscope.
For immunofluorescence (IF), skin samples were embedded in optimal cutting temperature compound, frozen, and sectioned. IF was performed on 7 μm skin sections as previously described (Mavilio et al., 2006) using laminin 332-β3 6F12 monoclonal antibody (mAb; Acris Antibodies), 332-γ2 D4B5 mAb (Chemicon), 332-α3 BM165 mAb (a gift from Patricia Rousselle, IBCP), 332-α6 450-30A mAb and 332-β4 450-9D mAb (Thermo Fisher Scientific), rabbit purified anti-p63α immunoglobulin G (IgG; PRIMM) (Di Iorio et al., 2005), K10 and K14 guinea pig antisera (Progen), K9 sc-58743 mAb (Santa Cruz Biotechnology), elastin MAB2503 mAb (Millipore), and human involucrin mAb (Leica Biosystems). Alexa Fluor 488 goat anti-mouse or Alexa Fluor 568 goat-anti rabbit (Life Technologies) conjugated secondary antibodies were used for detection. Cell nuclei were stained with DAPI.
Fluorescent signals were monitored under a Zeiss confocal microscope LSM510meta with a Zeiss EC Plan-Neofluar × 40/1.3 oil immersion objective, and analyses were done with the LSM510 Confocal Analyzer (Zeiss). Elastin staining was monitored using an Axio Imager A1 with a Zeiss EC-Plan Neofluar x40, and analyses were done using Axiovision Rel. 4.8 software.
In Situ Hybridization
The probe sequence was obtained by PCR reaction on Claudio’s genomic DNA using 5′-AGTAACGCCATTTTGCAAGG-3′ and 5′-AACAGAAGCGAGAAGCGAAC-3′ primers cloned in pCRII-topoVector (TOPO TA cloning kit; Promega). In situ hybridization was performed as previously described (Brancaccio et al., 2004). Digoxigenin-labeled cRNAs were synthesized using the DIG RNA labeling kit (Roche) according to the manufacturer’s instructions. The antisense RNA probe was transcribed with T7 polymerase, and the control sense probe was transcribed with SP6 polymerase.
Analysis of RV Integration Sites
Integration sites were cloned by LM-PCR as previously described (Recchia et al., 2006). Genomic DNA was digested with MseI and PstI, and ligated to an MseI double-strand linker. LM-PCR was performed with nested primers specific for the LTR and the linker. PCR products were shotgun cloned by the TOPO TA cloning kit (Invitrogen/Life Technologies) into libraries of integration junctions, which were sequenced to saturation. Sequences were mapped onto the human genome by the BLAT genome browser (UCSC Human Genome Project Working Draft, Feb 2009, hg19; http://www.genome.ucsc.edu).
Acknowledgments
This work was supported by the Ministero Istruzione Università e Ricerca, the Italian Ministry of Health and European Community Seventh Framework Program “Optimization of Stem Cell Therapy for Degenerative Epithelial and Muscle Diseases” (OptiStem, HEALTH-F5-2009-223098), and POR-FESR 2007-13-Tecnopolo.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- Aiuti A., Cattaneo F., Galimberti S., Benninghoff U., Cassani B., Callegaro L., Scaramuzza S., Andolfi G., Mirolo M., Brigida I. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N. Engl. J. Med. 2009;360:447–458. doi: 10.1056/NEJMoa0805817. [DOI] [PubMed] [Google Scholar]
- Aiuti A., Bacchetta R., Seger R., Villa A., Cavazzana-Calvo M. Gene therapy for primary immunodeficiencies: Part 2. Curr. Opin. Immunol. 2012;24:585–591. doi: 10.1016/j.coi.2012.07.012. [DOI] [PubMed] [Google Scholar]
- Barbaro V., Testa A., Di Iorio E., Mavilio F., Pellegrini G., De Luca M. C/EBPdelta regulates cell cycle and self-renewal of human limbal stem cells. J. Cell Biol. 2007;177:1037–1049. doi: 10.1083/jcb.200703003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrandon Y., Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc. Natl. Acad. Sci. USA. 1987;84:2302–2306. doi: 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P., Cao X., Frenette P.S., Mao J.J., Robey P.G., Simmons P.J., Wang C.Y. The meaning, the sense and the significance: translating the science of mesenchymal stem cells into medicine. Nat. Med. 2013;19:35–42. doi: 10.1038/nm.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boztug K., Schmidt M., Schwarzer A., Banerjee P.P., Díez I.A., Dewey R.A., Böhm M., Nowrouzi A., Ball C.R., Glimm H. Stem-cell gene therapy for the Wiskott-Aldrich syndrome. N. Engl. J. Med. 2010;363:1918–1927. doi: 10.1056/NEJMoa1003548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio A., Minichiello A., Grachtchouk M., Antonini D., Sheng H., Parlato R., Dathan N., Dlugosz A.A., Missero C. Requirement of the forkhead gene Foxe1, a target of sonic hedgehog signaling, in hair follicle morphogenesis. Hum. Mol. Genet. 2004;13:2595–2606. doi: 10.1093/hmg/ddh292. [DOI] [PubMed] [Google Scholar]
