Abstract
Background
This study was undertaken to characterize the vaginal microbiota throughout normal human pregnancy using sequence-based techniques. We compared the vaginal microbial composition of non-pregnant patients with a group of pregnant women who delivered at term.
Results
A retrospective case–control longitudinal study was designed and included non-pregnant women (n = 32) and pregnant women who delivered at term (38 to 42 weeks) without complications (n = 22). Serial samples of vaginal fluid were collected from both non-pregnant and pregnant patients. A 16S rRNA gene sequence-based survey was conducted using pyrosequencing to characterize the structure and stability of the vaginal microbiota. Linear mixed effects models and generalized estimating equations were used to identify the phylotypes whose relative abundance was different between the two study groups. The vaginal microbiota of normal pregnant women was different from that of non-pregnant women (higher abundance of Lactobacillus vaginalis, L. crispatus, L. gasseri and L. jensenii and lower abundance of 22 other phylotypes in pregnant women). Bacterial community state type (CST) IV-B or CST IV-A characterized by high relative abundance of species of genus Atopobium as well as the presence of Prevotella, Sneathia, Gardnerella, Ruminococcaceae, Parvimonas, Mobiluncus and other taxa previously shown to be associated with bacterial vaginosis were less frequent in normal pregnancy. The stability of the vaginal microbiota of pregnant women was higher than that of non-pregnant women; however, during normal pregnancy, bacterial communities shift almost exclusively from one CST dominated by Lactobacillus spp. to another CST dominated by Lactobacillus spp.
Conclusion
We report the first longitudinal study of the vaginal microbiota in normal pregnancy. Differences in the composition and stability of the microbial community between pregnant and non-pregnant women were observed. Lactobacillus spp. were the predominant members of the microbial community in normal pregnancy. These results can serve as the basis to study the relationship between the vaginal microbiome and adverse pregnancy outcomes.
Keywords: Community stability, Longitudinal sampling, Pregnancy, Vaginal microbiome, Lactobacillus, Dynamics
Background
The human vagina and the bacterial communities that reside therein represent a finely balanced mutualistic association [1]. Since the report (and discovery) of Lactobacillus (Döderlein Bacillus) as common inhabitants of the human vagina in 1892 by Gustav Döderlein, it is common wisdom that Lactobacillus is a keystone genus in the vagina [2-4]. The presence of Lactobacillus spp. is associated with a healthy state and is thought to protect reproductive age women from non-indigenous pathogens [5-26], certainly by contributing to the maintenance of a low vaginal pH (<4.5) through the production of lactic acid [24,27-34]. The vaginal microbiota is unique as it undergoes major compositional changes throughout a women’s lifespan from birth, to puberty and menopause [35-41]. Very little is known about the composition of the vaginal microbiota throughout these transitional stages, but it appears that sex steroid hormones play major roles in driving the composition and stability of the vaginal microbiota [39,42-49].
The development of culture-independent profiling methods to detect fastidious or non-cultivable organisms through the analysis of the sequence of marker genes, such as the 16S rRNA gene, has precipitated a revolution in biology and medicine, by spurring projects such as the National Institutes of Health (NIH)-funded Human Microbiome Project [50-56], the European MetaHit project [57] and the creation of the International Human Microbiome Consortium. Culture-based analyses have been used for decades and have contributed critical knowledge about the microbes inhabiting the human body, including the vagina, and the understanding of infectious diseases that affect the genital tract [17,58-71]. However, cultivation techniques are laborious, time-consuming, and quantitative microbiology of polymicrobial infection or complex ecosystems is challenging when trying to accurately assess the contribution of each organism to the microbial population structure [72-74]. Moreover, many organisms cannot be cultured because the essential requirements for growth are not known [72,75,76]. Advances in cultivation techniques continue to occur and are sometimes informed by the results of sequence-based methods [73,77-79].
Culture-independent characterization of bacterial communities can be generated using the amplification and sequencing of the 16S rRNA gene [80-83] or metagenomics approaches in which the sequences of the bacterial community genes and genomes are obtained [51,57,76-78,84-88]. However, 16S rRNA gene profiling is widespread and has been used for the discovery of important clinically relevant organisms which had resisted cultivation for decades [76,77,89-91]. This method is also affordable and rapid, and results are tractable from an analytical point of view. The use of molecular culture-independent techniques has increased the knowledge about the complexity of the microbial ecosystem of multiple body sites, including the human vagina [21,26,40,41,47,76,92-110].
Most of the data published to date on the human vagina microbial ecosystem focused on healthy asymptomatic non-pregnant women of reproductive age [100,109,111,112]. These studies have established that at least six types of vaginal microbiota exist, referred to as community state types (CSTs) [100,109,112,113]. Four of these CSTs are most often dominated by one of four Lactobacillus spp. commonly found in the vagina (L. crispatus, L. iners, L. jensenii and L. gasseri), while the remaining two lack substantial numbers of Lactobacillus spp. and are composed of a diverse array of anaerobic bacteria including species associated with bacterial vaginosis such as Prevotella, Megasphaera, Gardnerella vaginalis, Sneathia and Atopobium vaginae[13,96,102,105,114-122]. While these two states are found in otherwise healthy asymptomatic women, they are often associated with high Nugent scores [123], a Gram stain method used in the diagnosis of bacterial vaginosis in research settings [61,71,124-126]. High Nugent scores or changes in the vaginal microbiota have been associated with increased risk of sexually transmitted infections [20,127-138], including HIV [10,14,22,99,139-150], preterm birth [62,108,151-203], and adverse perinatal outcomes such as post-abortal sepsis [204], early and late miscarriage [165,205,206], recurrent abortion [205], histological chorioamnionitis [160,164] and postpartum endometritis [183,207].
Interestingly, in some women the vaginal microbiota is remarkably dynamic (it can change over a short period of time from Lactobacillus dominated CSTs to CSTs lacking a substantial number of Lactobacillus spp.), while in other women it is relatively stable [100,112]. Menstruation and sexual activity have been shown to have negative effects on the stability of the vaginal microbiota [26,42,43,112,208-210]. The secretory phase of the menstrual cycle, which is characterized by high concentrations of estrogen and progesterone, appears to be more stable in terms of microbial community composition [112].
Knowledge of the vaginal microbiota throughout pregnancy is sparse, and only a few studies have examined the vaginal microbiota in pregnant women using culture-independent methods [211-213], and none analyzed samples collected longitudinally throughout pregnancy from the same women using 16S rRNA gene sequence-based methods. Using a Gram stain scoring system, cultivation and terminal restriction fragment length polymorphism, Verstraelen et al. demonstrated the importance of L. crispatus and L. gasseri in maintaining stability in a population of Dutch women sampled once in each trimester [211]. The consensus from previous studies is that Lactobacillus spp. predominate the vaginal microbiota during pregnancy; this observation is consistent with the results of a recent 16S rRNA gene sequence-based cross-sectional study reported by Aagaard and colleagues [213]. None of these studies examined the degree of stability in the vaginal microbiota during pregnancy using 16S rRNA gene sequence analysis. Stability and resilience of ecosystems are now recognized to be important in understanding the fitness of the community, as well as the response to perturbations [56,214-220]. Therefore, studies of the microbiota in several body sites are characterizing stability and resilience, as well as how they relate to health and disease [221-233].
The purpose of this study was to characterize the changes in the composition of the vaginal microbiota of pregnant women followed longitudinally (over the duration of pregnancy). The control group consisted of non-pregnant women who were frequently sampled. Here we report the use of 16S rRNA gene sequence-based methods to characterize the vaginal microbiota of normal pregnant women and the differences observed between these and non-pregnant subjects. The two major findings were that the microbial composition of the vaginal microbiota in normal pregnancy is different from that of non-pregnant women; moreover we demonstrate, for the first time, that the vaginal microbiota during pregnancy is more stable than in the non-pregnant state.
Methods
Study design
This was a prospective longitudinal cohort study to characterize changes in the vaginal microbiota in normal pregnant and non-pregnant women. A normal pregnancy was defined as a woman with no obstetrical, medical or surgical complications, who agreed to participate in this study, provided written signed informed consent, and delivered at term (38 to 42 weeks) without complications. Non-pregnant women were of reproductive age and free of clinical disease [112]. These patients were enrolled in a prospective study designed to describe the vaginal microbiota as a function of time. Details of this study have been previously reported [112].
Study procedures
Pregnant women who agreed to participate in the longitudinal study had a speculum examination at each visit and a sample of vaginal fluid was collected under direct visualization from the posterior vaginal fornix by an obstetrician or a midwife using a Dacron swab (Medical Packaging Swab-Pak™, Camarillo, CA, USA). Samples were collected every 4 weeks until 24 weeks of gestation, and every 2 weeks until the last prenatal visit. Samples were stored at -70°C until assayed. Non-pregnant patients were self-collected sampled twice weekly for 16 weeks using validated methods previously described [112,234]. All samples were Gram-stained and analyzed using the Nugent score [61]. The use of samples from the longitudinal study of pregnant women was approved by the Human Investigations Committee of Wayne State University and the Institutional Review Board of the Eunice Kennedy Shriver National Institute of Child Health and Human Development. The data from non-pregnant women are derived from a previous study [112] and are publicly available in the sequence read archive (accession no. SRA026073). The metadata associated with the sequence data are available in dbGap (dbGap study no. phs000261).
DNA extraction, amplification and pyrosequencing of barcoded 16S rRNA genes
Genomic DNA was extracted from archived vaginal swab specimens. Procedures for the extraction of genomic DNA from frozen vaginal swabs have been developed, validated and previously published [109]. Briefly, frozen vaginal swabs were immersed in 1 ml pre-warmed (55°C) cell lysis buffer, composed of 0.05 M potassium phosphate buffer containing 50 μl lyzosyme (10 mg/ml), 6 μl mutanolysin (25,000 U/ml; Sigma-Aldrich, St. Louis, MO, USA) and 3 μl lysostaphin (4,000 U/ml in sodium acetate; Sigma-Aldrich). The mixture was incubated for 1 hour at 37°C followed by the addition of 10 μl proteinase K (20 mg/ml), 100 μl 10% SDS, and 20 μl RNase A (20 mg/ml), and the mixture was incubated for 1 hour at 55°C. The samples were then transferred to a FastPrep Lysing Matrix B tube (MP Biomedicals, Santa Ana, CA, USA) and microbial cells were lysed by mechanical disruption using a bead beater (FastPrep instrument, MP Biomedicals) set at 6.0 m/s for 30 seconds. The lysate was processed using the ZR Fecal DNA extraction kit (ZYMO Research, Irvine, CA, USA) according to the manufacturer’s recommendation and omitting the lysis steps (steps 1 to 3). The kit included a column (Zymo-Sin IV-HRC spin filter) specifically designed to remove PCR inhibitors from DNA samples. The DNA was eluted into 100 μl TE buffer, pH 8.0. This procedure provided between 2.5 and 5 μg of high quality whole genomic DNA from vaginal swabs.
Universal primers 27F (Forward) and 338R (Reverse) were used for PCR amplification of the V1-V2 hypervariable regions of 16S rRNA genes [112]. The 338R primer included a unique sequence tag to barcode each sample. The primers were as follows: 27F-5′-GCCTTGCCAGCCCGCTCAGTCAGAGTTTGATCCTGGCTCAG-3′ and 338R-5′-GCCTC CCTCGCGCCATCAGNNNNNNNNCATGCTGCCTCCCGTAGGAGT-3′, where the italicized sequences are the 454 Life Sciences FLX sequencing primers B and A in 27 F and 338R, respectively, and the bold font denotes the universal 16S rRNA gene primers 27 F and 338R. The 8-bp barcode within primer 338R is denoted by eight Ns. Using 96 barcoded 338R primers [109], the V1-V2 regions of 16S rRNA genes were amplified in 96 well microtiter plates using AmpliTaq Gold DNA polymerase (Applied Biosystems, NY, USA) and 50 ng template DNA in a total reaction volume of 50 μl. Reactions were run in a PTC-100 thermal controller (BioRad, Hercules, CA, USA) using the following cycling parameters: 5 minutes denaturation at 95°C, followed by 20 cycles of 30 seconds at 95°C (denaturing), 30 seconds at 56°C (annealing) and 90 seconds at 72°C (elongation), with a final extension at 72°C for 7 minutes. Negative controls without a template were included for each barcoded primer pair. The presence of amplicons was confirmed by gel electrophoresis on a 2% agarose gel and staining with SYBRGreen (Life Technologies, Carlsbad, CA, USA). PCR products were quantified using the Quant-iT™ PicoGreen® dsDNA assay (Life Technologies). Equimolar amounts (100 ng) of the PCR amplicons were mixed in a single tube. Amplification primers and reaction buffer were removed from each sample using the Agencourt AMPure Kit (Beckman-Coulter, Pasadena, CA, USA). The purified amplicon mixtures were sequenced by 454 FLX Titanium pyrosequencing using 454 Life Sciences® primer A by the Genomics Resource Center at the Institute for Genome Sciences, University of Maryland School of Medicine using protocols recommended by the manufacturer as amended by the Center and previously described [109].
Sequence analysis
Sequences were binned by samples using the sample-specific barcode sequences and trimmed by removal of the barcode and primer sequences. Sequence read quality check was performed using a bioinformatics pipeline that is in accordance with NIH Human Microbiome Project (HMP) standard operating procedures [109]. Briefly, raw sequence reads were filtered to meet the following criteria: 1) minimum and maximum read length of 200 bp and 400 bp; 2) no ambiguous base calls; 3) no homopolymeric runs longer than 8 bp; 4) a read was discarded if the average quality value was less than q25 within a sliding window of 50 bp; 5) a read was discarded if it was identified as a putative chimeric sequence by UCHIME [235]. The sequences that passed the above filtering procedure were denoised in order to correct for potential sequencing errors at 99% level using UCLUST [235]. Sequences were then taxonomically classified using pplacer version v1.1.alpha08 [236]. pplacer makes taxonomic assignment using a linear time maximum-likelihood method (or alternatively a Bayesian phylogenetic placement method) using a community specific reference tree. Version 0.2 of the vaginal community 16S rRNA gene reference tree was employed. Overall, 86% of all sequence reads that passed quality control criteria in this study were classified to the species level, and 57% of the reads were taxonomically assigned to the genus Lactobacillus.
Statistical analysis
In microbiology, the abundance of bacteria is measured in a logarithmic scale (base 10), given the wide range of bacterial abundance and the exponential nature of bacterial growth under certain circumstances (for example, in vitro). Therefore, it is the norm to compare microbial abundance over time using the difference of logs, log10 (p) - log10 (q), which is the same as the log fold change log10 (p/q), where p and q are relative abundances of a given microorganism in two samples.
Estimating changes in abundance of a complex microbial ecosystem within a patient at two time points becomes more challenging, as several microorganism types (phylotypes) need to be considered. In order to address this challenge, we assessed the dissimilarity between two community states (in other words, how divergent two community states are) using the Jensen-Shannon metric [237]. The term “community state” in microbial ecology refers to the relative abundance of all phylotypes at a particular time point in a subject; in our case, a sample of vaginal fluid.
