Summary
Previous studies have suggested that breast cancer stem cells (BCSCs) mediate metastasis, are resistant to radiation and chemotherapy, and contribute to relapse. Although several BCSC markers have been described, it is unclear whether these markers identify the same or independent BCSCs. Here, we show that BCSCs exist in distinct mesenchymal-like (epithelial-mesenchymal transition [EMT]) and epithelial-like (mesenchymal-epithelial transition [MET]) states. Mesenchymal-like BCSCs characterized as CD24−CD44+ are primarily quiescent and localized at the tumor invasive front, whereas epithelial-like BCSCs express aldehyde dehydrogenase (ALDH), are proliferative, and are located more centrally. The gene-expression profiles of mesenchymal-like and epithelial-like BCSCs are remarkably similar across different molecular subtypes of breast cancer, and resemble those of distinct basal and luminal stem cells found in the normal breast. We propose that the plasticity of BCSCs that allows them to transition between EMT- and MET-like states endows these cells with the capacity for tissue invasion, dissemination, and growth at metastatic sites.
Highlights
-
•
BCSCs exist in EMT and MET states with distinct marker and gene-expression profiles
-
•
CSCs from different subtypes of breast cancer express common EMT/MET genes
-
•
EMT and MET BCSC profiles resemble normal mammary basal and luminal stem cells
-
•
BCSCs display plasticity that enables them to transition between EMT and MET states
Breast cancer stem cells (BCSCs) are thought to mediate metastasis and resistance to radiation and chemotherapy. Here, Liu et al. show that BCSCs exist in distinct mesenchymal-like (CD24−CD44+) and epithelial-like (ALDH+) states with different proliferative and invasive capacities, suggesting it may be clinically important to target alternative CSC states.
Introduction
Recent studies have provided strong support for the cancer stem cell (CSC) hypothesis, which suggests that many cancers, including breast cancer, are driven by a subpopulation of cells that display stem cell properties. These cells may mediate metastasis and, by virtue of their relative resistance to chemotherapy and radiation, contribute to treatment relapse. Although some studies have indicated a close association between CSCs and the acquisition of an epithelial-mesenchymal transition (EMT) state (Mani et al., 2008), other studies have suggested that EMT and CSC states are mutually exclusive (Tsuji et al., 2008). The process of EMT plays an important role in embryogenesis as well as in a number of biological processes associated with cancer progression (Thiery et al., 2009). During EMT, epithelial cells lose cell-cell contacts, undergo cytoskeletal remodeling resulting in loss of polarity, and acquire a mesenchymal morphology (Moreno-Bueno et al., 2008). Importantly, EMT is reversible, and the epithelial phenotype generated through mesenchymal-epithelial transition (MET) is characterized by expression of E-cadherin and establishment of cell polarity. Interestingly, a number of pathways that are known to regulate CSCs, including Notch, hedgehog, Wingless (Wnt), transforming growth factor-β (TGFβ), and nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB), also are capable of inducing EMT (Shin et al., 2010; Takebe et al., 2011; Yoo et al., 2011). However, other pathways that regulate CSCs, including those involving bone morphogenetic proteins (BMPs) and human epidermal growth factor receptor (HER) signaling, promote MET (Korkaya et al., 2012; Samavarchi-Tehrani et al., 2010). Further studies are needed to more fully define the relationship among EMT, MET, and CSCs.
The development of biomarkers to identify BCSCs by our group and others, as well as validation of in vitro and mouse models, has facilitated the isolation and characterization of BCSC from both murine and human tumors (Al-Hajj et al., 2003; Dontu et al., 2003; Ginestier et al., 2007). In human breast cancer, tumor-initiating cells were first identified by virtue of their expression of the cell surface marker profile CD24−CD44+. In primary breast xenografts, cells expressing these markers were enriched for their ability to initiate tumors in immunodeficient nonobese diabetic (NOD)/severe combined immunodeficiency (SCID) mice (Al-Hajj et al., 2003). More recently, we have shown that both normal and malignant breast stem cells that express the enzyme aldehyde dehydrogenase (ALDH), as assessed by the ALDEFLUOR assay, are also enriched for tumor-initiating characteristics (Ginestier et al., 2007). Furthermore, in primary breast xenografts, CD24−CD44+ and ALDH identified overlapping, but nonidentical cell populations, each capable of initiating tumors in NOD/SCID mice (Ginestier et al., 2007). Tumor cells that simultaneously expressed both CSC markers (i.e., CD24−CD44+ and ALDH+) displayed the greatest tumor-initiating capacity, generating tumors in NOD/SCID mice from as few as 20 cells (Ginestier et al., 2007). Subsequently, CD44, CD24, and ALDH were reported to be expressed in CSCs from a wide variety of carcinomas, including those of the pancreas, colon, lung, ovary, and prostate gland (Eramo et al., 2008; Huang et al., 2009; Kryczek et al., 2012; Li et al., 2007; Prince et al., 2007). In addition to carcinomas, these markers have also proven useful for isolating CSCs from hematologic malignancies (Storms et al., 1999) and sarcomas. This suggests that CSCs across a wide variety of malignancies may share marker expression as well as biological characteristics. However, it remains unclear whether tumors contain multiple types of CSCs and whether CSC markers identify distinct CSC populations.
Here, we show that BCSCs exist in distinct EMT and MET states characterized by expression of distinct CSC markers. We find that the gene-expression profiles of mesenchymal-like BCSC resemble those of basal stem cells, whereas the profiles of epithelial-like BCSCs resemble those of luminal stem cells in the normal breast. Furthermore, we show that BCSCs are endowed with a plasticity that enables them to transition between these EMT and MET states. Based on these studies, we propose that reversible transitions between mesenchymal-like and epithelial-like stem cell states in a process regulated by the tumor microenvironment are necessary for these tumor cells to invade and form metastasis at distant sites.
Results
CD24−CD44+ and ALDH Identify Anatomically Distinct BCSCs with Distinct Gene-Expression Profiles Independent of Molecular Subtype
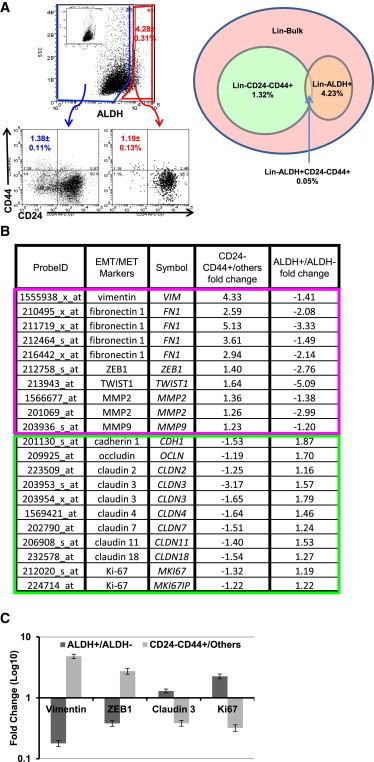
Previous studies by our group identified CD24−CD44+ and ALDH as BCSC markers, with cells displaying either marker being capable of initiating tumors in NOD/SCID mice (Al-Hajj et al., 2003; Ginestier et al., 2007). To determine whether these BCSC markers identify the same or distinct cell populations, and to localize these populations within human breast cancers, we performed immunofluorescence utilizing antibodies against CD44, CD24, and ALDH1, the ALDH isoform most commonly expressed in human breast cancers (Ginestier et al., 2007). As assessed by immunofluorescence, CD24−CD44+ and ALDH1 identified largely nonoverlapping cell populations in primary human breast cancers (Figure 1A). Furthermore, these cell populations were located in anatomically distinct areas. CD24−CD44+ cells were located primarily at the tumor-invasive edge adjacent to the tumor stroma (Figure 1A). In contrast, ALDH1+ cells were located more centrally, consistent with our previous studies demonstrating that ALDH+ CSCs are generated by hypoxia (Conley et al., 2012), in tumor internal zones. A small population of cells that simultaneously expressed CD24−CD44+ and ALDH1 were identified (Figure 1A).