- Bushman F., Lewinski M., Ciuffi A., Barr S., Leipzig J., Hannenhalli S., Hoffmann C. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- Carulli S., Contin R., De Rosa L., Pellegrini G., De Luca M. The long and winding road that leads to a cure for epidermolysis bullosa. Regen. Med. 2013;8:467–481. doi: 10.2217/rme.13.33. [DOI] [PubMed] [Google Scholar]
- Cattoglio C., Maruggi G., Bartholomae C., Malani N., Pellin D., Cocchiarella F., Magnani Z., Ciceri F., Ambrosi A., von Kalle C. High-definition mapping of retroviral integration sites defines the fate of allogeneic T cells after donor lymphocyte infusion. PLoS ONE. 2010;5:e15688. doi: 10.1371/journal.pone.0015688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoglio C., Pellin D., Rizzi E., Maruggi G., Corti G., Miselli F., Sartori D., Guffanti A., Di Serio C., Ambrosi A. High-definition mapping of retroviral integration sites identifies active regulatory elements in human multipotent hematopoietic progenitors. Blood. 2010;116:5507–5517. doi: 10.1182/blood-2010-05-283523. [DOI] [PubMed] [Google Scholar]
- Cavazza A., Moiani A., Mavilio F. Mechanisms of retroviral integration and mutagenesis. Hum. Gene Ther. 2013;24:119–131. doi: 10.1089/hum.2012.203. [DOI] [PubMed] [Google Scholar]
- De Luca M., Pellegrini G., Green H. Regeneration of squamous epithelia from stem cells of cultured grafts. Regen. Med. 2006;1:45–57. doi: 10.2217/17460751.1.1.45. [DOI] [PubMed] [Google Scholar]
- Di Iorio E., Barbaro V., Ruzza A., Ponzin D., Pellegrini G., De Luca M. Isoforms of DeltaNp63 and the migration of ocular limbal cells in human corneal regeneration. Proc. Natl. Acad. Sci. USA. 2005;102:9523–9528. doi: 10.1073/pnas.0503437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine J.D., Eady R.A., Bauer E.A., Bauer J.W., Bruckner-Tuderman L., Heagerty A., Hintner H., Hovnanian A., Jonkman M.F., Leigh I. The classification of inherited epidermolysis bullosa (EB): Report of the Third International Consensus Meeting on Diagnosis and Classification of EB. J. Am. Acad. Dermatol. 2008;58:931–950. doi: 10.1016/j.jaad.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Gallico G.G., 3rd, O’Connor N.E., Compton C.C., Kehinde O., Green H. Permanent coverage of large burn wounds with autologous cultured human epithelium. N. Engl. J. Med. 1984;311:448–451. doi: 10.1056/NEJM198408163110706. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S., Garrigue A., Wang G.P., Soulier J., Lim A., Morillon E., Clappier E., Caccavelli L., Delabesse E., Beldjord K. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe S.J., Mansour M.R., Schwarzwaelder K., Bartholomae C., Hubank M., Kempski H., Brugman M.H., Pike-Overzet K., Chatters S.J., de Ridder D. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langbein L., Heid H.W., Moll I., Franke W.W. Molecular characterization of the body site-specific human epidermal cytokeratin 9: cDNA cloning, amino acid sequence, and tissue specificity of gene expression. Differentiation. 1993;55:57–71. doi: 10.1111/j.1432-0436.1993.tb00033.x. [DOI] [PubMed] [Google Scholar]
- Maruggi G., Porcellini S., Facchini G., Perna S.K., Cattoglio C., Sartori D., Ambrosi A., Schambach A., Baum C., Bonini C. Transcriptional enhancers induce insertional gene deregulation independently from the vector type and design. Mol. Ther. 2009;17:851–856. doi: 10.1038/mt.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui C., Pereira P., Wang C.K., Nelson C.F., Kutzkey T., Lanigan C., Woodley D., Morohashi M., Welsh E.A., Hoeffler W.K. Extent of laminin-5 assembly and secretion effect junctional epidermolysis bullosa phenotype. J. Exp. Med. 1998;187:1273–1283. doi: 10.1084/jem.187.8.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavilio F., Pellegrini G., Ferrari S., Di Nunzio F., Di Iorio E., Recchia A., Maruggi G., Ferrari G., Provasi E., Bonini C. Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nat. Med. 2006;12:1397–1402. doi: 10.1038/nm1504. [DOI] [PubMed] [Google Scholar]