The Jensen-Shannon divergence between two community states, p and q, is the average of the Kullback–Leibler divergences DKL(p,a) and DKL(q,a):
where a is the mean of p and q and DKL(p,q) is the Kullback–Leibler divergence defined as:
and where p = (p1, …. , pn) and q = (q1, … , qn). In essence, the Kullback–Leibler divergence DKL(p,q) calculates the mean log fold changes log (pi/qi). While the Kullback–Leibler divergence measure is widely used, it has one drawback: its value becomes infinite if one of the components of q is zero. In contrast, the Jensen-Shannon divergence always yields a value between 0 and 1. A Jensen-Shannon divergence score of 0 means that two community states are the same. In contrast, a Jensen-Shannon divergence scores of 1 means that the two community states are completely different. The square root of the Jensen-Shannon divergence is called Jensen-Shannon distance.
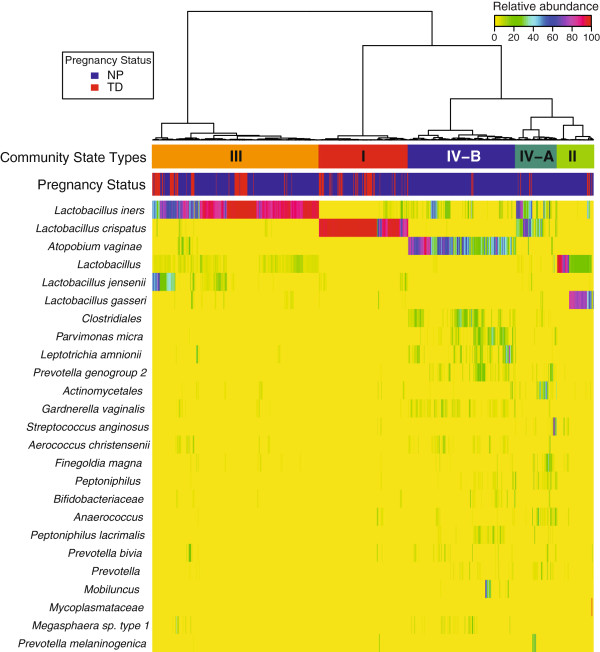
The term “community state type” (CST) is used in microbial ecology to describe a group of community states with similar microbial phylotype composition and abundance [109,112]. Such grouping is desirable in order to reduce dimensionality. Utilizing Jensen-Shannon divergence as a measure of dissimilarity among community states and hierarchical clustering with Ward linkage, five CSTs in the combined dataset of pregnant and non-pregnant women have been identified (Figure 1). Three of the CSTs (CST I, II, III) are dominated by Lactobacillus spp. and the remaining two (CST IV-A, IV-B) consist of community states with substantially lower number of Lactobacillus spp. than the other CSTs.
Figure 1.
Heatmap of percentage abundance of microbial taxa found in the vaginal microbial communities of 22 normal pregnant women who delivered at term and 32 non-pregnant women sampled longitudinally. Ward linkage hierarchical clustering of Jensen-Shannon metric identified five community state types (CST I, II, III, IV-A and IV-B). The upper color bar shows the five community state types while the lower color bar shows the pregnancy status of each sample (NP: Non Pregnant; TD: Term Delivery).
Comparison of community state type frequencies in the non-pregnant state and normal pregnancy
In order to assess significance of differences in frequencies of CSTs between pregnant and non-pregnant women, we considered one CST at a time and created an indicator variable Y, with Y = 1 for samples that belonged to the CST of interest and 0 otherwise. We regressed the CST indicator variable on the pregnancy status using generalized estimation equations (GEE) considering that the response is correlated within patients. The model fitting was performed using the geepack package [238] in R (version 2.15), specifying a binomial distribution for the dependent variable (CST indicator), and assuming an exchangeable correlation structure (the response correlation within a subject is similar between all pairs of time points). The odds ratio of belonging to a given CST given that the woman is pregnant was reported together with the significance of the effect (determined by default via a Wald test in geepack). P-values false discovery rate adjustment for multiple comparisons across the five CSTs was performed and a q-value <0.05 was deemed significant.
Identification of phylotypes accounting for differences in the structure of vaginal microbiota between the non-pregnant state and normal pregnancy
In order to assess which phylotypes account for the differences in the structure of microbial communities, we modeled relative abundance of one phylotype at a time as a function of pregnancy status, then selected those phylotypes for which there was a significant effect of pregnancy status. Only phylotypes present (based on at least one read count) in 25% or more for the samples were considered in this analysis.
Read count data obtained from a longitudinal experiment design are typically modeled using GEE or linear mixed-effects models by assuming a Poisson or negative binomial distribution of the response. The choice of a Poisson distribution will be justified when the counts variance equals the counts mean, while the negative binomial distribution will be preferred when the mean variance equality cannot be safely assumed.
Several phylotypes were not detected in a large proportion of samples; hence, the frequency of 0 count values in the dataset is larger than expected under a Poisson or negative binomial distribution. For such circumstances, models that can allow for zeroes inflation are more suitable.
In general, the zero-inflated version of a distribution D (for example, negative binomial) of a random variable Y has a probability function of the form:
where fD(y) is the probability function of the distribution D, fZID(y) is the probability function of the zero inflated version of D with an additional parameter π as the proportion of additional zeros and I(x) is the indicator function equal to 1 if x is true and equal to 0 otherwise. From the above equation, the probability of y = 0 is equal to π + (1 - π)fD(0), while the probability of y > 0 is (1 - π)fD(y). Zero inflated models for count data have been used in statistics for at least 20 years [239].
To ensure a proper fit of the count data of each phylotype, we have utilized zero-inflated negative binomial mixed-effects (ZINBLME) models in addition to the simpler negative binomial linear mixed effects (NBLME) and Poisson linear mixed effects (PLME) models. These three types of models were fitted to each phylotype and the model with lowest Akaike Information Criterion (AIC) value was retained. The significance P-value for the association between the microbial relative abundance and the group variable was computed only for the best model (smallest AIC).
The mixed effects modeling of the reads count data (dependent variable) on the pregnancy status (independent variable) was performed using the NLMIXED procedure in SAS (version 9.3; SAS, Cary, NC, USA) as discussed elsewhere [240-242]. All three types of models (PLME, NBLME and ZINBLME) had included an offset term (the log of the total number of reads in a given sample) to allow for a comparison in the relative abundance (and not absolute counts) between groups. The random effect in the ZINBLME models was allowed only on the non-zero inflation component (negative binomial mean).
For each of the three types of models, the reported coefficient represents the difference in mean log relative abundance between in samples from pregnant and non-pregnant women that was further converted into a fold change. The P-value of the model with the best fit (smallest AIC) was retained and false discovery rate adjustment was applied across the phylotypes. A q-value <0.1 and fold change >1.5 was used to claim significance.
Results
Characteristics of the study population
The clinical and demographic characteristics of the pregnant population are displayed in Table 1. The clinical and demographic characteristics of non-pregnant subjects have been previously reported [112]. The present study included 32 non-pregnant women and 22 pregnant women who had a term delivery without complications (gestational age at delivery from 38 to 42 weeks). Non-pregnant women self-sampled with a frequency of twice a week for 16 weeks. The median (interquartile range (IQR)) number of samples with available sequence data was 27.5 samples per participant (IQR: 20.7 to 29). Pregnant women had a median of 6.5 samples per pregnancy (IQR: 6.0 to 7.0).
Table 1.
Descriptive characteristics of the pregnant woman enrolled in the longitudinal study (n=22)
| Mean | SD a | Minimun | Maximum | |
|---|---|---|---|---|
| Age (years) |
24.2 |
5.2 |
19 |
35 |
| Raceb |
|
|
|
|
| African American |
19 (86%) |
|
|
|
| White |
2 (9%) |
|
|
|
| Hispanic |
1 (5%) |
|
|
|
| Body Mass Index (BMI; kg/m2) |
29.8 |
4.9 |
20.2 |
39.9 |
| Nulliparity |
10 (45%) |
|
|
|
| Cesarean delivery |
2/22 (9%) |
|
|
|
| Gestational age at delivery (weeks) |
39.8 |
1.0 |
38.1 |
42.1 |
| Birthweight (grams) |
3320 |
290 |
2645 |
4090 |
| Apgar at 1 minute (median) |
9.0 |
|
7 |
9 |
| Apgar at 5 minutes (median) |
9.0 |
|
5 |
9 |
| Nugent score >=7c | 2 (9%) |
aStandard Deviation.
bNon-pregnant women: African American (50%), White (40.6%), Hispanic and others (9.4%).
cIn at least one sample of a given subject – According to reference 61, Nugent score above 7 correspond to a diagnosis of bacterial vaginosis.
Characterization of the microbial taxa as a function of depth of coverage
We characterized the vaginal microbiota using pyrosequencing of barcoded 16S RNA genes. The dataset consisted of 2,946,507 high-quality sequences, with an average length of 240 bp. The median number of sequences per sample was 2,878 (IQR: 2,446 to 4,171). Taxonomic assignment of the sequences identified a total of 143 taxa in the vaginal microbiota of the women studied; all 143 taxa were observed both in non-pregnant as well as pregnant women who delivered at term. The taxonomic assignments of vaginal bacterial community members are shown in Additional file 1: Table S1.
The vaginal microbiota in the non-pregnant state and normal pregnancy
To study the vaginal bacterial communities of pregnant versus non-pregnant women, we hierarchically clustered the vectors of relative abundances of bacterial phylotypes (one per sample) using the Jensen-Shannon divergence metric and Ward linkage [112]. In this study, we refer to a community state as a vector of relative abundances of bacterial phylotypes for a given sample. Community states were clustered into five groups with similar bacterial composition and abundance (Figure 1), referred to as CSTs according to the nomenclature established by Gajer and colleagues [112].
Three of these CSTs were most often dominated by L. crispatus (CST I), L. gasseri (CST II) and L. iners (CST III). Communities that clustered in CST IV-A or IV-B lacked a substantial number of Lactobacillus spp. and differed in taxa composition. For example, CST IV-A was characterized by a roughly equal number of Peptoniphilus, Anaerococcus, Corynebacterium, Finegoldia, Prevotella and a few other taxa. In contrast, those of CST IV-B had higher relative abundance of the genus Atopobium and were characterized by the presence of L. iners (low relative abundance), Prevotella, Sneathia, Gardnerella, Ruminococcaceae, Parvimonas, Mobiluncus and other taxa previously shown to be associated with bacterial vaginosis [96]. These findings are consistent with previous observations indicating that there is no single “core” microbiota of the human vagina [109]. The relationship between Nugent score and CST was demonstrated. It is noteworthy that CST IV-B was strongly associated with a high Nugent score (defined as 7 to 10) (P = 0.013 using a mixed effect model; odds ratio = 24.3).
Table 2 shows the counts of samples assigned to each CST and corresponding percentages stratified by pregnancy status. A dramatic difference in the distribution of frequency of CSTs between non-pregnant and pregnant patients who delivered at term was observed (a decrease of 95% in the odds of observing CST IV-B in pregnant women compared to non-pregnant women).
Table 2.
Distribution of samples in each community state-type as a function of pregnancy status (non-pregnant vs normal)
| CST/Pregnancy status | I | II | III | IV-A | IV-B | Total |
|---|---|---|---|---|---|---|
| Non-pregnant women |
129 (17%) |
68 (8.9%) |
268 (35.2%) |
79 (10.4%) |
217 (28.5%) |
761 |
| Normal pregnancy | 53 (38.1%) | 6 (4.3%) | 72 (51.8%) | 5 (3.6%) | 3 (2.2%) | 139 |
Since Table 2 was generated using correlated samples, standard methods (for example, Fisher tests) cannot be applied to assess significance of differences in frequencies of each CST between pregnant and non-pregnant women. Instead, for each CST (T), a logistic regression GEE model was fitted with the binary response variable (T versus non-T) used as a dependent variable and the pregnancy status used as an independent variable. The coefficients, odds ratios, P-values, and q-values for the five GEE models are shown in Table 3. The frequency of CST IV-B (most often dominated by Atopobium) was significantly lower in pregnant compared to non-pregnant women. The relative abundance of CST I (dominated by L. crispatus) was borderline significantly different between pregnant and non-pregnant women (based on unadjusted P = 0.0507 at the 5% significance level).
Table 3.
Coefficient estimates, odds ratios, p-values and q-values for the association between each community state type with the pregnancy status
| Community state type a | Estimate b | Odds ratio | p-value | q-value c |
|---|---|---|---|---|
| IV-B |
-3.06 |
0.047 |
0.00000 |
0.00001 |
| I |
1.09 |
2.986 |
0.05076 |
0.12689 |
| III |
0.76 |
2.136 |
0.11344 |
0.18907 |
| IV-A |
-1.23 |
0.292 |
0.16958 |
0.21198 |
| II | -0.73 | 0.482 | 0.48193 | 0.48193 |
a Community state type: a group of community states with similar microbial phylotype composition and abundance identified via unsupervised clustering (Figure 1).
b Estimate: the value of the coefficient in the logistic regression model for a binary variable indicating whether (1) or not (0) a given sample was assigned in the community state named in column 1. The value of the coefficient represents the log of the odds ratio that the sample belongs to the community state indicated in column 1 given that the sample belongs to a pregnant woman (as opposed to a non-pregnant woman).
c q-value: the False Discovery Rate adjusted p-value across all 5 community types that were tested.
Constancy of the vaginal microbiota in pregnant and non-pregnant women
Figure 2 shows the profiles of CSTs for pregnant women who delivered at term as a function of gestation time. The CST profiles of pregnant and non-pregnant women are somewhat similar (given smaller number of samples per pregnant woman) except that CST IV-B is rarely present in pregnant women. In particular, none of the pregnant women persist in this CST, which lacks substantial number of Lactobacillus, whereas communities of seven non-pregnant women persist in CST IV-B for 16 weeks [112].
Figure 2.
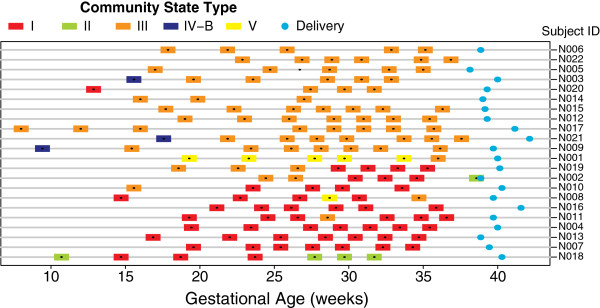
Profiles of community state types for pregnant women who delivered at term as a function of gestational age. Gestational age at delivery is indicated by blue solid circles.