Figure 1.
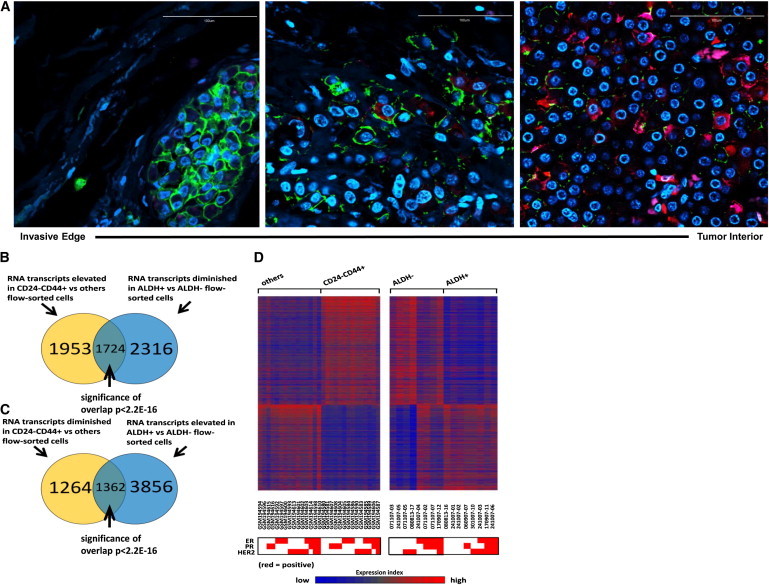
CD24−CD44+ and ALDH+ Identify Anatomically Distinct BCSCs with Distinct EMT and MET Gene-Expression Profiles in Primary Breast Cancers
(A) Localization of CD24 (magenta), CD44 (green), ALDH1 (red), and DAPI (blue) in clinical samples of human invasive breast carcinoma as assessed by immunofluorescence staining. Bar: 100 μm.
(B) Venn diagram of the intersection between genes elevated in CD24−CD44+ compared with other flow-sorted cells (others) and genes diminished in ALDH+ compared with ALDH− cells (|fold change| > 1.5 for each comparison).
(C) Venn diagram of the intersection between genes diminished in CD24−CD44+ versus others and genes elevated in ALDH+ versus ALDH−.
(D) Heatmap of genes with opposite expression patterns between CD24−CD44+ and ALDH+ (from B and C). Each row represents an RNA transcript; each column represents a sample (red, high expression). The molecular subtype based on expression of ER, PR, and HER2 is displayed below the heatmap for each patient sample.
See also Figure S1.
To characterize the BCSC populations identified by these markers, we isolated CD24−CD44+- or ALDH-expressing cell populations from a total of 45 primary human breast cancers and performed gene-expression analyses using Affymetrix microarrays. Of 3,677 RNA transcripts that were elevated in the CD24−CD44+ population, 1,724 were diminished in the ALDH+ population (Figure 1B). Conversely, of the 5,218 mRNA transcripts that were elevated in the ALDH+ population, 1,362 were diminished in the CD24−CD44+ population (Figure 1C). This suggests that a large number of genes display reciprocal expression patterns within CD24−CD44+ and ALDH+ populations. Our group and others have shown that normal and malignant stem cells have the capacity to form spherical colonies in vitro under anchorage-independent conditions (Dontu et al., 2003). Therefore, we compared the gene-expression patterns of CD24−CD44+ and ALDH+ fractionated and tumorsphere cultured cells. Even though many genes demonstrated reciprocal expression patterns in the CD24−CD44+ and ALDH+ populations, a set of genes expressed in CD24−CD44+ and ALDH+ populations displayed overlap with those from tumorsphere populations generated from unsorted cells (Figure S1 available online). This suggests that despite significant differences between cells characterized by CD24−CD44+ or ALDH expression, both of these populations share characteristics with tumorsphere-forming cells, a property of stemness. These results, together with prior studies demonstrating the tumor-initiating capacity of both CD24−CD44+ and ALDH+ populations, suggest that these CSC markers identify two stem cell populations in human breast cancers.
Breast cancers can be subdivided by gene-expression patterns into categories with distinct biological and clinical characteristics. These include luminal breast cancers, which express the steroid hormone receptors estrogen receptor (ER) and progesterone receptor (PR); HER2+ breast cancers characterized by amplification of the HER2 gene; and basal breast cancers that are triple negative (TN) in that they fail to express hormone receptors or display HER2 amplification. Of interest, although whole-tumor gene-expression profiles are distinct across these tumor types, the CD24−CD44+ and ALDH+ BCSC populations isolated from these tumors showed a high concordance in gene expression that was independent of the molecular subtypes of breast cancer (Figure 1D). Together, these studies suggest that human breast cancers contain BCSCs with distinct gene-expression profiles characterized by either CD24−CD44+ or ALDH+. Furthermore, although tumor-profiling studies of bulk tumor populations revealed distinct molecular subtypes, the gene-expression patterns of the ALDH+ and CD24−CD44+ BCSCs displayed a remarkable similarity across molecular subtypes of breast cancer (Figure 1D).
CD24−CD44+ Identifies Mesenchymal-like CSCs, and ALDH+ Identifies Epithelial-Like BCSCs
Several recent studies have suggested a strong correlation between CSCs and the EMT, while other studies have suggested that EMT and CSC states are mutually exclusive (Tsuji et al., 2008). To shed light on this controversy, we compared expression of EMT- and MET-associated genes in sorted CD24−CD44+ and ALDH+ tumor cells isolated from primary human breast cancers. As shown in Figure 2, EMT-associated genes, including vimentin, zinc finger E-box-binding homeobox 1 (ZEB1), ZEB2, β-catenin (CTNNB1), and matrix metalloproteinase-1 (MMP9), were significantly enriched in the CD24−CD44+ populations, whereas expression of these EMT genes was correspondingly decreased in the ALDH+ population. Conversely, genes associated with the alternative epithelial-like state, such as cadherin, occludin, claudins, and desmoplakin, were elevated in the ALDH+ population and correspondingly diminished in the CD24−CD44+ cell population. These results were confirmed by quantitative RT-PCR (qRT-PCR; Figure 2D). The proliferation marker Ki67 was preferentially expressed in ALDH+ compared with CD24−CD44+ cells, which is consistent with our previous finding demonstrating an association between ALDH1 and Ki67 expression in primary human breast cancers (Ginestier et al., 2007). These studies suggest that in primary human breast cancers, the CD24−CD44+ markers identify mesenchymal-like CSCs, whereas ALDH expression identifies epithelial-like proliferative CSCs.
Figure 2.