- McMillan J.R., McGrath J.A., Pulkkinen L., Kon A., Burgeson R.E., Ortonne J.P., Meneguzzi G., Uitto J., Eady R.A. Immunohistochemical analysis of the skin in junctional epidermolysis bullosa using laminin 5 chain specific antibodies is of limited value in predicting the underlying gene mutation. Br. J. Dermatol. 1997;136:817–822. [PubMed] [Google Scholar]
- Mills A.A., Zheng B., Wang X.J., Vogel H., Roop D.R., Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Pellegrini G., Traverso C.E., Franzi A.T., Zingirian M., Cancedda R., De Luca M. Long-term restoration of damaged corneal surfaces with autologous cultivated corneal epithelium. Lancet. 1997;349:990–993. doi: 10.1016/S0140-6736(96)11188-0. [DOI] [PubMed] [Google Scholar]
- Pellegrini G., Golisano O., Paterna P., Lambiase A., Bonini S., Rama P., De Luca M. Location and clonal analysis of stem cells and their differentiated progeny in the human ocular surface. J. Cell Biol. 1999;145:769–782. doi: 10.1083/jcb.145.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini G., Ranno R., Stracuzzi G., Bondanza S., Guerra L., Zambruno G., Micali G., De Luca M. The control of epidermal stem cells (holoclones) in the treatment of massive full-thickness burns with autologous keratinocytes cultured on fibrin. Transplantation. 1999;68:868–879. doi: 10.1097/00007890-199909270-00021. [DOI] [PubMed] [Google Scholar]
- Pellegrini G., Dellambra E., Golisano O., Martinelli E., Fantozzi I., Bondanza S., Ponzin D., McKeon F., De Luca M. p63 identifies keratinocyte stem cells. Proc. Natl. Acad. Sci. USA. 2001;98:3156–3161. doi: 10.1073/pnas.061032098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini G., Rama P., Matuska S., Lambiase A., Bonini S., Pocobelli A., Colabelli R.G., Spadea L., Fasciani R., Balestrazzi E. Biological parameters determining the clinical outcome of autologous cultures of limbal stem cells. Regen. Med. 2013;8:553–567. doi: 10.2217/rme.13.43. [DOI] [PubMed] [Google Scholar]
- Quaglino D., Fornieri C., Botti B., Davidson J.M., Pasquali-Ronchetti I. Opposing effects of ascorbate on collagen and elastin deposition in the neonatal rat aorta. Eur. J. Cell Biol. 1991;54:18–26. [PubMed] [Google Scholar]
- Rama P., Matuska S., Paganoni G., Spinelli A., De Luca M., Pellegrini G. Limbal stem-cell therapy and long-term corneal regeneration. N. Engl. J. Med. 2010;363:147–155. doi: 10.1056/NEJMoa0905955. [DOI] [PubMed] [Google Scholar]
- Recchia A., Bonini C., Magnani Z., Urbinati F., Sartori D., Muraro S., Tagliafico E., Bondanza A., Stanghellini M.T., Bernardi M. Retroviral vector integration deregulates gene expression but has no consequence on the biology and function of transplanted T cells. Proc. Natl. Acad. Sci. USA. 2006;103:1457–1462. doi: 10.1073/pnas.0507496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochat A., Kobayashi K., Barrandon Y. Location of stem cells of human hair follicles by clonal analysis. Cell. 1994;76:1063–1073. doi: 10.1016/0092-8674(94)90383-2. [DOI] [PubMed] [Google Scholar]
- Ronfard V., Rives J.M., Neveux Y., Carsin H., Barrandon Y. Long-term regeneration of human epidermis on third degree burns transplanted with autologous cultured epithelium grown on a fibrin matrix. Transplantation. 2000;70:1588–1598. doi: 10.1097/00007890-200012150-00009. [DOI] [PubMed] [Google Scholar]
- Ryan M.C., Lee K., Miyashita Y., Carter W.G. Targeted disruption of the LAMA3 gene in mice reveals abnormalities in survival and late stage differentiation of epithelial cells. J. Cell Biol. 1999;145:1309–1323. doi: 10.1083/jcb.145.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo M., Pinto F., Crum C.P., McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Uitto J., Li Q., Urban Z. The complexity of elastic fibre biogenesis in the skin—a perspective to the clinical heterogeneity of cutis laxa. Exp. Dermatol. 2013;22:88–92. doi: 10.1111/exd.12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A., Kaghad M., Wang Y., Gillett E., Fleming M.D., Dötsch V., Andrews N.C., Caput D., McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R.T., Tabin C., Sharpe A., Caput D., Crum C., McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]