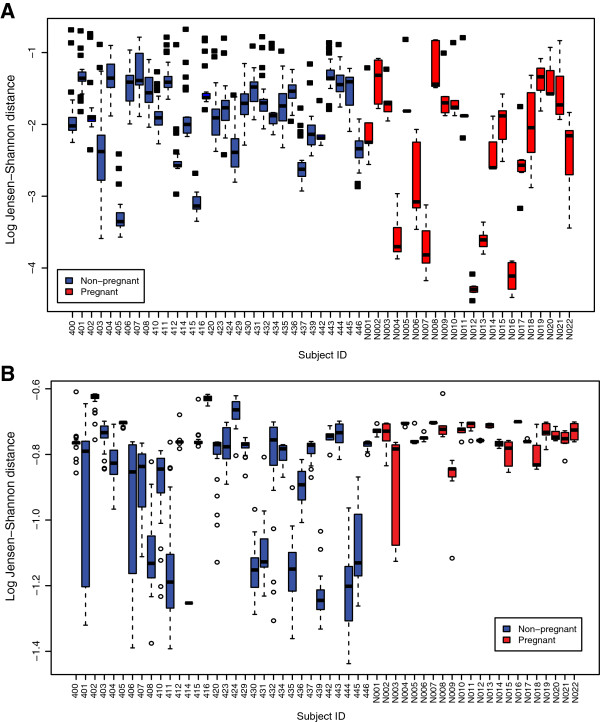
Vaginal bacterial communities of most pregnant and non-pregnant women persist in one CST with some intermittent transitions to other CSTs. Is there a difference in constancy of vaginal bacterial communities between pregnant and non-pregnant women? To address this question, we used an approach in which we computed the mean community state within a subject (mean relative abundance of each bacterial phylotype across all samples of a subject), and then the Jensen-Shannon distance was computed between each community state and the mean community state for each subject. These distances are shown in Figure 3A. This is a measure of instability: the larger the distance, the higher the instability of the microbial community within a subject (in other words, community composition changes often over time). To test if the instability was different between pregnant and non-pregnant women, we modeled the log of these Jensen-Shannon distances using a GEE model. The mean within-subject log Jensen-Shannon distance of pregnant women was significantly lower than that for non-pregnant women (difference in means -0.473 log units; that is, 1.6-fold lower Jensen-Shannon distance, P < 0.001). This means that vaginal bacterial communities are significantly more stable in pregnant than in non-pregnant women. However, the results indicate that, during pregnancy, the structure of the bacterial community undergoes some change. To characterize the nature of the changes during pregnancy, we evaluated the ability of a community to shift to CST IV (A or B) by computing the Jensen-Shannon distance between each community state and the mean community state of all samples assigned to CST IV-A and CST IV-B (mean relative abundance of each bacterial phylotype across all samples in CST IV-A and CST IV-B). We modeled the log of these Jensen-Shannon distances using a GEE model and found that the mean log Jensen-Shannon distance of pregnant women was significantly higher (further away from CST IV-A or CST IV-B) than that for non-pregnant women (difference in means 0.13 log units; that is, 1.14-fold, P < 0.001) (Figure 3B). Altogether, these results indicate that bacterial communities in pregnancy do shift from one CST dominated by Lactobacillus spp. to another CST dominated by Lactobacillus spp., but rarely to CST IV-A or CST IV-B.
Figure 3.
Estimates of community change over time in non-pregnant and pregnant women who delivered at term. (A) Jensen-Shannon distances between each community state and the mean community state for each subject. The larger the distance, the higher the instability of the microbial community within a subject. The mean within-subject log Jensen-Shannon distance of pregnant women was significantly lower than that for non-pregnant women (difference in means -0.473 log units; that is, 1.6-fold lower Jensen-Shannon distance, P < 0.001). (B) Jensen-Shannon distance between each community state and the mean community state of all samples assigned to CST IV-A and CST IV-B. The higher the distance, the less frequently a community enters CST IV-A or CST IV-B. The mean log Jensen-Shannon distance of pregnant women was significantly higher than that for non-pregnant women (difference in means 0.13 log units; that is, 1.14-fold, P < 0.001).
Identification of phylotypes accounting for differences in the structure of vaginal microbiota between the non-pregnant state and normal pregnancy
Table 3 provides evidence that the vaginal microbiota in women who deliver at term is different from the vaginal microbiota of non-pregnant women. Nonetheless, this analysis does not identify explicitly the phylotypes responsible for differences in the structure of the vaginal microbiota between pregnant and non-pregnant women.
In order to identify phylotypes whose relative abundances were significantly different between pregnant and non-pregnant women, we used statistical models that: 1) were designed for count data modeling (assuming Poisson and negative binomial distributions); and 2) allowed correlated observations from the same individuals (for example, linear mixed effect models); while 3) allowing for extra zeroes in the data since some phylotypes were frequently undetected. Three types of models were fitted for each phylotype, including PLME, NBLME and ZINBLME models. The model type with the smallest AIC value was retained for each phylotype and the P-value for group variable (pregnant versus non-pregnant) was computed only for this model. Only phylotypes that were present in at least 25% of all samples were included in the analysis, restricting the number of phylotypes to 28. Table 4 shows the AIC statistics for all three types of models for each phylotype, as well as the estimate, confidence interval and P-value for the best (smallest AIC) model. Of interest, out of the 28 phylotypes tested, the relative abundance of 26 was significantly different between the two groups (q-value <0.1 and fold change >1.5). Four of the significant phylotypes (L. vaginalis, L. crispatus, L. gasseri and L. jensenii) were more abundant in pregnant than non-pregnant women (Additional file 2: Figures S1 show box plots of the relative abundances of all significant phylotypes listed in Table 4). The NBLME model provided the optimal fit for a majority of phylotypes, indicating that there is over-dispersion in the sequence count data and, hence, the Poisson distribution may be too restrictive for the observed count data. This finding is in agreement with previous observations [243]. About a quarter of the significant phylotypes showed zero inflation; therefore, the zero-inflation version of the negative binomial model (ZINBLME) provided the optimum fit based on AIC values. The SAS code and input dataset used to generate the results presented in Table 4 are provided in Additional files 3 and 4, respectively.
Table 4.
Differential relative abundance of microbial phylotypes between pregnant and non-pregnant women and statistics for the phylotype level analysis
| Phylotypes | PLME AIC a,d | NBLME AIC b,d | ZINBLME AIC c,d | Best AIC d | Estimate | Lower 95% CI | Upper 95% CI | Fold change | p-value | q-value e | |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Phylotypes less abundant in pregnancy | |||||||||||
| |
Clostridiales Family XI Incertae Sedis |
3488.3 |
2996.9 |
2998.9 |
NBLME |
-7.095 |
-9.020 |
-5.169 |
-1205.6 |
0.0001 |
0.0002 |
| |
Anaerococcus vaginalis |
3043.8 |
2748.2 |
2759.9 |
NBLME |
-5.873 |
-7.260 |
-4.486 |
-355.4 |
0.0001 |
0.0002 |
| |
Anaerococcus |
5252.6 |
3804.3 |
3806.3 |
NBLME |
-5.557 |
-6.605 |
-4.509 |
-259.0 |
0.0001 |
0.0002 |
| |
Prevotella genogroup 2 |
5510.3 |
4498.2 |
4504.2 |
NBLME |
-5.019 |
-5.655 |
-4.384 |
-151.3 |
0.0001 |
0.0002 |
| |
Peptoniphilus |
4707.8 |
4431.3 |
4433.3 |
NBLME |
-4.921 |
-5.670 |
-4.171 |
-137.1 |
0.0001 |
0.0002 |
| |
Streptococcus anginosus |
3285.4 |
2565.8 |
2583.8 |
NBLME |
-4.629 |
-5.748 |
-3.511 |
-102.4 |
0.0001 |
0.0002 |
| |
Actinomycetales |
5672.8 |
5110.1 |
5112.1 |
NBLME |
-4.546 |
-5.281 |
-3.811 |
-94.2 |
0.0001 |
0.0002 |
| |
Leptotrichia amnionii |
7299.3 |
3889.1 |
3893.3 |
NBLME |
-4.490 |
-5.359 |
-3.621 |
-89.1 |
0.0001 |
0.0002 |
| |
Finegoldia magna |
4687.1 |
4300.6 |
4302.6 |
NBLME |
-4.174 |
-4.958 |
-3.391 |
-65.0 |
0.0001 |
0.0002 |
| |
Prevotella |
4540 |
4094.7 |
4096.7 |
NBLME |
-3.870 |
-4.502 |
-3.238 |
-48.0 |
0.0001 |
0.0002 |
| |
Clostridiales |
5864.5 |
4852.6 |
NA |
NBLME |
-3.373 |
-4.274 |
-2.472 |
-29.2 |
0.0001 |
0.0002 |
| |
Atopobium |
3853.1 |
3275 |
3261.6 |
ZINBLME |
-3.268 |
-3.943 |
-2.593 |
-26.3 |
0.0001 |
0.0002 |
| |
Bacteria |
3167.5 |
3033 |
3040.1 |
NBLME |
-3.083 |
-3.921 |
-2.245 |
-21.8 |
0.0001 |
0.0002 |
| |
Prevotella.bivia |
4178.9 |
3043 |
3045 |
NBLME |
-3.038 |
-4.089 |
-1.986 |
-20.9 |
0.0001 |
0.0002 |
| |
Eggerthella |
3149.7 |
3083.7 |
3065.2 |
ZINBLME |
-1.936 |
-2.813 |
-1.060 |
-6.9 |
0.0001 |
0.0002 |
| |
Gardnerella vaginalis |
5472 |
5105.1 |
5076.7 |
ZINBLME |
-1.760 |
-2.253 |
-1.266 |
-5.8 |
0.0001 |
0.0002 |
| |
Dialister |
4048.5 |
3939.6 |
3940.9 |
NBLME |
-1.399 |
-2.147 |
-0.651 |
-4.1 |
0.0003 |
0.0004 |
| |
Ureaplasma |
2819 |
2700.6 |
2707.8 |
NBLME |
-1.153 |
-1.817 |
-0.490 |
-3.2 |
0.0007 |
0.0010 |
| |
Lactobacillus |
10572 |
9170.2 |
9172.2 |
NBLME |
-0.726 |
-1.169 |
-0.283 |
-2.1 |
0.0013 |
0.0017 |
| |
Atopobium vaginae |
12734 |
6971.9 |
7024.5 |
PLME |
-2.381 |
-3.946 |
-0.816 |
-10.8 |
0.0029 |
0.0037 |
| |
Parvimonas micra |
4512.2 |
3835.1 |
3821.7 |
ZINBLME |
-4.202 |
-7.609 |
-0.795 |
-66.8 |
0.0157 |
0.0183 |
| |
Bifidobacteriaceae |
4056.8 |
3989.9 |
3991.9 |
NBLME |
-0.660 |
-1.429 |
0.110 |
-1.9 |
0.0927 |
0.0998 |
|
Phylotypes more abundant in pregnancy | |||||||||||
| |
Lactobacillus vaginalis |
2489.2 |
2467.9 |
2458.2 |
ZINBLME |
1.704 |
1.190 |
2.218 |
5.5 |
0.0001 |
0.0002 |
| |
Lactobacillus jensenii |
6544.1 |
5564.1 |
5549.7 |
ZINBLME |
1.549 |
1.453 |
1.645 |
4.7 |
0.0001 |
0.0002 |
| |
Lactobacillus crispatus |
11702 |
8094 |
8263.2 |
NBLME |
0.754 |
0.212 |
1.295 |
2.1 |
0.0064 |
0.0078 |
| |
Lactobacillus gasseri |
6917.3 |
4412.4 |
NA |
NBLME |
1.193 |
0.214 |
2.172 |
3.3 |
0.0170 |
0.0190 |
|
Non-significantly different phylotypes | |||||||||||
| |
Lactobacillus iners |
18755 |
12576 |
12604 |
NBLME |
0.165 |
-0.136 |
0.466 |
1.2 |
0.2824 |
0.2929 |
| Aerococcus christensenii | 4321.2 | 3986.9 | 3957.1 | ZINBLME | -0.425 | -1.251 | 0.401 | -1.5 | 0.3132 | 0.3132 | |
aPLME: Poisson Linear Mixed Effects Model.
bNBLME: Negative Binomial Linear Mixed Effects.
cZINBLME: Zero-Inflated Negative Binomial Mixed-Effects Model.
dAIC: Akaike Information Criterion.
eq-value is p-value after adjustment for false-discovery rate (0.1).
Some of the selected phylotypes are defined at the genus and some at the species level (for example, Anaerococcus and Anaerococcus vaginalis), respectively. A genus level phylotype corresponds to a set of sequences that could not be reliably identified at the species level for any known species of the given genus. Thus, in the case of Anaerococcus and Anaerococcus vaginalis, the first phylotype corresponds to reads that cannot be taxonomically assigned to any known species of Anaerococcus and might represent uncharacterized species of Anaerococcus, whereas the phylotype Anaerococcus vaginalis consists of reads that are classified as corresponding to that species.
Discussion
Principal findings of the study
Using sequence-based methods (rather than cultivation techniques) to characterize the vaginal microbiota in a longitudinal study of normal pregnant women and non-pregnant women, we established that: 1) at the bacterial community level, CST IV-B (characterized by high relative abundance of species of Atopobium as well as the presence of Prevotella, Sneathia, Gardnerella, Ruminococcaceae, Parvimonas, Mobiluncus and other taxa previously shown to be associated with bacterial vaginosis) was rarely observed in pregnant women who delivered at term; 2) the vaginal microbiota of normal pregnant women who deliver at term was different from that of non-pregnant women (higher abundance of L. vaginalis, L. crispatus, L. gasseri and L. jensenii and lower abundance of 22 other phylotypes in normal pregnancy); 3) the stability of the vaginal microbiota of pregnant women was higher than that of non-pregnant women; and 4) during normal pregnancy, bacterial communities do shift from one CST dominated by Lactobacillus spp. to another CST dominated by Lactobacillus spp. but rarely to CST IV-A or CST IV-B.
The vaginal microbiota of normal pregnant women
This is the first longitudinal study of the vaginal microbiota in normal pregnancy where samples have been frequently collected and microbial composition has been characterized using high-throughput pyrosequencing of the 16S rRNA gene. Previous studies have used a cross-sectional approach [213] and sparse sampling [212]. Some have used low resolution microbiological and molecular techniques [211,212] to characterize the microbial communities. The methodology used in the present study provides a less biased, in-depth characterization of the bacterial composition and abundance of the vaginal microbiota. The major finding of this study is that normal pregnant women maintain (throughout the entire pregnancy) vaginal CSTs dominated by Lactobacillus spp. This is in contrast with the observations made in the non-pregnant state, in which there were fluctuations between CSTs lacking a substantial number of Lactobacillus spp. and those that are dominated by members of this genus [112].
In a previous study, we focused on non-pregnant women and characterized five different CSTs (CST I to V); CST I, II, III and V were characterized by a predominance of Lactobacillus spp. CST IV was characterized by a low abundance of Lactobacillus spp. and a predominance of other phylotypes, mainly of anaerobic bacteria. This CST was further subdivided into IV-A and IV-B based on hierarchical clustering [112]. The major difference between the two is that CST IV-B has a higher abundance of Atopobium, while CST IV-A has a more even microbial composition including the following phylotypes: Peptoniphilus, Anaerococcus, Corynebacterium, Finegoldia and Prevotella. We have also reported that CST IV-A and CST IV-B were more common in certain ethnic groups (African-American and Hispanic) and were associated with a higher vaginal pH and high Nugent score [109]. In the current study focusing on pregnant women, we identified five of the six CSTs previously described: I, II, III, IV-A and IV-B. We did not find CST V. The most likely explanation for this is that the majority of women enrolled in the present study were African-American, and CST V was previously observed in only 1% of such women [109]. Given the sample size of the current study (n = 22 pregnant women) and the ethnic composition (90% African-American), the lack of representation of CST V is not unexpected. Therefore, these findings do not mean that other studies of the microbiota of pregnant women using a different population would not identify CST V.