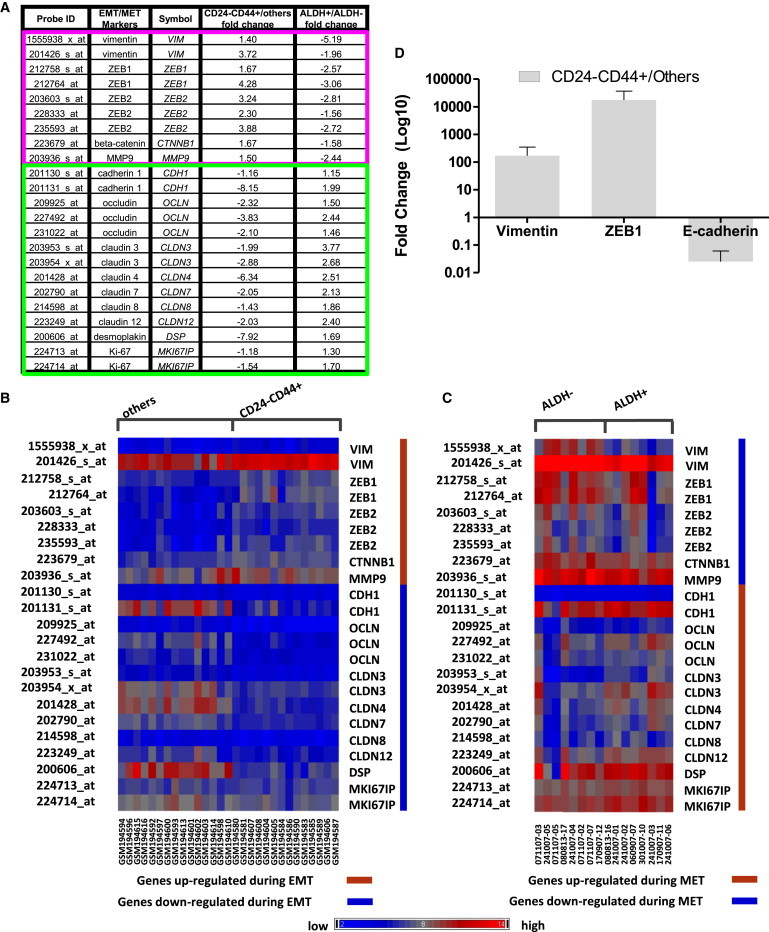
Specific EMT Markers Have Opposite Expression Patterns in CD24−CD44+ and ALDH+ Cells from Clinical Patient Breast Tumors
(A) Expression of EMT/MET markers in CD24−CD44+ versus others and ALDH+ versus ALDH−. Except for MMP9 (probe 203926_s_at p = 0.138), Ki67 (probe 224713_at p = 0.229), all other EMT markers listed are significant at p < 0.05 for CD24−CD44+ versus others. Except for ZEB1 (probe 212764_at p = 0.106), ZEB2 (probe 228333_at p = 0.336), CDH1 (probe 201131_s_at p = 0.209), OCLN (probe 209925_at p = 0.203, probe 231022_at p = 0.203), CLDN8 (probe 214598_at p = 0.15), and DSP (probe 200606_at p = 0.46), all other EMT markers listed are significant with p < 0.05 for ALDH+ versus ALDH−.
(B) For CD24−CD44+ versus others flow-sorted profile data sets, heatmap of EMT markers (|fold change| > 1.5).
(C) As in (A), but for ALDH+ versus ALDH− profile data sets.
(D) Gene-expression levels measured by qRT-PCR confirm the results obtained with Affymetrix array HU133 Plus 2.0. Error bars represent mean ± SD. The fold changes are significant with p < 0.05 for CD24−CD44+ versus others.
BCSCs Display Plasticity that Enables Them to Transition between Epithelial-like and Mesenchymal-like States
We utilized established breast cancer cell lines as well as primary tumor xenografts to characterize the biological characteristics of mesenchymal EMT-like and epithelial MET-like CSC populations. Our group and others have previously demonstrated that these cell lines contain subpopulations expressing CD24−CD44+ or ALDH, both of which are enriched for tumor-initiating potential (Al-Hajj et al., 2003; Ginestier et al., 2007). We performed flow cytometry to examine overlap of expression of these markers. As was the case with primary breast tumors, breast cancer cell line SUM149 contained CD24−CD44+- and ALDH-expressing cell populations with minimal overlap (Figure 3A). Similar populations displaying CD24−CD44+ and ALDH+ were found in luminal MCF7 and basal HCC1954 cell lines, as well as in the primary tumor xenograph UM2, which is of the basal molecular subtype. Furthermore, as was the case with primary breast cancers, gene-expression profiling of breast cancer cell lines of various molecular subtypes (SUM149, HCC1954, and MCF7) and primary xenografts (MC1 and UM2), gene-expression profiling revealed that CD24−CD44+ cells were enriched for expression of mesenchymal genes, and ALDH-expressing cells demonstrated a reciprocal epithelial gene-expression pattern (Figures 2A–2C and S2). These results were confirmed by qRT-PCR (Figures 2D and S3).
Figure 3.
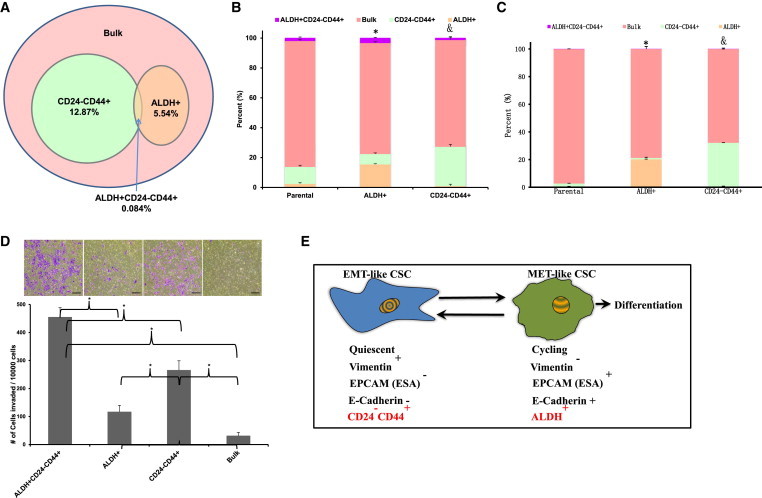
BCSC Display a Cellular Plasticity that Allows Them to Transit between EMT and MET States
Basal breast cancer cell lines SUM149 and MCF7 were immunostained with CD24 and CD44 antibodies, and subsequently with ALDEFLUOR. The four cell subpopulations defined by the ALDEFLUOR and CD24−CD44+ phenotypes were separated by fluorescence-activated cell sorting (FACS).
(A) The percentages shown in the diagram show the representation of the cell subpopulations in the total tumor cell population and the overlap between the ALDEFLUOR phenotype and the CD24−CD44+ phenotype.
(B) The unsorted SUM149 cells and the sorted CD24−CD44+ and ALDH+ populations as described in (A) were placed in culture for 10 days, and the resulting cells were reanalyzed by FACS for ALDEFLUOR, CD24, and CD44. ∗p < 0.05 (percentage of ALDH+ cells generated from the ALDH+ population compared with that generated from parental cells). &p < 0.05 (percentage of CD24−CD44+ cells generated from the CD24−CD44+ population compared with that generated from parental cells).
(C) The unsorted MCF7 cells and the sorted CD24−CD44+ and ALDH+ populations as described in (A) were placed in culture for 10 days, and the resulting cells were reanalyzed by FACS for ALDEFLUOR, CD24, and CD44. ∗p < 0.05 (percentage of ALDH+ cells generated from ALDH+ population compared with that generated from parental cells). &p < 0.05 (percentage of CD24−CD44+ cells generated from the CD24−CD44+ population compared with that generated from parental cells).
(D) The sorted four populations of SUM149 as described in (A) were assessed for invasive capacity utilizing the Matrigel invasion assay. Scale bar, 200 μm.
(E) Hypothetical models show the characteristics of two different states of BCSCs. ∗p < 0.05; error bars represent mean ± SD.
See also Figure S6.
To determine the degree of cellular plasticity between EMT- and MET-like CSC states, we isolated CD24−CD44+- and ALDH-expressing cell populations from the basal breast carcinoma cell line SUM149 and determined the stability of these phenotypes in culture. Purified CD24−CD44+- or ALDH-expressing cells generated heterogeneous populations recapitulating the proportion of CD24−CD44+ and ALDH+ BCSCs present in the original cell line (Figure 3B). Cellular plasticity in vitro was also demonstrated with the luminal cell line MCF7. This suggests that BCSCs in multiple molecular breast cancer subtypes can reversibly transit between epithelial-like and mesenchymal-like states.