Stability of the vaginal microbiota during pregnancy
During normal pregnancy, bacterial communities are more stable than in the non-pregnant state; however, some changes do occur. For example, bacterial communities commonly transitioned from one Lactobacillus-dominated CST to another, but rarely to CST IV-A or CST IV-B. This is a reflection of the importance of Lactobacillus spp. in the vaginal ecosystem during pregnancy. Such an interesting feature can be interpreted to represent an adaptation of the microbial community and the host to maximize reproductive fitness. We propose that the enhanced stability confers greater resilience and has a protective role against ascending infection of the genital tract, which is risk factor for preterm delivery [244-246] and other conditions such as a sonographic short cervix [247-249], cervical insufficiency [250-254], preterm labor in twin gestations [255-257], vaginal bleeding in the third trimester [258], placenta previa [259,260], or some cases of fetal death [261-265]. The mechanisms by which bacterial community stability promotes health in the vaginal niche remain to be determined.
Is the vaginal microbiota unique during pregnancy?
Our findings indicate that there are phylotypes with relative abundance that differ between pregnant and non-pregnant women. Specifically, four phylotypes (L. vaginalis, L. jensenii, L. crispatus and L. gasseri) had higher relative abundance in pregnant than in non-pregnant women. We identified another 22 phylotypes that had lower relative abundance in pregnant than non-pregnant women (Table 4); many of these phylotypes are associated with CST IV-A and CST IV-B. Interestingly, the relative abundance of L. iners was not significantly different between the two groups. This finding might reflect a lack of optimal protection by this common Lactobacillus sp. [109] and deserves further investigation. Aagaard and colleagues [213] have proposed that there is a microbiota signature of pregnancy based upon a cross-sectional study of pregnant (n = 24) and non-pregnant women (n = 60). Using a random forest algorithm, pregnancy was well predicted by relative abundances of different phylotypes in vaginal fluid. At this point, even though there are differences in microbial compositions between the pregnant and non-pregnant state, there is no evidence that these differences are specific to pregnant women. Further, it is unclear if a microbial signature of pregnancy could have utility for diagnostic purposes.
It is possible that the composition of the vaginal microbiome associated with pregnancy may have functional (that is, metabolic, immune) implications for the host [266]. An alternative interpretation is that changes in the microbiota are a consequence of the physiological state of pregnancy. During the course of the menstrual cycle, stability of microbial communities is higher at the time when estrogen concentrations are high (14 and 21 days) [112]. This has been attributed to the effect of estrogens on the maturation of the vaginal epithelium, resulting in the accumulation of glycogen on the upper layer of the epithelium [267-270]. Glycogen is a carbon source metabolized to lactic acid by Lactobacillus spp., causing a low vaginal pH [24,26,29]. Further research is required to determine if the relationship between high estrogens and increased stability is causal.
Strengths and limitations
The major strengths of this work are: 1) the longitudinal nature of the study, which allows characterization of the vaginal microbiota over time; 2) the frequent sampling protocol - this allowed characterization of the dynamics of the bacterial communities in pregnancy to an extent not done before; 3) the quality of the sequence-based techniques (16S rRNA) which reduced bias over other methods, including cultivation techniques; 4) the analytical methods that took into consideration changes over time on the same subject, therefore increasing the power of detection of differences between clinical groups; and 5) inclusion of relevant clinical groups: non-pregnant and normal pregnant women. These strengths allowed meaningful differences to be found among these clinical groups. The use of primer 27 F could be a limitation of this study; this primer may have underestimated the true relative abundance of 16S rRNA genes of Bifidobacteriaceae in general, and those of the genus G. vaginalis, a bacterium commonly found in the vagina of women who experience bacterial vaginosis. The selection of optimal PCR primers is a subject of considerable ongoing discussion in the field of microbiome studies. Unfortunately, there is no consensus, nor a perfect set of primers. In this study, we followed the recommendations of the NIH-funded Human Microbiome Project (http://www.hmpdacc.org/). Another potential limitation of the study is the sample size, which included 22 pregnant women who delivered at term. Yet, despite the apparently limited sample size, the identification of significant differences provides evidence that the study of the vaginal microbiota during pregnancy can yield important insights into the relationship between the structure and dynamics of microbial communities and pregnancy outcome. Further studies are required to confirm these findings, extend the observations and elucidate the role of microorganisms in adverse pregnancy outcome.
Conclusion
This is the first longitudinal study of the human vaginal microbiota in pregnancy. We demonstrate differences in the vaginal bacterial community structure between normal pregnant and non-pregnant women and show that pregnancy is characterized by a greater degree of stability than observed in non-pregnant women. We established the baseline stability patterns of the vaginal microbiota in pregnancy. This could serve as the basis to study the relationship between the vaginal microbiota and adverse pregnancy outcomes. The characterization of the vaginal microbiota in pregnancy has the potential to yield information of prognostic, diagnostic and therapeutic value.
Abbreviations
AIC: Akaike Information Criterion; bp: base pair; CST: community state type; GEE: generalized estimation equation; IQR: interquartile range; NBLME: negative binomial linear mixed effect; NIH: National Institutes of Health; PCR: polymerase chain reaction; PLME: Poisson linear mixed effect; ZINBLME: zero-inflated negative binomial mixed-effect.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RR, SSH, PG, AT and JR conceived the study. RR, SSH, LN, MG, RFL, PC, JM and TC performed the clinical sampling and samples management. DWF performed DNA extraction, 16S rRNA gene amplification and sequencing. DWF, PG and JR processed the sequence data. PG and AT performed the statistical analyses. RR, SSH, PG, AT and JR wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Taxonomic assignments, total 16S rRNA gene sequences assigned to each taxa, and metadata.
Box plots of relative abundances of all phylotypes that have statistically significantly different relative abundance between pregnant and non-pregnant women.
SAS code used to generate the results presented in Table 4.
Input dataset for SAS code used to generate the results presented in Table 4.
Contributor Information
Roberto Romero, Email: romeror@mail.nih.gov.
Sonia S Hassan, Email: hassans2@mail.nih.gov.
Pawel Gajer, Email: pgajer@gmail.com.
Adi L Tarca, Email: atarca@med.wayne.edu.
Douglas W Fadrosh, Email: dougfadrosh@gmail.com.
Lorraine Nikita, Email: lnikita@med.wayne.edu.
Marisa Galuppi, Email: mgaluppi@med.wayne.edu.
Ronald F Lamont, Email: pauline.mills@nwlh.nhs.uk.
Piya Chaemsaithong, Email: pchaemsa@med.wayne.edu.
Jezid Miranda, Email: jmiranda@med.wayne.edu.
Tinnakorn Chaiworapongsa, Email: tchaiwor@med.wayne.edu.
Jacques Ravel, Email: jravel@som.umaryland.edu.
Acknowledgements
This work was funded, in part, by the Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH, DHHS. The authors wish to acknowledge the contributions of the patients who volunteered for these studies, the medical and healthcare personnel involved in the research effort, and colleagues who contributed to the discussions which eventually led to the conduct of the study. We are particularly grateful to Dr Sharon Hillier of the University of Pittsburgh, Dr Jack Sobel of Wayne State University, Dr David Relman of Stanford University and Dr Sorin Draghici of Wayne State University.
References
- Ma B, Forney LJ, Ravel J. Vaginal microbiome: rethinking health and disease. Annu Rev Microbiol. 2012;66:371–389. doi: 10.1146/annurev-micro-092611-150157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döderlein A. Das Scheidensekret und seine Bedeutung fuerdas Puerperalfieber. Die Arten des Scheidensekretes. 1892;11:699. [Google Scholar]
- Thomas S. Döderlein Bacillus: lactobacillus acidophilus. J Infect Dis. 1928;43:218–227. [Google Scholar]
- Rogosa M, Sharpe ME. Species differentiation of human vaginal lactobacilli. J Gen Microbiol. 1960;23:197–201. doi: 10.1099/00221287-23-1-197. [DOI] [PubMed] [Google Scholar]
- Redondo-Lopez V, Cook RL, Sobel JD. Emerging role of lactobacilli in the control and maintenance of the vaginal bacterial microflora. Rev Infect Dis. 1990;12:856–872. doi: 10.1093/clinids/12.5.856. [DOI] [PubMed] [Google Scholar]
- Klebanoff SJ, Coombs RW. Viricidal effect of Lactobacillus acidophilus on human immunodeficiency virus type 1: possible role in heterosexual transmission. J Exp Med. 1991;174:289–292. doi: 10.1084/jem.174.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier SL, Krohn MA, Klebanoff SJ, Eschenbach DA. The relationship of hydrogen peroxide-producing lactobacilli to bacterial vaginosis and genital microflora in pregnant women. Obstet Gynecol. 1992;79:369–373. doi: 10.1097/00006250-199203000-00008. [DOI] [PubMed] [Google Scholar]
- Hillier SL, Krohn MA, Rabe LK, Klebanoff SJ, Eschenbach DA. The normal vaginal flora, H2O2-producing lactobacilli, and bacterial vaginosis in pregnant women. Clin Infect Dis. 1993;16(Suppl 4):S273–S281. doi: 10.1093/clinids/16.supplement_4.s273. [DOI] [PubMed] [Google Scholar]
- Curtis H. What is normal vaginal flora? Genitourin Med. 1997;73:230. doi: 10.1136/sti.73.3.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewankambo N, Gray RH, Wawer MJ, Paxton L, McNaim D, Wabwire-Mangen F. et al. HIV-1 infection associated with abnormal vaginal flora morphology and bacterial vaginosis. Lancet. 1997;350:546–550. doi: 10.1016/s0140-6736(97)01063-5. [DOI] [PubMed] [Google Scholar]
- Gupta K, Stapleton AE, Hooton TM, Roberts PL, Fennell CL, Stamm WE. Inverse association of H2O2-producing lactobacilli and vaginal Escherichia coli colonization in women with recurrent urinary tract infections. J Infect Dis. 1998;178:446–450. doi: 10.1086/515635. [DOI] [PubMed] [Google Scholar]
- Sobel JD. Is there a protective role for vaginal flora? Curr Infect Dis Rep. 1999;1:379–383. doi: 10.1007/s11908-999-0045-z. [DOI] [PubMed] [Google Scholar]
- Pybus V, Onderdonk AB. Microbial interactions in the vaginal ecosystem, with emphasis on the pathogenesis of bacterial vaginosis. Microbes Infect. 1999;1:285–292. doi: 10.1016/s1286-4579(99)80024-0. [DOI] [PubMed] [Google Scholar]
- Martin HL, Richardson BA, Nyange PM, Lavreys L, Hillier SL, Chohan B. et al. Vaginal lactobacilli, microbial flora, and risk of human immunodeficiency virus type 1 and sexually transmitted disease acquisition. J Infect Dis. 1999;180:1863–1868. doi: 10.1086/315127. [DOI] [PubMed] [Google Scholar]
- Donders GG, Bosmans E, Dekeersmaecker A, Vereecken A, Van Bulck B, Spitz B. Pathogenesis of abnormal vaginal bacterial flora. Am J Obstet Gynecol. 2000;182:872–878. doi: 10.1016/s0002-9378(00)70338-3. [DOI] [PubMed] [Google Scholar]
- van De Wijgert JH, Mason PR, Gwanzura L, Mbizvo MT, Chirenje ZM, Iliff V. et al. Intravaginal practices, vaginal flora disturbances, and acquisition of sexually transmitted diseases in Zimbabwean women. J Infect Dis. 2000;181:587–594. doi: 10.1086/315227. [DOI] [PubMed] [Google Scholar]
- Larsen B, Monif GR. Understanding the bacterial flora of the female genital tract. Clin Infect Dis. 2001;32:e69–e77. doi: 10.1086/318710. [DOI] [PubMed] [Google Scholar]
- Vasquez A, Jakobsson T, Ahrne S, Forsum U, Molin G. Vaginal lactobacillus flora of healthy Swedish women. J Clin Microbiol. 2002;40:2746–2749. doi: 10.1128/JCM.40.8.2746-2749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocana VS, Nader-Macias ME. Vaginal lactobacilli: self- and co-aggregating ability. Br J Biomed Sci. 2002;59:183–190. doi: 10.1080/09674845.2002.11783657. [DOI] [PubMed] [Google Scholar]
- Wiesenfeld HC, Hillier SL, Krohn MA, Landers DV, Sweet RL. Bacterial vaginosis is a strong predictor of Neisseria gonorrhoeae and Chlamydia trachomatis infection. Clin Infect Dis. 2003;36:663–668. doi: 10.1086/367658. [DOI] [PubMed] [Google Scholar]
- De Backer E, Verhelst R, Verstraelen H, Alqumber MA, Burton JP, Tagg JR. et al. Quantitative determination by real-time PCR of four vaginal Lactobacillus species. Gardnerella vaginalis and Atopobium vaginae indicates an inverse relationship between L. gasseri and L. iners. BMC Microbiol. 2007;7:115. doi: 10.1186/1471-2180-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SK, Hida K, Shukair S, Wang YY, Figueiredo A, Cone R. et al. Human immunodeficiency virus type 1 is trapped by acidic but not by neutralized human cervicovaginal mucus. J Virol. 2009;83:11196–11200. doi: 10.1128/JVI.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppaluoto PA. Bacterial vaginosis: what is physiological in vaginal bacteriology? An update and opinion. Acta Obstetricia Et Gynecologica Scandinavica. 2011;90:1302–1306. doi: 10.1111/j.1600-0412.2011.01279.x. [DOI] [PubMed] [Google Scholar]