Previous studies have suggested a relationship between the EMT state and the ability of cells to mediate tissue invasion and metastasis. In order to compare the invasive properties of mesenchymal-like and epithelial-like BCSCs, we utilized a Matrigel invasion assay. As shown in Figure 3D, tumor cells that displayed either BCSC marker were more invasive than bulk tumor populations. Within CSC marker-positive cell populations, CD24−CD44+ cells were significantly more invasive than ALDH-expressing cells, and cells that displayed all three stem cell markers demonstrated the greatest invasive capacity.
Together, these studies suggest a model in which BCSCs may exist in alternative mesenchymal-like and epithelial-like states, as depicted in Figure 3E. CD24−CD44+ BCSCs display a mesenchymal phenotype with high invasive capacity and low proliferative potential. Conversely, ALDH-expressing BCSCs are characterized by an epithelial-like proliferative state. BCSCs display a plasticity that allows them to transition between these states.
Mesenchymal and Epithelial BCSCs Resemble Basal and Luminal Stem Cells in the Normal Mammary Gland
The relationship between BCSC populations and their counterparts in the normal mammary gland remains controversial. The markers alpha 6 integrin (CD49f) and epithelial cell adhesion molecule (EPCAM) have been utilized to identify a hierarchy of normal mammary cell differentiation. When used in combination, these markers identify four populations in the human mammary gland: EPCAM+CD49f− epithelial cells, EPCAM+CD49f+ luminal progenitor cells, EPCAM−CD49f+ stem cells, and EPCAM−CD49f− stromal cells (Visvader, 2009). We utilized the nontransformed MCF10A human mammary cell line to determine the relationship between these markers and the CD24−CD44+ and ALDH+ markers used to characterize CSCs. Consistent with previous reports, when placed in tissue culture, EPCAM+CD49f+ cells displayed an epithelial morphology (Figure 4A) characterized by expression of E-cadherin (Figure 4B), whereas EPCAM−CD49f+ cells displayed a mesenchymal morphology (Figure 4A) characterized by expression of the EMT marker vimentin (Figure 4C). However, these phenotypes were not stable in culture, and EPCAM+CD49f− cells generated double-positive cells and vice versa (Figure 4A). To determine the relationship between cell populations identified by expression of these markers and the BCSC markers CD24−CD44+ and ALDH, we performed serial flow cytometry using both sets of markers. EPCAM−CD49f+ mesenchymal-like cells were enriched for expression of CD24−CD44+ (Figure 4D), whereas EPCAM+CD49f+ epithelial-like cells were enriched for ALDH expression as assessed by the ALDEFLUOR assay (Figure 4D). Furthermore, these populations were distinct, with little overlap (Figure 4E). As was the case for breast cancer cells, gene-expression analysis of sorted CD24−CD44+ and ALDH+ populations isolated from the nontransformed cell line MCF10A demonstrated that these cells express genes characteristic of mesenchymal (basal) and epithelial phenotypes, respectively (Figure 4F). We verified expression of a subset of these genes by qRT-PCR (Figure S4).
Figure 4.
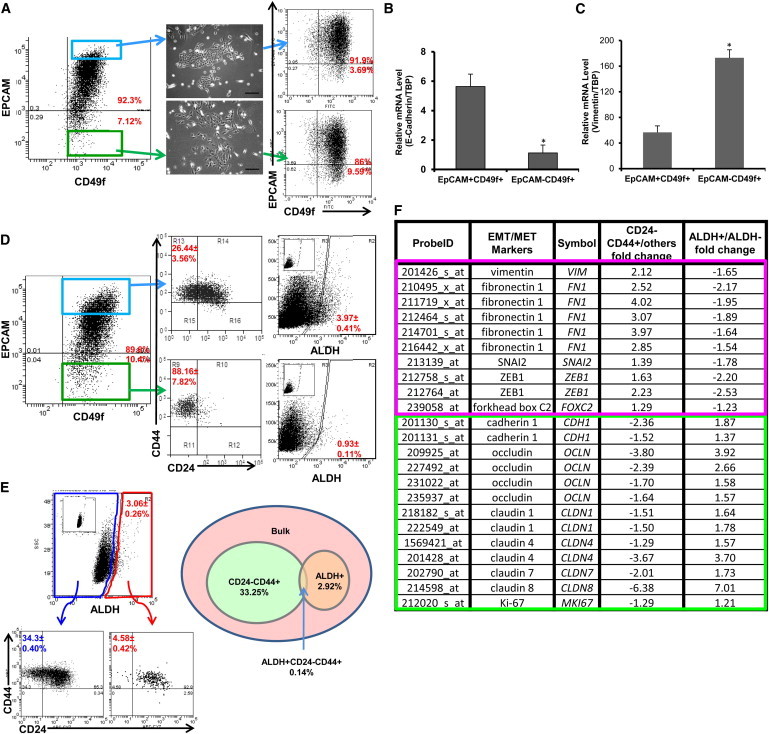
Identification of Mesenchymal-like and Epithelial-like States in Nontransformed MCF10A Cells
(A) Cells were immunostained with EPCAM and CD49f antibodies. EPCAM+CD49f+ and EPCAM−CD49f+ cells were separated by FACS and plated in culture for 10 days. FACS analysis for EPCAM and CD49f was then repeated. Scale bar, 200 μm.
(B and C) Total RNA was extracted from the sorted EPCAM+CD49f+ cells and EPCAM−CD49f+ cells, and the expression level of E-cadherin, vimentin, and TATA-binding protein (TBP) was measured by qRT-PCR. ∗p < 0.05; error bars represent mean ± SD.
(D) Cells were immunostained with EPCAM and CD49f antibodies, and subsequently stained with CD24/CD44 or ALDEFLUOR. Inset displays the negative control; cells incubated with DEAB, the specific inhibitor of ALDH, were used to establish the baseline fluorescence of these cells.
(E) Cells were immunostained with CD24 and CD44 antibodies, and subsequently with ALDEFLUOR. The four cell subpopulations defined by the ALDEFLUOR and CD24−CD44+ phenotypes were separated by FACS. The percentages shown in the diagram depict cell subpopulations and the overlap between the ALDEFLUOR-positive phenotype and the CD24−CD44+ phenotype. Inset displays the negative control; cells incubated with DEAB, the specific inhibitor of ALDH, were used to establish the baseline fluorescence of these cells.
(F) Total RNA was extracted from the four populations as described in (E) and used for Affymetrix array (HU133 Plus 2.0) analysis. The fold change for EMT/MET markers was compared among ALDH+, ALDH−, CD24−CD44+, and others.
See also Figure S4.
ALDH+-Expressing Normal Breast Cells Display High Proliferative Potential and Multilineage Differentiation Capacity
We utilized primary human breast tissue obtained from reduction mammoplasties to determine the gene-expression profiles of cell populations expressing these markers. As was the case for MCF10A cells, EPCAM−CD49f+ cells isolated from normal breast expressed mesenchymal markers, whereas EPCAM+CD49f+ cells primarily expressed epithelial markers (Figure 5).
Figure 5.
Characterization of CD24−CD44+ Cells and ALDH+ Epithelial Cells Isolated from Normal Breast Tissues
(A) Cells were immunostained with CD24 and CD44 antibodies, and subsequently stained with ALDEFLUOR. The four cell subpopulations defined by the ALDEFLUOR and CD24−CD44+ phenotypes were separated by FACS. The percentages in the diagram represent the cell subpopulations as a representative of the total tumor cell population and the overlap between the ALDEFLUOR phenotype and the CD24−CD44+ phenotype. Inset displays the negative control; cells incubated with DEAB, the specific inhibitor of ALDH, were used to establish the baseline fluorescence of these cells.
(B) Expression of EMT/MET markers in CD24−CD44+ versus others and ALDH+ versus ALDH− of normal breast as assessed by Affymetrix array HU133 Plus 2.0.
(C) To confirm the gene-expression results for EMT/MET markers, we measured the mRNA expression level for Vimentin, ZEB1, Claudin 3, and Ki67 in a set of breast tumor cells sorted for ALDH+/ALDH−, CD24−CD44+/others by qRT-PCR. Gene-expression levels measured by qRT-PCR confirm the results obtained with the Affymetrix array HU133 Plus 2.0. The fold changes are significant with p < 0.05 for CD24−CD44+ versus others or ALDH+ versus ALDH−. Error bars represent mean ± SD.
To assess the self-renewal capacity and differentiation potential of different cell populations, flow-cytometry-sorted populations of primary human mammary cells obtained from breast reduction surgery were transplanted into the cleared fat pads of NOD/SCID mice that had been humanized by addition of human mammary fibroblasts (Liu et al., 2006). Within the CD49f+ population, both EPCAM+ and EPCAM− cells were able to generate mammary structures containing both epithelial and myoepithelial cell lineages (Figure 6A). To further characterize these populations, we determined the relationship between expression of CD49f and EPCAM with that of the CSC markers CD24−CD44+ and ALDH+. As was the case with MCF10A cells, EPCAM−CD49f+ cells were enriched for CD24−CD44+, whereas EPCAM+CD49f+ cells were enriched for ALDH expression (Figure 6B) with little overlap between these populations. Gene-expression analysis of sorted CD24−CD44+ and ALDH+ cells demonstrated that these cell populations expressed genes characteristic of mesenchymal and epithelial phenotypes, respectively (Figure 5B), and this was verified by qRT-PCR (Figure 5C). Despite the observation that ALDH+ cells were predominantly contained within the EPCAM+CD49f+ population, this ALDH+ population constituted only 6% of total EPCAM+CD49f+ cells. Furthermore, within the EPCAM+CD49f+ population, only ALDH+ cells were able to generate bilineage colonies in vitro (Figures 6C and S5A) and generate ductal/alveolar structures in 3D Matrigel cultures (Figures 6D and S5B), consistent with a recent report (Shehata et al., 2012). These experiments expand on previous work by suggesting that in the normal mammary gland, the EPCAM+CD49f+ cell population is heterogeneous and consists of an ALDH+ subcomponent with high proliferative capacity and multilineage differentiating potential, and an ALDH− component at is luminal restricted with lower proliferative potential.
Figure 6.
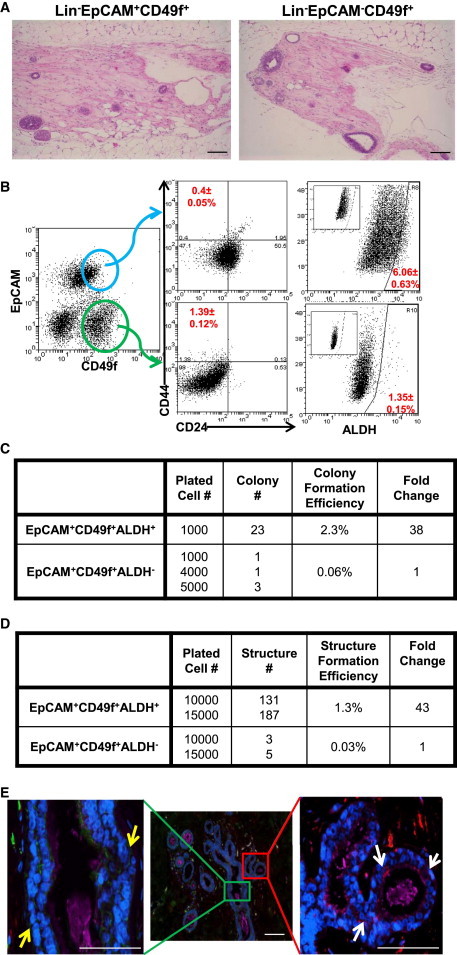
Normal Human Breast Contains Distinct Basal and Luminal Stem Cells that Express EMT and MET Markers, Respectively
(A) Cells isolated from reduction mammoplasty tissue were immunostained with lineage markers (Lin−), EPCAM, and CD49f antibodies, and were first gated based on viability (DAPI−) and lineage markers. The sorted cells (Lin−EPCAM+CD49f+ and Lin−EPCAM−CD49f+ cells) were injected into the No. 4 mammary gland fat pads, which were previously humanized with normal human breast fibroblasts. After about 2 months, the mice were sacrificed and the outgrowths that were generated in the fat pads were assessed by hematoxylin and eosin staining. Scale bar, 100 μm.
(B) Cells were immunostained with EPCAM and CD49f antibodies followed by CD24/CD44 or ALDEFLUOR. Inset displays the negative control: cells incubated with DEAB, the specific inhibitor of ALDH, were used to establish the baseline fluorescence of these cells.
(C) Cells were immunostained with Lin−, EPCAM, and CD49f antibodies, and subsequently with ALDEFLUOR. The cells were first gated based on viability (DAPI−) and lineage markers. The sorted cells (Lin−EPCAM+CD49f+ALDH+ cells and Lin−EPCAM+CD49f+ALDH− cells) were grown in differentiating conditions on collagen-coated plates for 12 days and the number of colonies generated was assessed.
(D) Sorted cells (Lin−EPCAM+CD49f+ALDH+ and Lin−EPCAM+CD49f+ALDH− cells) were cultured in 3D Matrigel culture for 3 weeks and the number of branched structures generated was assessed.
(E) Localization of CD24 (magenta), CD44 (green), ALDH1 (red), and DAPI (blue) in normal breast tissue as assessed by immunofluorescence staining. Yellow arrow, CD24−CD44+ cells; white arrow, ALDH1+ cells. Scale bar, 100 μm.
See also Figure S5.
We utilized immunofluorescence to localize cell populations identified by the stem cell markers in normal human breast tissue. CD24−CD44+ cells were located in a basal location within ductal structures frequently associated with ductal branch points (Figure 6E). In contrast, ALDH1+ cells were located in an abluminal location within lobules (Figure 6E). The quantitation of each cell population is shown in Figure S5C. These results suggest that the human mammary gland contains two stem cell populations, each of which is capable of self-renewal and multilineage differentiation. EPCAM−CD49f+ cells that display the BCSC marker CD24−CD44+ express basal/mesenchymal-like genes and are located in the basal layer of mammary ducts. In contrast, EPCAM+CD49f+ALDH+ cells are highly proliferative, display an epithelial-like gene-expression profile, and are present in an abluminal location in mammary lobules.
Discussion
Utilizing primary breast cancer tissue as well as primary xenografts and established cell lines, we demonstrate that BCSCs exist in distinct mesenchymal-like and epithelial-like states that can be identified by specific marker expression and display distinct gene-expression profiles. Mesenchymal-like CSCs characterized as CD24−CD44+ are primarily quiescent and are located at the tumor-invasive front, whereas epithelial-like CSCs expressing ALDH are proliferative and located more centrally within tumors. Despite the fact that different molecular subtypes of breast cancer can be distinguished based on their gene-expression profiles, the EMT- and MET-like BCSC gene-expression profiles are remarkably similar across these different molecular subtypes. In addition to suggesting a potential common cell of origin for different breast cancer subtypes, these results also have important clinical implications. Currently, breast tumors are subdivided into subtypes based on molecular profiling, which reflects the expression of steroid hormone receptors (ER and PR) as well as HER2 gene amplification. This classification dictates the therapeutic modality utilized, such as hormone therapy, HER2 blockade, or chemotherapy. The demonstration of common gene-expression profiles in BCSCs across molecular subtypes suggests that agents targeting BCSCs may have utility across molecular subtypes of breast cancer. Such CSC-targeting therapies could be added to the current subtype-specific therapies that target bulk tumor populations. Our recent report that HER2 drives luminal BCSCs in the absence of HER2 gene amplification supports this approach (Ithimakin et al., 2013).