- Linhares IM, Summers PR, Larsen B, Giraldo PC, Witkin SS. Contemporary perspectives on vaginal pH and lactobacilli. Am J Obstet Gynecol. 2011;204(120):e1–e5. doi: 10.1016/j.ajog.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Mirmonsef P, Gilbert D, Zariffard MR, Hamaker BR, Kaur A, Landay AL. et al. The effects of commensal bacteria on innate immune responses in the female genital tract. Am J Reprod Immunol. 2011;65:190–195. doi: 10.1111/j.1600-0897.2010.00943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin SS, Ledger WJ. Complexities of the uniquely human vagina. Science Translational Medicine. 2012;4:132fs11. doi: 10.1126/scitranslmed.3003944. [DOI] [PubMed] [Google Scholar]
- Kashket ER. Bioenergitcs of lactic acid bacteria: cytoplasmic pH and osmotolerance. FEMS Microbiol. 1987;46:233–244. [Google Scholar]
- Russell JB, Diez-Gonzalez F. The effects of fermentation acids on bacterial growth. Adv Microb Physiol. 1998;39:205–234. doi: 10.1016/s0065-2911(08)60017-x. [DOI] [PubMed] [Google Scholar]
- Boskey ER, Telsch KM, Whaley KJ, Moench TR, Cone RA. Acid production by vaginal flora in vitro is consistent with the rate and extent of vaginal acidification. Infect Immun. 1999;67:5170–5175. doi: 10.1128/iai.67.10.5170-5175.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alakomi HL, Skytta E, Saarela M, Mattila-Sandholm T, Latva-Kala K, Helander IM. Lactic acid permeabilizes gram-negative bacteria by disrupting the outer membrane. Appl Environ Microbiol. 2000;66:2001–2005. doi: 10.1128/aem.66.5.2001-2005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saling E, Schreiber M, al-Taie T. A simple, efficient and inexpensive program for preventing prematurity. J Perinat Med. 2001;29:199–211. doi: 10.1515/JPM.2001.029. [DOI] [PubMed] [Google Scholar]
- Boskey ER, Cone RA, Whaley KJ, Moench TR. Origins of vaginal acidity: high D/L lactate ratio is consistent with bacteria being the primary source. Hum Reprod (Oxford, England) 2001;16:1809–1813. doi: 10.1093/humrep/16.9.1809. [DOI] [PubMed] [Google Scholar]
- Hoyme UB, Saling E. Efficient prematurity prevention is possible by pH-self measurement and immediate therapy of threatening ascending infection. Eur J Obstet Gyn R B. 2004;115:148–153. doi: 10.1016/j.ejogrb.2004.02.038. [DOI] [PubMed] [Google Scholar]
- O’Hanlon DE, Moench TR, Cone RA. In vaginal fluid, bacteria associated with bacterial vaginosis can be suppressed with lactic acid but not hydrogen peroxide. BMC Infect Dis. 2011;11:200. doi: 10.1186/1471-2334-11-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschlag MR, Alpert S, Onderdonk AB, Thurston P, Drude E, McCormack WM. et al. Anaerobic microflora of the vagina in children. Am J Obstet Gynecol. 1978;131:853–856. doi: 10.1016/s0002-9378(16)33130-1. [DOI] [PubMed] [Google Scholar]
- Hillier SL, Lau RJ. Vaginal microflora in postmenopausal women who have not received estrogen replacement therapy. Clin Infect Dis. 1997;25(Suppl 2):S123–S126. doi: 10.1086/516221. [DOI] [PubMed] [Google Scholar]
- Alvarez-Olmos MI, Barousse MM, Rajan L, Van Der Pol BJ, Fortenberry D, Orr D. et al. Vaginal lactobacilli in adolescents: presence and relationship to local and systemic immunity, and to bacterial vaginosis. Sex Transm Dis. 2004;31:393–400. doi: 10.1097/01.olq.0000130454.83883.e9. [DOI] [PubMed] [Google Scholar]
- Farage M, Maibach H. Lifetime changes in the vulva and vagina. Arch Gynecol Obstet. 2006;273:195–202. doi: 10.1007/s00404-005-0079-x. [DOI] [PubMed] [Google Scholar]
- Gupta S, Kumar N, Singhal N, Kaur R, Manektala U. Vaginal microflora in postmenopausal women on hormone replacement therapy. Indian J Pathol Microbiol. 2006;49:457–461. [PubMed] [Google Scholar]
- Yamamoto T, Zhou X, Williams CJ, Hochwalt A, Forney LJ. Bacterial populations in the vaginas of healthy adolescent women. J Pediatr Adolesc Gynecol. 2009;22:11–18. doi: 10.1016/j.jpag.2008.01.073. [DOI] [PubMed] [Google Scholar]
- Hickey RJ, Zhou X, Pierson JD, Ravel J, Forney LJ. Understanding vaginal microbiome complexity from an ecological perspective. Transl Res. 2012;160:267–282. doi: 10.1016/j.trsl.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SR, Petzold CR, Galask RP. Qualitative and quantitative changes of the vaginal microbial flora during the menstrual cycle. Am J Reprod Im Mic. 1985;9:1–5. doi: 10.1111/j.1600-0897.1985.tb00331.x. [DOI] [PubMed] [Google Scholar]
- Eschenbach DA, Thwin SS, Patton DL, Hooton TM, Stapleton AE, Agnew K. et al. Influence of the normal menstrual cycle on vaginal tissue, discharge, and microflora. Clin Infect Dis. 2000;30:901–907. doi: 10.1086/313818. [DOI] [PubMed] [Google Scholar]
- Devillard E, Burton JP, Hammond JA, Lam D, Reid G. Novel insight into the vaginal microflora in postmenopausal women under hormone replacement therapy as analyzed by PCR-denaturing gradient gel electrophoresis. Eur J Obstet Gyn R B. 2004;117:76–81. doi: 10.1016/j.ejogrb.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Heinemann C, Reid G. Vaginal microbial diversity among postmenopausal women with and without hormone replacement therapy. Can J Microbiol. 2005;51:777–781. doi: 10.1139/w05-070. [DOI] [PubMed] [Google Scholar]
- Bezirtzoglou E, Voidarou C, Papadaki A, Tsiotsias A, Kotsovolou O, Konstandi M. Hormone therapy alters the composition of the vaginal microflora in ovariectomized rats. Microb Ecol. 2008;55:751–759. doi: 10.1007/s00248-007-9317-z. [DOI] [PubMed] [Google Scholar]
- Hyman RW, Herndon CN, Jiang H, Palm C, Fukushima M, Bernstein D. et al. The dynamics of the vaginal microbiome during infertility therapy with in vitro fertilization-embryo transfer. J Assist Reprod Genet. 2012;29:105–115. doi: 10.1007/s10815-011-9694-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazi YF, Saleem S, Kazi N. Investigation of vaginal microbiota in sexually active women using hormonal contraceptives in Pakistan. BMC Urol. 2012;12:22. doi: 10.1186/1471-2490-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotman RM, Ravel J, Bavoil PM, Gravitt PE, Ghanem KG. Microbiome, sex hormones, and immune responses in the reproductive tract: challenges for vaccine development against sexually transmitted infections. Vaccine. 2013. Epub 2013/10/19. [DOI] [PMC free article] [PubMed]
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA. et al. The NIH human microbiome project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, Wortman JR. et al. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan S, Kong HH, Segre JA. Species-level analysis of DNA sequence data from the NIH Human Microbiome Project. PloS one. 2012;7:e47075. doi: 10.1371/journal.pone.0047075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relman DA. Microbiology: learning about who we are. Nature. 2012;486:194–195. doi: 10.1038/486194a. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel CA, Amsel R, Holmes KK. Diagnosis of bacterial vaginosis by direct gram stain of vaginal fluid. J Clin Microbiol. 1983;18:170–177. doi: 10.1128/jcm.18.1.170-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschenbach DA, Hillier S, Critchlow C, Stevens C, DeRouen T, Holmes KK. Diagnosis and clinical manifestations of bacterial vaginosis. Am J Obstet Gynecol. 1988;158:819–828. doi: 10.1016/0002-9378(88)90078-6. [DOI] [PubMed] [Google Scholar]
- Krohn MA, Hillier SL, Eschenbach DA. Comparison of methods for diagnosing bacterial vaginosis among pregnant women. J Clin Microbiol. 1989;27:1266–1271. doi: 10.1128/jcm.27.6.1266-1271.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent RP, Krohn MA, Hillier SL. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation. J Clin Microbiol. 1991;29:297–301. doi: 10.1128/jcm.29.2.297-301.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier SL, Krohn MA, Nugent RP, Gibbs RS. Characteristics of three vaginal flora patterns assessed by gram stain among pregnant women. Vaginal Infections and Prematurity Study Group. Am J Obstet Gynecol. 1992;166:938–944. doi: 10.1016/0002-9378(92)91368-k. [DOI] [PubMed] [Google Scholar]
- Hay PE, Morgan DJ, Ison CA, Bhide SA, Romney M, McKenzie P. et al. A longitudinal study of bacterial vaginosis during pregnancy. Br J Obstet Gynaecol. 1994;101:1048–1053. doi: 10.1111/j.1471-0528.1994.tb13580.x. [DOI] [PubMed] [Google Scholar]
- Rosenstein IJ, Morgan DJ, Sheehan M, Lamont RF, Taylor-Robinson D. Bacterial vaginosis in pregnancy: distribution of bacterial species in different gram-stain categories of the vaginal flora. J Med Microbiol. 1996;45:120–126. doi: 10.1099/00222615-45-2-120. [DOI] [PubMed] [Google Scholar]
- Schwebke JR, Hillier SL, Sobel JD, McGregor JA, Sweet RL. Validity of the vaginal gram stain for the diagnosis of bacterial vaginosis. Obstet Gynecol. 1996;88:573–576. doi: 10.1016/0029-7844(96)00233-5. [DOI] [PubMed] [Google Scholar]
- Donders GG. Gram stain method shows better sensitivity than clinical criteria for detection of bacterial vaginosis in surveillance of pregnant, low-income women in a clinical setting. Infect Dis Obstet Gynecol. 1999;7:273–275. doi: 10.1002/(SICI)1098-0997(1999)7:6<273::AID-IDOG3>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curzik D, Drazancic A, Hrgovic Z. Nonspecific aerobic vaginitis and pregnancy. Fetal Diagn Ther. 2001;16:187–192. doi: 10.1159/000053906. [DOI] [PubMed] [Google Scholar]
- Marrazzo JM, Koutsky LA, Eschenbach DA, Agnew K, Stine K, Hillier SL. Characterization of vaginal flora and bacterial vaginosis in women who have sex with women. J Infect Dis. 2002;185:1307–1313. doi: 10.1086/339884. [DOI] [PubMed] [Google Scholar]
- Wilson JD, Ralph SG, Rutherford AJ. Rates of bacterial vaginosis in women undergoing in vitro fertilisation for different types of infertility. BJOG. 2002;109:714–717. doi: 10.1111/j.1471-0528.2002.01297.x. [DOI] [PubMed] [Google Scholar]
- Donders GG, Vereecken A, Bosmans E, Dekeersmaecker A, Salembier G, Spitz B. Definition of a type of abnormal vaginal flora that is distinct from bacterial vaginosis: aerobic vaginitis. BJOG. 2002;109:34–43. doi: 10.1111/j.1471-0528.2002.00432.x. [DOI] [PubMed] [Google Scholar]
- Brotman RM, Ravel J, Cone RA, Zenilman JM. Rapid fluctuation of the vaginal microbiota measured by Gram stain analysis. Sex Transm Infect. 2010;86:297–302. doi: 10.1136/sti.2009.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken LR. Separation and purification of bacteria from soil. Appl Environ Microbiol. 1985;49:1482–1487. doi: 10.1128/aem.49.6.1482-1487.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P, Goebel BM, Pace NR. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra A, Palcu CT, Sobel JD, Akins RA. Bacterial vaginosis: culture- and PCR-based characterizations of a complex polymicrobial disease’s pathobiology. Curr Infect Dis Rep. 2007;9:485–500. doi: 10.1007/s11908-007-0074-4. [DOI] [PubMed] [Google Scholar]
- Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M. et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- Torsvik V, Ovreas L. Microbial diversity and function in soil: from genes to ecosystems. Curr Opin Microbiol. 2002;5:240–245. doi: 10.1016/s1369-5274(02)00324-7. [DOI] [PubMed] [Google Scholar]
- Martin DH, Zozaya M, Lillis R, Miller J, Ferris MJ. The microbiota of the human genitourinary tract: trying to see the forest through the trees. Trans Am Clin Climatol Assoc. 2012;123:242–256. [PMC free article] [PubMed] [Google Scholar]
- Ward DM, Weller R, Bateson MM. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- Zoetendal EG, Akkermans AD, De Vos WM. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64:3854–3859. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD. et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;65:4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y. Identification and Quantification of Abundant Species from Pyrosequences of 16S rRNA by Consensus Alignment. Proceedings (IEEE Int Conf Bioin Biomed) 2011;2010:153–157. doi: 10.1109/BIBM.2010.5706555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol R. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosino JF, Highlander S, Luna RA, Gibbs RA, Versalovic J. Metagenomic pyrosequencing and microbial identification. Clin Chem. 2009;55:856–866. doi: 10.1373/clinchem.2008.107565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallen MJ, Loman NJ, Penn CW. High-throughput sequencing and clinical microbiology: progress, opportunities and challenges. Curr Opin Microbiol. 2010;13:625–631. doi: 10.1016/j.mib.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosalbes MJ, Abellan JJ, Durban A, Perez-Cobas AE, Latorre A, Moya A. Metagenomics of human microbiome: beyond 16s rDNA. Clin Microbiol Infec. 2012;18(Suppl 4):47–49. doi: 10.1111/j.1469-0691.2012.03865.x. [DOI] [PubMed] [Google Scholar]
- Hugenholtz P, Pace NR. Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends Biotechnol. 1996;14:190–197. doi: 10.1016/0167-7799(96)10025-1. [DOI] [PubMed] [Google Scholar]
- Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J Microbiol Meth. 2003;55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Weng L, Rubin EM, Bristow J. Application of sequence-based methods in human microbial ecology. Genome Res. 2006;16:316–322. doi: 10.1101/gr.3676406. [DOI] [PubMed] [Google Scholar]
- Pavlova SI, Kilic AO, Kilic SS, So JS, Nader-Macias ME, Simoes JA. et al. Genetic diversity of vaginal lactobacilli from women in different countries based on 16S rRNA gene sequences. J Appl Microbiol. 2002;92:451–459. doi: 10.1046/j.1365-2672.2002.01547.x. [DOI] [PubMed] [Google Scholar]
- Zhou X, Bent SJ, Schneider MG, Davis CC, Islam MR, Forney LJ. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology. 2004;150:2565–2573. doi: 10.1099/mic.0.26905-0. [DOI] [PubMed] [Google Scholar]
- Ferris MJ, Masztal A, Martin DH. Use of species-directed 16S rRNA gene PCR primers for detection of Atopobium vaginae in patients with bacterial vaginosis. J Clin Microbiol. 2004;42:5892–5894. doi: 10.1128/JCM.42.12.5892-5894.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhelst R, Verstraelen H, Claeys G, Verschraegen G, Delanghe J, Van Simaey L. et al. Cloning of 16S rRNA genes amplified from normal and disturbed vaginal microflora suggests a strong association between Atopobium vaginae. Gardnerella vaginalis and bacterial vaginosis. BMC Microbiol. 2004;4:16. doi: 10.1186/1471-2180-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353:1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- Hyman RW, Fukushima M, Diamond L, Kumm J, Giudice LC, Davis RW. Microbes on the human vaginal epithelium. Proc Natl Acad Sci USA. 2005;102:7952–7957. doi: 10.1073/pnas.0503236102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JE, Goh SH, Money DM, Doyle M, Li A, Crosby WL. et al. Characterization of vaginal microflora of healthy, nonpregnant women by chaperonin-60 sequence-based methods. Am J Obstet Gynecol. 2005;193:682–692. doi: 10.1016/j.ajog.2005.02.094. [DOI] [PubMed] [Google Scholar]
- Sha BE, Chen HY, Wang QJ, Zariffard MR, Cohen MH, Spear GT. Utility of Amsel criteria, Nugent score, and quantitative PCR for Gardnerella vaginalis, Mycoplasma hominis, and Lactobacillus spp. for diagnosis of bacterial vaginosis in human immunodeficiency virus-infected women. J Clin Microbiol. 2005;43:4607–4612. doi: 10.1128/JCM.43.9.4607-4612.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Brown CJ, Abdo Z, Davis CC, Hansmann MA, Joyce P. et al. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 2007;1:121–133. doi: 10.1038/ismej.2007.12. [DOI] [PubMed] [Google Scholar]
- Witkin SS, Linhares IM, Giraldo P. Bacterial flora of the female genital tract: function and immune regulation. Best Pract Res Clin Obstet Gynaecol. 2007;21:347–354. doi: 10.1016/j.bpobgyn.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Fredricks DN, Fiedler TL, Thomas KK, Oakley BB, Marrazzo JM. Targeted PCR for detection of vaginal bacteria associated with bacterial vaginosis. J Clin Microbiol. 2007;45:3270–3276. doi: 10.1128/JCM.01272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley BB, Fiedler TL, Marrazzo JM, Fredricks DN. Diversity of human vaginal bacterial communities and associations with clinically defined bacterial vaginosis. Appl Environ Microbiol. 2008;74:4898–4909. doi: 10.1128/AEM.02884-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zozaya-Hinchliffe M, Martin DH, Ferris MJ. Prevalence and abundance of uncultivated Megasphaera-like bacteria in the human vaginal environment. Appl Environ Microbiol. 2008;74:1656–1659. doi: 10.1128/AEM.02127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trama JP, Pascal KE, Zimmerman J, Self MJ, Mordechai E, Adelson ME. Rapid detection of Atopobium vaginae and association with organisms implicated in bacterial vaginosis. Mol Cell Probes. 2008;22:96–102. doi: 10.1016/j.mcp.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Brotman RM, Ravel J. Ready or not: the molecular diagnosis of bacterial vaginosis. Clin Infect Dis. 2008;47:44–46. doi: 10.1086/588662. [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Fredricks DN. The human vaginal bacterial biota and bacterial vaginosis. Interdiscip Perspect Infect Dis. 2008;2008:750479. doi: 10.1155/2008/750479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RF, Sobel JD, Akins RA, Hassan SS, Chaiworapongsa T, Kusanovic JP. et al. The vaginal microbiome: new information about genital tract flora using molecular based techniques. BJOG. 2011;118:533–549. doi: 10.1111/j.1471-0528.2010.02840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL. et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fettweis JM, Serrano MG, Girerd PH, Jefferson KK, Buck GA. A new era of the vaginal microbiome: advances using next-generation sequencing. Chem Biodivers. 2012;9:965–976. doi: 10.1002/cbdv.201100359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Liu C, Mitchell CM, Fiedler TL, Thomas KK, Agnew KJ. et al. Temporal variability of human vaginal bacteria and relationship with bacterial vaginosis. PloS One. 2010;5:e10197. doi: 10.1371/journal.pone.0010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajer P, Brotman RM, Bai G, Sakamoto J, Schutte UM, Zhong X. et al. Temporal dynamics of the human vaginal microbiota. Science Translational Medicine. 2012;4:52–132. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Hansmann MA, Davis CC, Suzuki H, Brown CJ, Schutte U. et al. The vaginal bacterial communities of Japanese women resemble those of women in other racial groups. FEMS Immunol Med Microbiol. 2010;58:169–181. doi: 10.1111/j.1574-695X.2009.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus V, Onderdonk AB. Evidence for a commensal, symbiotic relationship between Gardnerella vaginalis and Prevotella bivia involving ammonia: potential significance for bacterial vaginosis. J Infect Dis. 1997;175:406–413. doi: 10.1093/infdis/175.2.406. [DOI] [PubMed] [Google Scholar]
- Pybus V, Onderdonk AB. A commensal symbiosis between Prevotella bivia and Peptostreptococcus anaerobius involves amino acids: potential significance to the pathogenesis of bacterial vaginosis. FEMS Immunol Med Microbiol. 1998;22:317–327. doi: 10.1111/j.1574-695X.1998.tb01221.x. [DOI] [PubMed] [Google Scholar]
- Verstraelen H, Verhelst R, Claeys G, Temmerman M, Vaneechoutte M. Culture-independent analysis of vaginal microflora: the unrecognized association of Atopobium vaginae with bacterial vaginosis. Am J Obstet Gynecol. 2004;191:1130–1132. doi: 10.1016/j.ajog.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Burton JP, Devillard E, Cadieux PA, Hammond JA, Reid G. Detection of Atopobium vaginae in postmenopausal women by cultivation-independent methods warrants further investigation. J Clin Microbiol. 2004;42:1829–1831. doi: 10.1128/JCM.42.4.1829-1831.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredricks DN, Marrazzo JM. Molecular methodology in determining vaginal flora in health and disease: its time has come. Curr Infect Dis Rep. 2005;7:463–470. doi: 10.1007/s11908-005-0049-2. [DOI] [PubMed] [Google Scholar]
- Bradshaw CS, Tabrizi SN, Fairley CK, Morton AN, Rudland E, Garland SM. The association of Atopobium vaginae and Gardnerella vaginalis with bacterial vaginosis and recurrence after oral metronidazole therapy. J Infect Dis. 2006;194:828–836. doi: 10.1086/506621. [DOI] [PubMed] [Google Scholar]
- Tamrakar R, Yamada T, Furuta I, Cho K, Morikawa M, Yamada H. et al. Association between Lactobacillus species and bacterial vaginosis-related bacteria, and bacterial vaginosis scores in pregnant Japanese women. BMC Infect Dis. 2007;7:128. doi: 10.1186/1471-2334-7-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard JP, Fenollar F, Henry M, Bretelle F, Raoult D. Molecular quantification of Gardnerella vaginalis and Atopobium vaginae loads to predict bacterial vaginosis. Clin Infect Dis. 2008;47:33–43. doi: 10.1086/588661. [DOI] [PubMed] [Google Scholar]
- Ling Z, Kong J, Liu F, Zhu H, Chen X, Wang Y. et al. Molecular analysis of the diversity of vaginal microbiota associated with bacterial vaginosis. BMC Genom. 2010;11:488. doi: 10.1186/1471-2164-11-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespers V, Menten J, Smet H, Poradosu S, Abdellati S, Verhelst R. et al. Quantification of bacterial species of the vaginal microbiome in different groups of women, using nucleic acid amplification tests. BMC Microbiol. 2012;12:83. doi: 10.1186/1471-2180-12-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joesoef MR, Hillier SL, Josodiwondo S, Linnan M. Reproducibility of a scoring system for gram stain diagnosis of bacterial vaginosis. J Clin Microbiol. 1991;29:1730–1731. doi: 10.1128/jcm.29.8.1730-1731.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney ML, Onderdonk AB. Nugent score related to vaginal culture in pregnant women. Obstet Gynecol. 2001;98:79–84. doi: 10.1016/s0029-7844(01)01402-8. [DOI] [PubMed] [Google Scholar]
- Burton JP, Reid G. Evaluation of the bacterial vaginal flora of 20 postmenopausal women by direct (Nugent score) and molecular (polymerase chain reaction and denaturing gradient gel electrophoresis) techniques. J Infect Dis. 2002;186:1770–1780. doi: 10.1086/345761. [DOI] [PubMed] [Google Scholar]
- Peters SE, Beck-Sague CM, Farshy CE, Gibson I, Kubota KA, Solomon F. et al. Behaviors associated with Neisseria gonorrhoeae and Chlamydia trachomatis: cervical infection among young women attending adolescent clinics. Clin Pediatr. 2000;39:173–177. doi: 10.1177/000992280003900307. [DOI] [PubMed] [Google Scholar]
- Cherpes TL, Meyn LA, Krohn MA, Lurie JG, Hillier SL. Association between acquisition of herpes simplex virus type 2 in women and bacterial vaginosis. Clin Infect Dis. 2003;37:319–325. doi: 10.1086/375819. [DOI] [PubMed] [Google Scholar]
- Schwebke JR. Gynecologic consequences of bacterial vaginosis. Obstet Gyn Clin N Am. 2003;30:685–694. doi: 10.1016/s0889-8545(03)00086-x. [DOI] [PubMed] [Google Scholar]
- Plitt SS, Garfein RS, Gaydos CA, Strathdee SA, Sherman SG, Taha TE. Prevalence and correlates of chlamydia trachomatis, neisseria gonorrhoeae, trichomonas vaginalis infections, and bacterial vaginosis among a cohort of young injection drug users in Baltimore, Maryland. Sex Transm Dis. 2005;32:446–453. doi: 10.1097/01.olq.0000154567.21291.59. [DOI] [PubMed] [Google Scholar]
- Kaul R, Nagelkerke NJ, Kimani J, Ngugi E, Bwayo JJ, Macdonald KS. et al. Prevalent herpes simplex virus type 2 infection is associated with altered vaginal flora and an increased susceptibility to multiple sexually transmitted infections. J Infect Dis. 2007;196:1692–1697. doi: 10.1086/522006. [DOI] [PubMed] [Google Scholar]
- Gibbs RS. Asymptomatic bacterial vaginosis: is it time to treat? Am J Obstet Gynecol. 2007;196:495–496. doi: 10.1016/j.ajog.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Fichorova RN. Impact of T. vaginalis infection on innate immune responses and reproductive outcome. J Reprod Immunol. 2009;83:185–189. doi: 10.1016/j.jri.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura K, Yoshimura M, Kobayashi T, Kubo T, Hachisuga T, Kashimura M. Can bacterial vaginosis help to find sexually transmitted diseases, especially chlamydial cervicitis? Int J Std Aids. 2009;20:108–111. doi: 10.1258/ijsa.2008.008249. [DOI] [PubMed] [Google Scholar]
- Mania-Pramanik J, Kerkar SC, Salvi VS. Bacterial vaginosis: a cause of infertility? Int J Std Aids. 2009;20:778–781. doi: 10.1258/ijsa.2009.009193. [DOI] [PubMed] [Google Scholar]
- Brotman RM, Klebanoff MA, Nansel TR, Yu KF, Andrews WW, Zhang J. et al. Bacterial vaginosis assessed by gram stain and diminished colonization resistance to incident gonococcal, chlamydial, and trichomonal genital infection. J Infect Dis. 2010;202:1907–1915. doi: 10.1086/657320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allsworth JE, Peipert JF. Severity of bacterial vaginosis and the risk of sexually transmitted infection. Am J Obstet Gynecol. 2011;205(113):e1–e6. doi: 10.1016/j.ajog.2011.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotman RM, Bradford LL, Conrad M, Gajer P, Ault K, Peralta L. et al. Association between Trichomonas vaginalis and vaginal bacterial community composition among reproductive-age women. Sex Transm Dis. 2012;39:807–812. doi: 10.1097/OLQ.0b013e3182631c79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha TE, Hoover DR, Dallabetta GA, Kumwenda NI, Mtimavalye LA, Yang LP. et al. Bacterial vaginosis and disturbances of vaginal flora: association with increased acquisition of HIV. AIDS. 1998;12:1699–1706. doi: 10.1097/00002030-199813000-00019. [DOI] [PubMed] [Google Scholar]
- Cu-Uvin S, Hogan JW, Caliendo AM, Harwell J, Mayer KH, Carpenter CC. Association between bacterial vaginosis and expression of human immunodeficiency virus type 1 RNA in the female genital tract. Clin Infect Dis. 2001;33:894–896. doi: 10.1086/322613. [DOI] [PubMed] [Google Scholar]
- Zariffard MR, Sha BE, Wang QJ, Chen HY, Bremer J, Cohen MH. et al. Relationship of U1 cell HIV-stimulatory activity to bacterial vaginosis and HIV genital tract virus load. AIDS Res Hum Retrovir. 2005;21:945–948. doi: 10.1089/aid.2005.21.945. [DOI] [PubMed] [Google Scholar]
- Myer L, Kuhn L, Stein ZA, Wright TC Jr, Denny L. Intravaginal practices, bacterial vaginosis, and women’s susceptibility to HIV infection: epidemiological evidence and biological mechanisms. Lancet Infect Dis. 2005;5:786–794. doi: 10.1016/S1473-3099(05)70298-X. [DOI] [PubMed] [Google Scholar]
- Watts DH, Fazzari M, Minkoff H, Hillier SL, Sha B, Glesby M. et al. Effects of bacterial vaginosis and other genital infections on the natural history of human papillomavirus infection in HIV-1-infected and high-risk HIV-1-uninfected women. J Infect Dis. 2005;191:1129–1139. doi: 10.1086/427777. [DOI] [PubMed] [Google Scholar]
- Watts DH, Springer G, Minkoff H, Hillier SL, Jacobson L, Moxley M. et al. The occurrence of vaginal infections among HIV-infected and high-risk HIV-uninfected women: longitudinal findings of the women’s interagency HIV study. J Acquir Immune Defic Syndr. 2006;43:161–168. doi: 10.1097/01.qai.0000242448.90026.13. [DOI] [PubMed] [Google Scholar]
- Spear GT, St John E, Zariffard MR. Bacterial vaginosis and human immunodeficiency virus infection. AIDS Res Ther. 2007;4:25. doi: 10.1186/1742-6405-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JS, Hitti J, Bukusi EA, Mwachari C, Muliro A, Nguti R. et al. Infectious correlates of HIV-1 shedding in the female upper and lower genital tracts. AIDS. 2007;21:755–759. doi: 10.1097/QAD.0b013e328012b838. [DOI] [PubMed] [Google Scholar]
- Rebbapragada A, Howe K, Wachihi C, Pettengell C, Sunderji S, Huibner S. et al. Bacterial vaginosis in HIV-infected women induces reversible alterations in the cervical immune environment. J Acquir Immune Defic Syndr. 2008;49:520–522. doi: 10.1097/QAI.0b013e318189a7ca. [DOI] [PubMed] [Google Scholar]