Recent studies have suggested a commonality between CSCs and the EMT state, whereas other studies have suggested that these states are mutually exclusive (Tsuji et al., 2008). Our work provides an explanation for these contradictory findings by demonstrating that BCSCs can exist in alternative epithelial-like and mesenchymal-like states. The mesenchymal-like state is associated with expression of mesenchymal markers, relative quiescence, and high invasive capacity, whereas the epithelial-like state is associated with expression of epithelial markers, establishment of cell polarity, and extensive proliferation. Furthermore, we demonstrate that BCSCs display a cellular plasticity that enables them to transition between epithelial-like and mesenchymal-like states. The transition between these states may be regulated by the tumor microenvironment. For example, TGFβ and IL-6 generated in the tumor microenvironment induce EMT, whereas inhibition of TGFβ signaling and stimulation of BMPs induce MET. BCSC signal transduction pathways involving Wnt and NFκB induce EMT, whereas HER2 induces MET (Liu et al., 2012a). In addition, microRNA circuits within BCSCs may further regulate EMT/MET transitions by coordinately regulating multiple pathways (Liu et al., 2012b). We demonstrate that mesenchymal-like BCSCs tend to be located at the tumor-invasive front, whereas ALDH+ epithelial-like BCSCs are located in interior hypoxic zones (Conley et al., 2012). Together, these findings suggest a model in which mesenchymal-like BCSCs mediate tumor invasion into the blood, where they survive due to anoikis resistance and form micrometastasis in distant organs. The tumor microenvironment at these sites induces a MET that is required for BCSC self-renewal. This generates additional BCSCs, as well as differentiated cells that recapitulate the cellular hierarchy at metastatic sites. The plasticity of CSCs that allows them to transition between mesenchymal-like and epithelial-like states may play a critical role in the ability of these cells to form metastasis at distant sites, as illustrated in Figure S6. Several recent studies that demonstrated the importance of cellular plasticity in metastasis support such a model (Biddle et al., 2011; Thompson and Haviv, 2011). This model is also supported by studies demonstrating that in patients with metastatic breast cancer, circulating tumor cells and bone marrow micrometastasis are highly enriched for cells expressing the EMT BCSC marker CD24−CD44+ (Sheridan et al., 2006). Furthermore, these cells do not express the proliferation marker Ki67, consistent with their quiescence. Conversely, macrometastases express the stem cell markers ALDH and CD24−CD44+ in proportions similar to those found in primary tumors.
The relationship between BCSCs and stem cells in the normal mammary gland has not been defined. Recent studies in mouse mammary glands suggested the existence of separate basal and luminal stem cells. Under normal developmental conditions, each stem cell population gives rise to cells restricted to either the basal or luminal lineage (Van Keymeulen et al., 2011). However, transplantation of these cells into cleared mammary fat pads of recipient mice uncovers their plasticity, since either one of these cell populations is capable of regenerating an entire mammary tree (Keller et al., 2012). The existence of separate luminal and basal stem cells has also been reported in the prostate, where both populations have the capacity to give rise to prostate carcinomas (Tokar et al., 2005). Our study suggests that the human mammary gland may display a similar organization, containing basal stem cells located within mammary ducts that are CD24−CD44+EPCAM−CD49f+, as well as luminal stem cells located in terminal lobules which are EPCAM+CD49f+ALDH+. It was previously suggested that basal and BRCA1-associated breast carcinomas originate from EPCAM+CD49f+ luminal progenitor cells (Proia et al., 2011; Molyneux et al., 2010). However, our studies indicate that the populations defined by expression of EPCAM and CD49f are heterogeneous. The majority of EPCAM+CD49f+ cells isolated from the normal mammary gland are indeed luminal progenitors. However, approximately 6% of this population, as defined by expression of ALDH, is characterized by extensive self-renewal capacity as well as the capacity to generate multilineage structures when transplanted into the humanized cleared fat pad of NOD/SCID mice. These results, as well as the gene-expression profile of this cell population, suggest that rather than being luminal progenitors, these cells are more accurately characterized as epithelial-like stem cells. Their location in terminal lobules is consistent with previous reports suggesting that “cap” cells in terminal lobules have stem cell properties (Shackleton et al., 2006; Smalley and Ashworth, 2003; Woodward et al., 2005). The self-renewing proliferative state of ALDH+ luminal stem cells may increase their susceptibility to carcinogenic mutations, making them a logical cell of origin for human breast cancer. Our previous study demonstrating expansion of lobules containing ALDH1-expressing cells associated with loss of heterozygosity in breast cancer 1 (BRCA1, early onset) mutation carriers is consistent with such a model (Liu et al., 2008). Furthermore, our studies showed that both basal and luminal breast cancer subtypes have these two different BCSC populations—one characterized as CD24−CD44+ and the other characterized by ALDH expression. Although these BCSC populations are found across molecular subtypes of breast cancer, their proportion may vary across these subtypes. Our group and others have shown that luminal breast cancers are enriched in ALDH-expressing CSCs relative to CD24−CD44+ cells. The preponderance of ALDH-expressing CSCs is most pronounced in HER2+ breast cancers. This is consistent with previous findings that HER2 is an important regulator of CSC self-renewal in both HER2+ (Korkaya et al., 2008) and luminal (Ithimakin et al., 2013) breast cancer. In contrast, basal breast cancers are enriched in CD24−CD44+ CSCs, and in claudin-low breast cancers the majority of cells are CD24−CD44+ (Prat et al., 2010). Interestingly, these tumors display a mesenchymal morphology and exhibit aggressive clinical behavior. The observation that different molecular subtypes of breast cancer are characterized by distinct mutations (Cancer Genome Atlas Network, 2012) suggests that both genetic and microenvironmental factors may contribute to breast BCSC phenotypes. The enrichment of ALDH-expressing CSCs in HER2+ breast cancer and CD24−CD44+ CSCs in basal/claudin-low breast cancers may contribute to the aggressive clinical behavior of these subtypes.
In summary, our studies suggest that BCSCs can exist in alternative mesenchymal-like and epithelial-like states that are reflective of their normal counterparts. The plasticity of BCSCs that allows them to undergo reversible EMT/MET transitions regulated by the tumor microenvironment may be necessary for successful metastatic colonization. A number of CSC-targeting agents are now entering clinical trials. Our studies suggest that it may be necessary to target alternative CSC states to eliminate this clinically important tumor cell population.
Experimental Procedures
Patients and Clinical Samples
We made the following comparisons on flow-sorted samples from breast cancer patients or breast cancer cell lines: (1) CD24−CD44+ versus all other flow-sorted cells obtained from GSE7513 in the Gene Expression Omnibus (GEO) (patients 32–56 years old), (2) ALDH+ versus ALDH− obtained from 16 breast cancer patients (35–60 years old), and (3) CD24−CD44+ versus all other flow-sorted cells and ALDH+ versus ALDH− from breast cell lines MCF10A, SUM149, and primary xenograft MC1, and normal breast epithelial cells from patients (18–40 years old).
Data Analysis
We analyzed gene-expression arrays using the Partek Genomics Suite. For each profile data set, we pretreated the data by Quantile normalization and made a log2-base conversion. Principal-component analysis was conducted to determine and remove outliers. The p values of genes were calculated by two-sample t test. Expression values were visualized as heatmaps by the intensity plot function in Partek.