- Hummelen R, Changalucha J, Butamanya NL, Cook A, Habbema JD, Reid G. Lactobacillus rhamnosus GR-1 and L. reuteri RC-14 to prevent or cure bacterial vaginosis among women with HIV. Int J Gynaecol Obstet. 2010;111:245–248. doi: 10.1016/j.ijgo.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Guedou FA, Van Damme L, Mirembe F, Solomon S, Becker M, Deese J. et al. Intermediate vaginal flora is associated with HIV prevalence as strongly as bacterial vaginosis in a cross-sectional study of participants screened for a randomised controlled trial. Sex Transm Infect. 2012;88:545–551. doi: 10.1136/sextrans-2011-050319. [DOI] [PubMed] [Google Scholar]
- Cohen CR, Lingappa JR, Baeten JM, Ngayo MO, Spiegel CA, Hong T. et al. Bacterial vaginosis associated with increased risk of female-to-male HIV-1 transmission: a prospective cohort analysis among African couples. PLoS Med. 2012;9:e1001251. doi: 10.1371/journal.pmed.1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschenbach DA, Gravett MG, Chen KC, Hoyme UB, Holmes KK. Bacterial vaginosis during pregnancy. An association with prematurity and postpartum complications. Scand J Urol Nephrol Suppl. 1984;86:213–222. [PubMed] [Google Scholar]
- Hardy PH, Hardy JB, Nell EE, Graham DA, Spence MR, Rosenbaum RC. Prevalence of six sexually transmitted disease agents among pregnant inner-city adolescents and pregnancy outcome. Lancet. 1984;2:333–337. doi: 10.1016/s0140-6736(84)92698-9. [DOI] [PubMed] [Google Scholar]
- Gravett MG, Nelson HP, DeRouen T, Critchlow C, Eschenbach DA, Holmes KK. Independent associations of bacterial vaginosis and Chlamydia trachomatis infection with adverse pregnancy outcome. JAMA. 1986;256:1899–1903. [PubMed] [Google Scholar]
- Gravett MG, Hummel D, Eschenbach DA, Holmes KK. Preterm labor associated with subclinical amniotic fluid infection and with bacterial vaginosis. Obstet Gynecol. 1986;67:229–237. doi: 10.1097/00006250-198602000-00013. [DOI] [PubMed] [Google Scholar]
- Minkoff H, Grunebaum A, Feldman J, Cummings M, McCormack WM. Relationship of vaginal pH and Papanicolaou smear results to vaginal flora and pregnancy outcome. Int J Gynaecol Obstet. 1987;25:17–23. doi: 10.1016/0020-7292(87)90179-2. [DOI] [PubMed] [Google Scholar]
- McGregor JA. Prevention of preterm birth: new initiatives based on microbial-host interactions. Obstet Gynecol Surv. 1988;43:1–14. [PubMed] [Google Scholar]
- McGregor JA, French JI, Lawellin D, Todd JK. Preterm birth and infection: pathogenic possibilities. Am J Reprod Im Mic. 1988;16:123–132. doi: 10.1111/j.1600-0897.1988.tb00181.x. [DOI] [PubMed] [Google Scholar]
- Romero R, Mazor M, Wu YK, Sirtori M, Oyarzun E, Mitchell MD. et al. Infection in the pathogenesis of preterm labor. Sem Perinatol. 1988;12:262–279. [PubMed] [Google Scholar]
- Silver HM, Sperling RS, St Clair PJ, Gibbs RS. Evidence relating bacterial vaginosis to intraamniotic infection. Am J Obstet Gynecol. 1989;161:808–812. doi: 10.1016/0002-9378(89)90406-7. [DOI] [PubMed] [Google Scholar]
- Martius J, Eschenbach DA. The role of bacterial vaginosis as a cause of amniotic fluid infection, chorioamnionitis and prematurity – a review. Arch Gynecol Obstet. 1990;247:1–13. doi: 10.1007/BF02390649. [DOI] [PubMed] [Google Scholar]
- Kurki T, Sivonen A, Renkonen OV, Savia E, Ylikorkala O. Bacterial vaginosis in early pregnancy and pregnancy outcome. Obstet Gynecol. 1992;80:173–177. [PubMed] [Google Scholar]
- Riduan JM, Hillier SL, Utomo B, Wiknjosastro G, Linnan M, Kandun N. Bacterial vaginosis and prematurity in Indonesia: association in early and late pregnancy. Am J Obstet Gynecol. 1993;169:175–178. doi: 10.1016/0002-9378(93)90157-e. [DOI] [PubMed] [Google Scholar]
- Eschenbach DA. Bacterial vaginosis and anaerobes in obstetric-gynecologic infection. Clin Infect Dis. 1993;16(Suppl 4):S282–S287. doi: 10.1093/clinids/16.supplement_4.s282. [DOI] [PubMed] [Google Scholar]
- Gibbs RS. Chorioamnionitis and bacterial vaginosis. Am J Obstet Gynecol. 1993;169:460–462. doi: 10.1016/0002-9378(93)90341-f. [DOI] [PubMed] [Google Scholar]
- Hay PE, Lamont RF, Taylor-Robinson D, Morgan DJ, Ison C, Pearson J. Abnormal bacterial colonisation of the genital tract and subsequent preterm delivery and late miscarriage. Br Med J. 1994;308:295–298. doi: 10.1136/bmj.308.6924.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst E, Goffeng AR, Andersch B. Bacterial vaginosis and vaginal microorganisms in idiopathic premature labor and association with pregnancy outcome. J Clin Microbiol. 1994;32:176–186. doi: 10.1128/jcm.32.1.176-186.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor JA, French JI, Jones W, Milligan K, McKinney PJ, Patterson E. et al. Bacterial vaginosis is associated with prematurity and vaginal fluid mucinase and sialidase: results of a controlled trial of topical clindamycin cream. Am J Obstet Gynecol. 1994;170:1048–1059. doi: 10.1016/s0002-9378(94)70098-2. Discussion 59–60. [DOI] [PubMed] [Google Scholar]
- Hillier SL, Nugent RP, Eschenbach DA, Krohn MA, Gibbs RS, Martin DH. et al. Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. The Vaginal Infections and Prematurity Study Group. N Engl J Med. 1995;333:1737–1742. doi: 10.1056/NEJM199512283332604. [DOI] [PubMed] [Google Scholar]
- Meis PJ, Goldenberg RL, Mercer B, Moawad A, Das A, McNellis D. et al. The preterm prediction study: significance of vaginal infections. National Institute of Child Health and Human Development Maternal-Fetal Medicine Units Network. Am J Obstet Gynecol. 1995;173:1231–1235. doi: 10.1016/0002-9378(95)91360-2. [DOI] [PubMed] [Google Scholar]
- Krohn MA, Hillier SL, Nugent RP, Cotch MF, Carey JC, Gibbs RS. et al. The genital flora of women with intraamniotic infection. Vaginal Infection and Prematurity Study Group. J Infect Dis. 1995;171:1475–1480. doi: 10.1093/infdis/171.6.1475. [DOI] [PubMed] [Google Scholar]
- Goldenberg RL, Thom E, Moawad AH, Johnson F, Roberts J, Caritis SN. The preterm prediction study: fetal fibronectin, bacterial vaginosis, and peripartum infection. NICHD Maternal Fetal Medicine Units Network. Obstet Gynecol. 1996;87:656–660. doi: 10.1016/0029-7844(96)00034-8. [DOI] [PubMed] [Google Scholar]
- Wennerholm UB, Holm B, Mattsby-Baltzer I, Nielsen T, Platz-Christensen J, Sundell G. et al. Fetal fibronectin, endotoxin, bacterial vaginosis and cervical length as predictors of preterm birth and neonatal morbidity in twin pregnancies. Br J Obstet Gynaecol. 1997;104:1398–1404. doi: 10.1111/j.1471-0528.1997.tb11010.x. [DOI] [PubMed] [Google Scholar]
- Newton ER, Piper J, Peairs W. Bacterial vaginosis and intraamniotic infection. Am J Obstet Gynecol. 1997;176:672–677. doi: 10.1016/s0002-9378(97)70568-4. [DOI] [PubMed] [Google Scholar]
- Chaim W, Mazor M, Leiberman JR. The relationship between bacterial vaginosis and preterm birth. A review. Arch Gynecol Obstet. 1997;259:51–58. doi: 10.1007/BF02505309. [DOI] [PubMed] [Google Scholar]
- Cotch MF, Pastorek JG 2nd, Nugent RP, Hillier SL, Gibbs RS, Martin DH. et al. Trichomonas vaginalis associated with low birth weight and preterm delivery. The Vaginal Infections and Prematurity Study Group. Sex Transm Dis. 1997;24:353–360. doi: 10.1097/00007435-199707000-00008. [DOI] [PubMed] [Google Scholar]
- Dammann O, Leviton A. Does prepregnancy bacterial vaginosis increase a mother’s risk of having a preterm infant with cerebral palsy? Dev Med Child Neurol. 1997;39:836–840. doi: 10.1111/j.1469-8749.1997.tb07554.x. [DOI] [PubMed] [Google Scholar]
- Sherman DJ, Tovbin J, Lazarovich T, Avrech O, Reif R, Hoffmann S. et al. Chorioamnionitis caused by gram-negative bacteria as an etiologic factor in preterm birth. Eur J Clin Microbiol. 1997;16:417–423. doi: 10.1007/BF02471905. [DOI] [PubMed] [Google Scholar]
- Kimberlin DF, Andrews WW. Bacterial vaginosis: association with adverse pregnancy outcome. Semin Perinatol. 1998;22:242–250. doi: 10.1016/s0146-0005(98)80012-8. [DOI] [PubMed] [Google Scholar]
- Gratacos E, Figueras F, Barranco M, Vila J, Cararach V, Alonso PL. et al. Spontaneous recovery of bacterial vaginosis during pregnancy is not associated with an improved perinatal outcome. Acta Obstetricia Et Gynecologica Scandinavica. 1998;77:37–40. [PubMed] [Google Scholar]
- Flynn CA, Helwig AL, Meurer LN. Bacterial vaginosis in pregnancy and the risk of prematurity: a meta-analysis. J Fam Pract. 1999;48:885–892. [PubMed] [Google Scholar]
- French JI, McGregor JA, Draper D, Parker R, McFee J. Gestational bleeding, bacterial vaginosis, and common reproductive tract infections: risk for preterm birth and benefit of treatment. Obstet Gynecol. 1999;93:715–724. doi: 10.1016/s0029-7844(98)00557-2. [DOI] [PubMed] [Google Scholar]
- Andrews WW, Hauth JC, Goldenberg RL. Infection and preterm birth. Am J Perinatol. 2000;17:357–365. doi: 10.1055/s-2000-13448. [DOI] [PubMed] [Google Scholar]
- Jacobsson B, Pernevi P, Chidekel L, Jorgen P-CJ. Bacterial vaginosis in early pregnancy may predispose for preterm birth and postpartum endometritis. Acta Obstetricia Et Gynecologica Scandinavica. 2002;81:1006–1010. doi: 10.1034/j.1600-0412.2002.811103.x. [DOI] [PubMed] [Google Scholar]
- Subtil D, Denoit V, Le Goueff F, Husson MO, Trivier D, Puech F. The role of bacterial vaginosis in preterm labor and preterm birth: a case–control study. Eur J Obstet Gynecol Reprod Biol. 2002;101:41–46. doi: 10.1016/s0301-2115(01)00515-2. [DOI] [PubMed] [Google Scholar]
- Goffinet F, Maillard F, Mihoubi N, Kayem G, Papiernik E, Cabrol D. et al. Bacterial vaginosis: prevalence and predictive value for premature delivery and neonatal infection in women with preterm labour and intact membranes. Eur J Obstet Gynecol Reprod Biol. 2003;108:146–151. doi: 10.1016/s0301-2115(02)00423-2. [DOI] [PubMed] [Google Scholar]
- Hauth JC, Macpherson C, Carey JC, Klebanoff MA, Hillier SL, Ernest JM. et al. Early pregnancy threshold vaginal pH and Gram stain scores predictive of subsequent preterm birth in asymptomatic women. Am J Obstet Gynecol. 2003;188:831–835. doi: 10.1067/mob.2003.184. [DOI] [PubMed] [Google Scholar]
- Leitich H, Bodner-Adler B, Brunbauer M, Kaider A, Egarter C, Husslein P. Bacterial vaginosis as a risk factor for preterm delivery: a meta-analysis. Am J Obstet Gynecol. 2003;189:139–147. doi: 10.1067/mob.2003.339. [DOI] [PubMed] [Google Scholar]
- Romero R, Chaiworapongsa T, Kuivaniemi H, Tromp G. Bacterial vaginosis, the inflammatory response and the risk of preterm birth: a role for genetic epidemiology in the prevention of preterm birth. Am J Obstet Gynecol. 2004;190:1509–1519. doi: 10.1016/j.ajog.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Goffinet F, Kayem G, Maillard F, Trebeden H, Cabrol D, Weill B. et al. Detection of interleukin 6 mRNA by RT-PCR in vaginal secretions: association with preterm delivery and neonatal infection in women with preterm labour and intact membranes. Eur J Obstet Gynecol Reprod Biol. 2005;123:167–173. doi: 10.1016/j.ejogrb.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Larsson PG, Bergstrom M, Forsum U, Jacobsson B, Strand A, Wolner-Hanssen P. Bacterial vaginosis. Transmission, role in genital tract infection and pregnancy outcome: an enigma. APMIS. 2005;113:233–245. doi: 10.1111/j.1600-0463.2005.apm_01.x. [DOI] [PubMed] [Google Scholar]
- Klebanoff MA, Hillier SL, Nugent RP, MacPherson CA, Hauth JC, Carey JC. et al. Is bacterial vaginosis a stronger risk factor for preterm birth when it is diagnosed earlier in gestation? Am J Obstet Gynecol. 2005;192:470–477. doi: 10.1016/j.ajog.2004.07.017. [DOI] [PubMed] [Google Scholar]
- Goldenberg RL, Culhane JF, Johnson DC. Maternal infection and adverse fetal and neonatal outcomes. Clin Perinatol. 2005;32:523–559. doi: 10.1016/j.clp.2005.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsson B, Mattsby-Baltzer I, Hagberg H. Interleukin-6 and interleukin-8 in cervical and amniotic fluid: relationship to microbial invasion of the chorioamniotic membranes. BJOG. 2005;112:719–724. doi: 10.1111/j.1471-0528.2005.00536.x. [DOI] [PubMed] [Google Scholar]
- Guaschino S, De Seta F, Piccoli M, Maso G, Alberico S. Aetiology of preterm labour: bacterial vaginosis. BJOG. 2006;113:46–51. doi: 10.1111/j.1471-0528.2006.01122.x. [DOI] [PubMed] [Google Scholar]
- Vogel I, Thorsen P, Hogan VK, Schieve LA, Jacobsson B, Ferre CD. The joint effect of vaginal Ureaplasma urealyticum and bacterial vaginosis on adverse pregnancy outcomes. Acta Obstetricia Et Gynecologica Scandinavica. 2006;85:778–785. doi: 10.1080/00016340500442423. [DOI] [PubMed] [Google Scholar]
- Thorsen P, Vogel I, Olsen J, Jeune B, Westergaard JG, Jacobsson B. et al. Bacterial vaginosis in early pregnancy is associated with low birth weight and small for gestational age, but not with spontaneous preterm birth: a population-based study on Danish women. J Matern-Fetal Neo M. 2006;19:1–7. doi: 10.1080/14767050500361604. [DOI] [PubMed] [Google Scholar]
- Azargoon A, Darvishzadeh S. Association of bacterial vaginosis, trichomonas vaginalis, and vaginal acidity with outcome of pregnancy. Arch Iran Med. 2006;9:213–217. [PubMed] [Google Scholar]
- Holst RM, Jacobsson B, Hagberg H, Wennerholm UB. Cervical length in women in preterm labor with intact membranes: relationship to intra-amniotic inflammation/microbial invasion, cervical inflammation and preterm delivery. Ultrasound Obst Gyn. 2006;28:768–774. doi: 10.1002/uog.3837. [DOI] [PubMed] [Google Scholar]
- Andrews WW, Klebanoff MA, Thom EA, Hauth JC, Carey JC, Meis PJ. et al. Midpregnancy genitourinary tract infection with Chlamydia trachomatis: association with subsequent preterm delivery in women with bacterial vaginosis and Trichomonas vaginalis. Am J Obstet Gynecol. 2006;194:493–500. doi: 10.1016/j.ajog.2005.08.054. [DOI] [PubMed] [Google Scholar]
- Leitich H, Kiss H. Asymptomatic bacterial vaginosis and intermediate flora as risk factors for adverse pregnancy outcome. Best Pract Res Clin Obstet Gynaecol. 2007;21:375–390. doi: 10.1016/j.bpobgyn.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Donders GG, Van Calsteren K, Bellen G, Reybrouck R, Van den Bosch T, Riphagen I. et al. Predictive value for preterm birth of abnormal vaginal flora, bacterial vaginosis and aerobic vaginitis during the first trimester of pregnancy. BJOG. 2009;116:1315–1324. doi: 10.1111/j.1471-0528.2009.02237.x. [DOI] [PubMed] [Google Scholar]
- Denney JM, Culhane JF. Bacterial vaginosis: a problematic infection from both a perinatal and neonatal perspective. Semin Fetal Neonatal Med. 2009;14:200–203. doi: 10.1016/j.siny.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Lamont RF, Taylor-Robinson D. The role of bacterial vaginosis, aerobic vaginitis, abnormal vaginal flora and the risk of preterm birth. BJOG. 2010;117:119–120. doi: 10.1111/j.1471-0528.2009.02403.x. Author reply 20–1. [DOI] [PubMed] [Google Scholar]
- Larsson PG, Platz-Christensen JJ, Dalaker K, Eriksson K, Fahraeus L, Irminger K. et al. Treatment with 2% clindamycin vaginal cream prior to first trimester surgical abortion to reduce signs of postoperative infection: a prospective, double-blinded, placebo-controlled, multicenter study. Acta Obstetricia Et Gynecologica Scandinavica. 2000;79:390–396. [PubMed] [Google Scholar]
- Llahi-Camp JM, Rai R, Ison C, Regan L, Taylor-Robinson D. Association of bacterial vaginosis with a history of second trimester miscarriage. Hum Reprod (Oxford, England) 1996;11:1575–1578. doi: 10.1093/oxfordjournals.humrep.a019440. [DOI] [PubMed] [Google Scholar]
- Ralph SG, Rutherford AJ, Wilson JD. Influence of bacterial vaginosis on conception and miscarriage in the first trimester: cohort study. Br Med J. 1999;319:220–223. doi: 10.1136/bmj.319.7204.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DH, Krohn MA, Hillier SL, Eschenbach DA. Bacterial vaginosis as a risk factor for post-cesarean endometritis. Obstet Gynecol. 1990;75:52–58. [PubMed] [Google Scholar]
- Shiraishi T, Fukuda K, Morotomi N, Imamura Y, Mishima J, Imai S. et al. Influence of menstruation on the microbiota of healthy women’s labia minora as analyzed using a 16S rRNA gene-based clone library method. Jpn J Infect Dis. 2011;64:76–80. [PubMed] [Google Scholar]
- Kyongo JK, Jespers V, Goovaerts O, Michiels J, Menten J, Fichorova RN. et al. Searching for lower female genital tract soluble and cellular biomarkers: defining levels and predictors in a cohort of healthy Caucasian women. PloS One. 2012;7:e43951. doi: 10.1371/journal.pone.0043951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell CM, Fredricks DN, Winer RL, Koutsky L. Effect of sexual debut on vaginal microbiota in a cohort of young women. Obstet Gynecol. 2012;120:1306–1313. doi: 10.1097/aog.0b013e31827075ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraelen H, Verhelst R, Claeys G, De Backer E, Temmerman M, Vaneechoutte M. Longitudinal analysis of the vaginal microflora in pregnancy suggests that L. crispatus promotes the stability of the normal vaginal microflora and that L. gasseri and/or L. iners are more conducive to the occurrence of abnormal vaginal microflora. BMC Microbiol. 2009;9:116. doi: 10.1186/1471-2180-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Rodriguez C, Romero-Gonzalez R, Albani-Campanario M, Figueroa-Damian R, Meraz-Cruz N, Hernandez-Guerrero C. Vaginal microbiota of healthy pregnant Mexican women is constituted by four Lactobacillus species and several vaginosis-associated bacteria. Infect Dis Obstet Gynecol. 2011;2011:851485. doi: 10.1155/2011/851485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard K, Riehle K, Ma J, Segata N, Mistretta TA, Coarfa C. et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PloS One. 2012;7:e36466. doi: 10.1371/journal.pone.0036466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relman DA. ‘Til death do us part’: coming to terms with symbiotic relationships. Forward. Nat Rev Microbiol. 2008;6:721–724. doi: 10.1038/nrmicro1990. [DOI] [PubMed] [Google Scholar]
- Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relman DA. The human microbiome: ecosystem resilience and health. Nutr Rev. 2012;70(Suppl 1):S2–S9. doi: 10.1111/j.1753-4887.2012.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC. et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker PI, Love DR, Ferguson LR. Role of gut microbiota in Crohn’s disease. Expet Rev Gastroenterol Hepatol. 2009;3:535–546. doi: 10.1586/egh.09.47. [DOI] [PubMed] [Google Scholar]
- Neuman MG, Nanau RM. Inflammatory bowel disease: role of diet, microbiota, life style. Transl Res. 2012;160:29–44. doi: 10.1016/j.trsl.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Knight R. Spatial and temporal variability of the human microbiota. Clin Microbiol Infec. 2012;18(Suppl 4):8–11. doi: 10.1111/j.1469-0691.2012.03861.x. [DOI] [PubMed] [Google Scholar]
- Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S. et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W. et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Aziz Q, Dore J, Emmanuel A, Guarner F, Quigley EM. Gut microbiota and gastrointestinal health: current concepts and future directions. Neurogastroent Motil. 2013;25:4–15. doi: 10.1111/nmo.12046. [DOI] [PubMed] [Google Scholar]
- Forney LJ, Gajer P, Williams CJ, Schneider GM, Koenig SS, McCulle SL. et al. Comparison of self-collected and physician-collected vaginal swabs for microbiome analysis. J Clin Microbiol. 2010;48:1741–1748. doi: 10.1128/JCM.01710-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsen FA, Kodner RB, Armbrust EV. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics. 2010;11:538. doi: 10.1186/1471-2105-11-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. Divergence measures based on the shannon entropy. IEEE Trans Inf Theory. 1991;37:145–151. [Google Scholar]
- Højsgaard S, Halekoh U, Yan J. The R package geepack for generalized estimating equations. J Stat Softw. 2006;15:1–11. [Google Scholar]
- Lambert D. Zero-inflated Poisson regression, with an application to defects in manufacturing. Technometrics. 1992;34:1–14. [Google Scholar]
- Littell CA, Milliken GA, Stroup WW, Wolfinger RD, Schabenberger O. Nonlinear Mixed Models. Cary, NY, USA: SAS Institute Inc: Edited by Littell CA, Milliken GA, Stroup WW, Wolfinger RD, Schabenberger O; 2006. pp. 567–635. ( In SAS for Mixed Models. 2nd edition). [Google Scholar]
- Erdman D, Jackson L, Sinko A. Zero-Inflated Poisson and Zero-Inflated Negative Binomial Models Using the COUNTREG Procedure. SAS Global Forum; 2008. [ http://www2.sas.com/proceedings/forum2008/322-2008.pdf] [Google Scholar]
- International Human Microbiome Consortium. [ http://www.human-microbiome.org/]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero R, Sirtori M, Oyarzun E, Avila C, Mazor M, Callahan R. et al. Infection and labor. V. Prevalence, microbiology, and clinical significance of intraamniotic infection in women with preterm labor and intact membranes. Am J Obstet Gynecol. 1989;161:817–824. doi: 10.1016/0002-9378(89)90409-2. [DOI] [PubMed] [Google Scholar]
- Romero R, Gomez R, Chaiworapongsa T, Conoscenti G, Kim JC, Kim YM. The role of infection in preterm labour and delivery. Paediatr Perinat Ep. 2001;15(Suppl 2):41–56. doi: 10.1046/j.1365-3016.2001.00007.x. [DOI] [PubMed] [Google Scholar]
- Goncalves LF, Chaiworapongsa T, Romero R. Intrauterine infection and prematurity. Ment Retard Dev D R. 2002;8:3–13. doi: 10.1002/mrdd.10008. [DOI] [PubMed] [Google Scholar]
- Gomez R, Romero R, Nien JK, Chaiworapongsa T, Medina L, Kim YM. et al. A short cervix in women with preterm labor and intact membranes: a risk factor for microbial invasion of the amniotic cavity. Am J Obstet Gynecol. 2005;192:678–689. doi: 10.1016/j.ajog.2004.10.624. [DOI] [PubMed] [Google Scholar]
- Hassan S, Romero R, Hendler I, Gomez R, Khalek N, Espinoza J. et al. A sonographic short cervix as the only clinical manifestation of intra-amniotic infection. J Perinat Med. 2006;34:13–19. doi: 10.1515/JPM.2006.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisbuch E, Hassan SS, Mazaki-Tovi S, Nhan-Chang CL, Kusanovic JP, Chaiworapongsa T. et al. Patients with an asymptomatic short cervix (< or =15 mm) have a high rate of subclinical intraamniotic inflammation: implications for patient counseling. Am J Obstet Gynecol. 2010;202(433):e1–e8. doi: 10.1016/j.ajog.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero R, Gonzalez R, Sepulveda W, Brandt F, Ramirez M, Sorokin Y. et al. Infection and labor. VIII. Microbial invasion of the amniotic cavity in patients with suspected cervical incompetence: prevalence and clinical significance. Am J Obstet Gynecol. 1992;167:1086–1091. doi: 10.1016/s0002-9378(12)80043-3. [DOI] [PubMed] [Google Scholar]
- Mays JK, Figueroa R, Shah J, Khakoo H, Kaminsky S, Tejani N. Amniocentesis for selection before rescue cerclage. Obstet Gynecol. 2000;95:652–655. doi: 10.1016/s0029-7844(99)00633-x. [DOI] [PubMed] [Google Scholar]
- Lee SE, Romero R, Park CW, Jun JK, Yoon BH. The frequency and significance of intraamniotic inflammation in patients with cervical insufficiency. Am J Obstet Gynecol. 2008;198(633):e1–e8. doi: 10.1016/j.ajog.2007.11.047. [DOI] [PubMed] [Google Scholar]
- Bujold E, Morency AM, Rallu F, Ferland S, Tetu A, Duperron L. et al. Bacteriology of amniotic fluid in women with suspected cervical insufficiency. J Obstet Gynaecol Can. 2008;30:882–887. doi: 10.1016/S1701-2163(16)32967-X. [DOI] [PubMed] [Google Scholar]
- Oh KJ, Lee SE, Jung H, Kim G, Romero R, Yoon BH. Detection of ureaplasmas by the polymerase chain reaction in the amniotic fluid of patients with cervical insufficiency. J Perinat Med. 2010;38:261–268. doi: 10.1515/JPM.2010.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero R, Shamma F, Avila C, Jimenez C, Callahan R, Nores J. et al. Infection and labor. VI. Prevalence, microbiology, and clinical significance of intraamniotic infection in twin gestations with preterm labor. Am J Obstet Gynecol. 1990;163:757–761. doi: 10.1016/0002-9378(90)91063-i. [DOI] [PubMed] [Google Scholar]
- Mazor M, Hershkovitz R, Ghezzi F, Maymon E, Horowitz S, Leiberman JR. Intraamniotic infection in patients with preterm labor and twin pregnancies. Acta Obstetricia Et Gynecologica Scandinavica. 1996;75:624–627. doi: 10.3109/00016349609054686. [DOI] [PubMed] [Google Scholar]
- Yoon BH, Park KH, Koo JN, Jun JK, Syn HC, Romero R. Intramniotic infection of twin pregnancies with preterm labor. Am J Obstet Gynecol. 1997;176:S35. [Google Scholar]
- Gomez R, Romero R, Nien JK, Medina L, Carstens M, Kim YM. et al. Idiopathic vaginal bleeding during pregnancy as the only clinical manifestation of intrauterine infection. J Matern-Fetal Neo M. 2005;18:31–37. doi: 10.1080/14767050500217863. [DOI] [PubMed] [Google Scholar]
- Park CW, Moon KC, Park JS, Jun JK, Yoon BH. The frequency and clinical significance of intra-uterine infection and inflammation in patients with placenta previa and preterm labor and intact membranes. Placenta. 2009;30:613–618. doi: 10.1016/j.placenta.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Madan I, Romero R, Kusanovic JP, Mittal P, Chaiworapongsa T, Dong Z. et al. The frequency and clinical significance of intra-amniotic infection and/or inflammation in women with placenta previa and vaginal bleeding: an unexpected observation. J Perinat Med. 2010;38:275–279. doi: 10.1515/JPM.2010.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorp JM Jr, Katz VL, Fowler LJ, Kurtzman JT, Bowes WA Jr. Fetal death from chlamydial infection across intact amniotic membranes. Am J Obstet Gynecol. 1989;161:1245–1246. doi: 10.1016/0002-9378(89)90675-3. [DOI] [PubMed] [Google Scholar]
- Goldenberg RL, Thompson C. The infectious origins of stillbirth. Am J Obstet Gynecol. 2003;189:861–873. doi: 10.1067/s0002-9378(03)00470-8. [DOI] [PubMed] [Google Scholar]
- Blackwell S, Romero R, Chaiworapongsa T, Kim YM, Bujold E, Espinoza J. et al. Maternal and fetal inflammatory responses in unexplained fetal death. J Matern-Fetal Neo M. 2003;14:151–157. doi: 10.1080/jmf.14.3.151.157. [DOI] [PubMed] [Google Scholar]
- Horvath B, Yang M, Manning FA. Intrauterine fetal death caused by Haemophilus influenzae infection: a case report. J Reprod Med. 2008;53:55–56. [PubMed] [Google Scholar]
- Vigliani M. Chorioamnionitis and intrauterine fetal death after second-trimester amniocentesis. Fetal Diagn Ther. 2009;26:216–218. doi: 10.1159/000257087. [DOI] [PubMed] [Google Scholar]
- Doh K, Barton PT, Korneeva I, Perni SC, Bongiovanni AM, Tuttle SL. et al. Differential vaginal expression of interleukin-1 system cytokines in the presence of Mycoplasma hominis and Ureaplasma urealyticum in pregnant women. Infect Dis Obstet Gynecol. 2004;12:79–85. doi: 10.1080/10647440400003667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paavonen J. Physiology and ecology of the vagina. Scand J Infect Dis Supplementum. 1983;40:31–35. [PubMed] [Google Scholar]
- Sjoberg I, Cajander S, Rylander E. Morphometric characteristics of the vaginal epithelium during the menstrual cycle. Gynecol Obstet Inves. 1988;26:136–144. doi: 10.1159/000293685. [DOI] [PubMed] [Google Scholar]
- Galask RP. Vaginal colonization by bacteria and yeast. Am J Obstet Gynecol. 1988;158:993–995. doi: 10.1016/0002-9378(88)90111-1. [DOI] [PubMed] [Google Scholar]
- Miller L, Patton DL, Meier A, Thwin SS, Hooton TM, Eschenbach DA. Depomedroxyprogesterone-induced hypoestrogenism and changes in vaginal flora and epithelium. Obstet Gynecol. 2000;96:431–439. doi: 10.1016/s0029-7844(00)00906-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Taxonomic assignments, total 16S rRNA gene sequences assigned to each taxa, and metadata.
Box plots of relative abundances of all phylotypes that have statistically significantly different relative abundance between pregnant and non-pregnant women.
SAS code used to generate the results presented in Table 4.
Input dataset for SAS code used to generate the results presented in Table 4.