Immunostaining
For ALDH1, CD24, CD44, and DAPI quadruple fluorescent staining, paraffin-embedded sections of breast tumors or normal breast tissues were deparaffinized in xylene and rehydrated in graded alcohol. Antigen enhancement was done by incubating the sections in citrate buffer pH 6.0 (Dakocytomation) as recommended by the manufacturer. CD24 antibody (Neomarkers), ALDH1 antibody (BD Biosciences), and CD44 antibody (Thermo Scientific) were used at a 1:50 dilution and incubated for 1 hr. Alexa Fluor 647-, Alexa Fluor 546-, and Alexa Fluor488-labeled secondary antibodies (Invitrogen) were used at a 1:200 dilution and incubated for 20 min. Nuclei were counterstained with DAPI/antifade (Invitrogen; blue color in the staining) and coverslipped. Sections were examined with a fluorescence microscope (EVOS FL; AMG).
Cell Culture
The breast cancer cell lines SUM149 (Asterland) (Neve et al., 2006) and MCF10A (ATCC) were cultured as previously described (Liu et al., 2012b).
Flow Cytometry
Core biopsies and normal breast tissues were obtained and placed immediately in cold RPMI-1640 supplemented with 10% heat-inactivated newborn calf serum (HINCS; Invitrogen). In summary, the samples were digested in collagenase and 1 × 106 single cells were resuspended, incubated for 15 min with anti-CD44, anti-CD24, and anti-lineage mixture antibodies (polyethyleneglycol-conjugated anti-CD2, anti-CD3, anti-CD10, anti-CD16, anti-CD18, anti-CD31, and anti-CD140B; PharMingen) using the manufacturer’s suggested concentrations. They were then flowed by the ALDEFLUOR assay (Stem Cell Technologies) as described previously (Ginestier et al., 2007) and analyzed by MoFlo Astrios flow cytometry. The negative control for antibodies included cells incubated with isotype control antibodies for each color. The negative control for the ALDEFLUOR assay was cells incubated with the specific inhibitor of ALDH, diethylaminobenzaldehyde (DEAB), which was used to establish the baseline fluorescence of these cells. Side and forward scatter were used to eliminate debris and cell doublets, and the Lin cells were further analyzed for expression of the CD44/CD24 and ALDH markers. The detailed procedure for cell lines and cells from primary xenografts was described previously (Liu et al., 2012b).
Invasion Assay
Assays were done in triplicate in invasion chambers precoated with reduced growth factor matrix from BD Biosciences. Cells were added to the upper chamber in 200 ml of serum-free medium. For the invasion assay, 20,000 cancer cells were seeded on the coated chamber, and the lower chamber was filled with 600 ml of medium (Cambrex) with FBS. After 37 hr of incubation, the cells on the underside of the upper chambers were stained with the blue stain in the Cell Invasion Assay Kit (ECM550; Chemicon) and counted by light microscopy.
Normal Breast-Derived Cell Implantation into the Cleared Fat Pads of NOD/SCID Mice
Three-week-old female NOD/SCID mice were anesthetized by an i.p. injection (Liu et al., 2006). The No. 4 inguinal mammary glands were cleared and humanized with 2.5 × 105 nonirradiated telomerase immortalized normal human mammary fibroblasts (a generous gift from John Stingl and Connie Eaves, Terry Fox Laboratory) and 2.5 × 105 irradiated (4 Gy) fibroblasts as previously described (Liu et al., 2006). A 60-day release estrogen pellet (0.18 mg/pellet; Innovative Research of America) was placed s.c. on the back of the mouse’s neck via a trocar. The sorted cells were then mixed with 2.5 × 105 normal human mammary fibroblasts, resuspended in 50 μl of 1:1 Matrigel/5% serum Ham’s F-12, and injected into each of the cleared fat pads. All of the implantation experiments were repeated five times using cells from different patients, with three mice implanted per patient sample.
3D Matrigel Culture
3D cultures in Matrigel were established as previously described (Weaver and Bissell, 1999). Briefly, we suspended the sorted cells in 1 ml of BD Matrigel Matrix (354234; BD Biosciences) and Ham’s F-12 medium (BioWhittaker) with 5% serum at a ratio of 1:1, and plated 1 ml of the mixture into one well of 24-well cold plates. Each group of cells was plated in quadruplicate. After the Matrigel was solidified, 0.5 ml of Ham’s F-12 medium (BioWhittaker) with 5% serum was added to the top of the Matrigel. The experiments were repeated with cells derived from at least three different patients.
Statistical Analysis
Results are presented as the mean ± SD for at least three repeated individual experiments for each group. The mean and SD were determined on the basis of an analysis of at least three replicates using Microsoft Excel. Statistical differences were determined by using ANOVA and Student’s t test for independent samples. A value of p < 0.05 was considered statistically significant.
Author Contributions
S.L., M.S.W., J.C.C., M. Z., S.T.W., D.B., and S.P.M. conceived and designed the research. S.L., R.M., L.S., S.P.M., S.H., A.A., R.D., C.G., and E.C. carried out the experiments. Y.C., S.L., M.S.W., S.G.C., J.C.C., and M.Z. analyzed the data and wrote the manuscript.
Acknowledgments
We thank Dr. Stephen Ethier for generously providing the breast cancer cell lines SUM149 and SUM159. We also thank the University of Michigan Flow Cytometry Core, Microarray Core (Joe Washburn and Craig Johnson), Vector Core, Molecular Imaging Core, and Pathology Core for invaluable assistance. This work was supported by CAS Stem Cell grant XDA01040410 and NSFC grant 81322033 (S.L.), NIH grants CA66233 and CA101860 (M.S.W.), and NIH grant U54CA149169 (S.T.W.). M.S.W. holds equity in OncoMed Pharmaceuticals, receives research support from MedImmune and Dompe, and is a scientific advisor to MedImmune, Verastem, Cerulean, and Paganini.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
Suling Liu, Email: suling@ustc.edu.cn.
Max S. Wicha, Email: mwicha@umich.edu.
Accession Numbers
The GEO accession number for the gene expression of ALDH+ and ALDH− patient breast cancer cells reported in this paper is GSE52327. The GEO accession number for the gene expression of CD24−CD44+ and ALDH+ cells reported in this paper is GSE52262.
Supplemental Information
References
- Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddle A., Liang X., Gammon L., Fazil B., Harper L.J., Emich H., Costea D.E., Mackenzie I.C. Cancer stem cells in squamous cell carcinoma switch between two distinct phenotypes that are preferentially migratory or proliferative. Cancer Res. 2011;71:5317–5326. doi: 10.1158/0008-5472.CAN-11-1059. [DOI] [PubMed] [Google Scholar]
- Conley S.J., Gheordunescu E., Kakarala P., Newman B., Korkaya H., Heath A.N., Clouthier S.G., Wicha M.S. Antiangiogenic agents increase breast cancer stem cells via the generation of tumor hypoxia. Proc. Natl. Acad. Sci. USA. 2012;109:2784–2789. doi: 10.1073/pnas.1018866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G., Abdallah W.M., Foley J.M., Jackson K.W., Clarke M.F., Kawamura M.J., Wicha M.S. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eramo A., Lotti F., Sette G., Pilozzi E., Biffoni M., Di Virgilio A., Conticello C., Ruco L., Peschle C., De Maria R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504–514. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- Ginestier C., Hur M.H., Charafe-Jauffret E., Monville F., Dutcher J., Brown M., Jacquemier J., Viens P., Kleer C.G., Liu S. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang E.H., Hynes M.J., Zhang T., Ginestier C., Dontu G., Appelman H., Fields J.Z., Wicha M.S., Boman B.M. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69:3382–3389. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ithimakin S., Day K.C., Malik F., Zen Q., Dawsey S.J., Bersano-Begey T.F., Quraishi A.A., Ignatoski K.W., Daignault S., Davis A. HER2 drives luminal breast cancer stem cells in the absence of HER2 amplification: implications for efficacy of adjuvant trastuzumab. Cancer Res. 2013;73:1635–1646. doi: 10.1158/0008-5472.CAN-12-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller P.J., Arendt L.M., Skibinski A., Logvinenko T., Klebba I., Dong S., Smith A.E., Prat A., Perou C.M., Gilmore H. Defining the cellular precursors to human breast cancer. Proc. Natl. Acad. Sci. USA. 2012;109:2772–2777. doi: 10.1073/pnas.1017626108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkaya H., Paulson A., Iovino F., Wicha M.S. HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene. 2008;27:6120–6130. doi: 10.1038/onc.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkaya H., Kim G.I., Davis A., Malik F., Henry N.L., Ithimakin S., Quraishi A.A., Tawakkol N., D’Angelo R., Paulson A.K. Activation of an IL6 inflammatory loop mediates trastuzumab resistance in HER2+ breast cancer by expanding the cancer stem cell population. Mol. Cell. 2012;47:570–584. doi: 10.1016/j.molcel.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczek I., Liu S., Roh M., Vatan L., Szeliga W., Wei S., Banerjee M., Mao Y., Kotarski J., Wicha M.S. Expression of aldehyde dehydrogenase and CD133 defines ovarian cancer stem cells. Int. J. Cancer. 2012;130:29–39. doi: 10.1002/ijc.25967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Heidt D.G., Dalerba P., Burant C.F., Zhang L., Adsay V., Wicha M., Clarke M.F., Simeone D.M. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- Liu S., Dontu G., Mantle I.D., Patel S., Ahn N.S., Jackson K.W., Suri P., Wicha M.S. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66:6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Ginestier C., Charafe-Jauffret E., Foco H., Kleer C.G., Merajver S.D., Dontu G., Wicha M.S. BRCA1 regulates human mammary stem/progenitor cell fate. Proc. Natl. Acad. Sci. USA. 2008;105:1680–1685. doi: 10.1073/pnas.0711613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.L., Clouthier S.G., Wicha M.S. Role of microRNAs in the regulation of breast cancer stem cells. J. Mammary Gland Biol. Neoplasia. 2012;17:15–21. doi: 10.1007/s10911-012-9242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.L., Patel S.H., Ginestier C., Ibarra I., Martin-Trevino R., Bai S.M., McDermott S.P., Shang L., Ke J., Ou S.J. MicroRNA93 regulates proliferation and differentiation of normal and malignant breast stem cells. PLoS Genet. 2012;8:e1002751. doi: 10.1371/journal.pgen.1002751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux G., Geyer F.C., Magnay F.A., McCarthy A., Kendrick H., Natrajan R., Mackay A., Grigoriadis A., Tutt A., Ashworth A. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Moreno-Bueno G., Portillo F., Cano A. Transcriptional regulation of cell polarity in EMT and cancer. Oncogene. 2008;27:6958–6969. doi: 10.1038/onc.2008.346. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve R.M., Chin K., Fridlyand J., Yeh J., Baehner F.L., Fevr T., Clark L., Bayani N., Coppe J.P., Tong F. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A., Parker J.S., Karginova O., Fan C., Livasy C., Herschkowitz J.I., He X., Perou C.M. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince M.E., Sivanandan R., Kaczorowski A., Wolf G.T., Kaplan M.J., Dalerba P., Weissman I.L., Clarke M.F., Ailles L.E. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc. Natl. Acad. Sci. USA. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proia T.A., Keller P.J., Gupta P.B., Klebba I., Jones A.D., Sedic M., Gilmore H., Tung N., Naber S.P., Schnitt S. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell. 2011;8:149–163. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P., Golipour A., David L., Sung H.K., Beyer T.A., Datti A., Woltjen K., Nagy A., Wrana J.L. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Shackleton M., Vaillant F., Simpson K.J., Stingl J., Smyth G.K., Asselin-Labat M.L., Wu L., Lindeman G.J., Visvader J.E. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Shehata M., Teschendorff A., Sharp G., Novcic N., Russell A., Avril S., Prater M., Eirew P., Caldas C., Watson C.J., Stingl J. Phenotypic and functional characterization of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 2012;14:R134. doi: 10.1186/bcr3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C., Kishimoto H., Fuchs R.K., Mehrotra S., Bhat-Nakshatri P., Turner C.H., Goulet R., Jr., Badve S., Nakshatri H. CD44+/CD24- breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 2006;8:R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S.Y., Rath O., Zebisch A., Choo S.M., Kolch W., Cho K.H. Functional roles of multiple feedback loops in extracellular signal-regulated kinase and Wnt signaling pathways that regulate epithelial-mesenchymal transition. Cancer Res. 2010;70:6715–6724. doi: 10.1158/0008-5472.CAN-10-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley M., Ashworth A. Stem cells and breast cancer: A field in transit. Nat. Rev. Cancer. 2003;3:832–844. doi: 10.1038/nrc1212. [DOI] [PubMed] [Google Scholar]
- Storms R.W., Trujillo A.P., Springer J.B., Shah L., Colvin O.M., Ludeman S.M., Smith C. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc. Natl. Acad. Sci. USA. 1999;96:9118–9123. doi: 10.1073/pnas.96.16.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe N., Warren R.Q., Ivy S.P. Breast cancer growth and metastasis: interplay between cancer stem cells, embryonic signaling pathways and epithelial-to-mesenchymal transition. Breast Cancer Res. 2011;13:211. doi: 10.1186/bcr2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Thompson E.W., Haviv I. The social aspects of EMT-MET plasticity. Nat. Med. 2011;17:1048–1049. doi: 10.1038/nm.2437. [DOI] [PubMed] [Google Scholar]
- Tokar E.J., Ancrile B.B., Cunha G.R., Webber M.M. Stem/progenitor and intermediate cell types and the origin of human prostate cancer. Differentiation. 2005;73:463–473. doi: 10.1111/j.1432-0436.2005.00047.x. [DOI] [PubMed] [Google Scholar]
- Tsuji T., Ibaragi S., Shima K., Hu M.G., Katsurano M., Sasaki A., Hu G.F. Epithelial-mesenchymal transition induced by growth suppressor p12CDK2-AP1 promotes tumor cell local invasion but suppresses distant colony growth. Cancer Res. 2008;68:10377–10386. doi: 10.1158/0008-5472.CAN-08-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Keymeulen A., Rocha A.S., Ousset M., Beck B., Bouvencourt G., Rock J., Sharma N., Dekoninck S., Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- Visvader J.E. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23:2563–2577. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver V.M., Bissell M.J. Functional culture models to study mechanisms governing apoptosis in normal and malignant mammary epithelial cells. J. Mammary Gland Biol. Neoplasia. 1999;4:193–201. doi: 10.1023/a:1018781325716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward W.A., Chen M.S., Behbod F., Rosen J.M. On mammary stem cells. J. Cell Sci. 2005;118:3585–3594. doi: 10.1242/jcs.02532. [DOI] [PubMed] [Google Scholar]
- Yoo Y.A., Kang M.H., Lee H.J., Kim B.H., Park J.K., Kim H.K., Kim J.S., Oh S.C. Sonic hedgehog pathway promotes metastasis and lymphangiogenesis via activation of Akt, EMT, and MMP-9 pathway in gastric cancer. Cancer Res. 2011;71:7061–7070. doi: 10.1158/0008-5472.CAN-11-1338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


