Abstract
The ability of plants to compete effectively for nitrogen (N) resources is critical to plant survival. However, controversy surrounds the importance of organic and inorganic sources of N in plant nutrition because of our poor ability to visualize and understand processes happening at the root–microbial–soil interface.
Using high-resolution nano-scale secondary ion mass spectrometry stable isotope imaging (NanoSIMS-SII), we quantified the fate of 15N over both space and time within the rhizosphere. We pulse-labelled the soil surrounding wheat (Triticum aestivum) roots with either
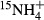 or 15N-glutamate and traced the movement of 15N over 24 h.
or 15N-glutamate and traced the movement of 15N over 24 h.Imaging revealed that glutamate was rapidly depleted from the rhizosphere and that most 15N was captured by rhizobacteria, leading to very high 15N microbial enrichment. After microbial capture, approximately half of the 15N-glutamate was rapidly mineralized, leading to the excretion of
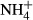 , which became available for plant capture. Roots proved to be poor competitors for 15N-glutamate and took up N mainly as
, which became available for plant capture. Roots proved to be poor competitors for 15N-glutamate and took up N mainly as 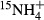 . Spatial mapping of 15N revealed differential patterns of 15N uptake within bacteria and the rapid uptake and redistribution of 15N within roots.
. Spatial mapping of 15N revealed differential patterns of 15N uptake within bacteria and the rapid uptake and redistribution of 15N within roots.In conclusion, we demonstrate the rapid cycling and transformation of N at the soil–root interface and that wheat capture of organic N is low in comparison to inorganic N under the conditions tested.
Keywords: amino acids, dissolved organic nitrogen, NanoSIMS, nitrogen cycling, nutrient uptake, rhizobacteria, rhizosphere architecture
Introduction
Nitrogen (N) is the primary nutrient limiting plant productivity and frequently represents a major constraint to food production world-wide. Consequently, there is increasing interest in plant-based strategies that may promote N use efficiency in crop plants. Conventionally, it was thought that non-N2-fixing plants acquire their N from the soil as nitrate (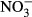 ) and ammonium (
) and ammonium (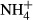 ; Loomis & Connor, 1992; Mosier et al., 2004). There is now strong evidence to suggest that, particularly in N-limiting environments, plants can directly acquire significant amounts of N in the form of organic N (e.g. amino acids and peptides) released during the breakdown of soil organic matter (Näsholm et al., 2009; Hill et al., 2011). However, the evidence to support the significance of organic N uptake in both natural and agricultural systems has drawn severe criticism (Sauheitl et al., 2009). This controversy is largely associated with limitations in experimental design (e.g. the inability to remove soil and microbial contamination from roots), the inappropriate interpretation of 15N and 13C isotopic data (e.g. lack of consideration of 15N pool dilution and N efflux from roots), poor consideration for differential carbon (C) and N fractionation in plants (e.g. metabolic and root–shoot partitioning) and the use of inappropriate N concentrations and timescales (Jones et al., 2005a,2005b; Rasmussen et al., 2010; Warren, 2012). While it is clear that plants can potentially take up and assimilate a wide range of organic N sources in sterile hydroponic culture (amino acids, peptides, proteins, urea, polyamines etc.; McKee, 1962; Jones & Darrah, 1994; Soper et al., 2011), it is the magnitude of the organic N flux versus
; Loomis & Connor, 1992; Mosier et al., 2004). There is now strong evidence to suggest that, particularly in N-limiting environments, plants can directly acquire significant amounts of N in the form of organic N (e.g. amino acids and peptides) released during the breakdown of soil organic matter (Näsholm et al., 2009; Hill et al., 2011). However, the evidence to support the significance of organic N uptake in both natural and agricultural systems has drawn severe criticism (Sauheitl et al., 2009). This controversy is largely associated with limitations in experimental design (e.g. the inability to remove soil and microbial contamination from roots), the inappropriate interpretation of 15N and 13C isotopic data (e.g. lack of consideration of 15N pool dilution and N efflux from roots), poor consideration for differential carbon (C) and N fractionation in plants (e.g. metabolic and root–shoot partitioning) and the use of inappropriate N concentrations and timescales (Jones et al., 2005a,2005b; Rasmussen et al., 2010; Warren, 2012). While it is clear that plants can potentially take up and assimilate a wide range of organic N sources in sterile hydroponic culture (amino acids, peptides, proteins, urea, polyamines etc.; McKee, 1962; Jones & Darrah, 1994; Soper et al., 2011), it is the magnitude of the organic N flux versus 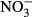 and
and 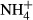 that is critical to determining its significance in natural systems. The simple demonstration that organic N is taken up intact from soil by the root also remains controversial (Sauheitl et al., 2009). Amino acids can be cleaved outside the root by extracellular enzymes or microorganisms releasing keto acids,
that is critical to determining its significance in natural systems. The simple demonstration that organic N is taken up intact from soil by the root also remains controversial (Sauheitl et al., 2009). Amino acids can be cleaved outside the root by extracellular enzymes or microorganisms releasing keto acids, 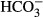 and
and 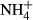 , all of which can be taken up by the root but by different transport mechanisms. Some of the most convincing evidence for the direct uptake of organic N has come from experiments involving the use of dual 13C- and 15N-labelled amino acids, where isotopic compound-specific tracking has show the flow of exogenously applied 13C-15N-glycine through the plant (Persson & Näsholm, 2001; Thornton, 2001).
, all of which can be taken up by the root but by different transport mechanisms. Some of the most convincing evidence for the direct uptake of organic N has come from experiments involving the use of dual 13C- and 15N-labelled amino acids, where isotopic compound-specific tracking has show the flow of exogenously applied 13C-15N-glycine through the plant (Persson & Näsholm, 2001; Thornton, 2001).
Our current understanding of N uptake in plants has been severely limited by the lack of suitable methods for the in situ imaging and quantification of isotopic N flow at the submillimetre scale. We have therefore had to rely on large bulk analysis of plants by conventional mass spectrometry, which prevents differentiation between adjacent tissues and between root cells and closely associated organisms (e.g. between epidermal and cortical cells, between roots and endophytic bacteria or between roots and their mycorrhizal symbionts). However, it is precisely at the root–soil interface and at this scale of resolution that experiments need to be conducted to resolve the controversy surrounding inorganic versus organic N nutrition. Secondary ion mass spectrometry (SIMS) enables imaging and isotopic discrimination for stable isotopes (e.g. 14N versus 15N), thereby providing the possibility to undertake spatially discrete studies of nutrient flow at the subcellular level. In a preliminary study, we showed the potential to spatially resolve 15N in wheat (Triticum aestivum) root cells through application of nano-scale SIMS (NanoSIMS; Clode et al., 2009). Here, we utilized a combination of traditional and NanoSIMS techniques to image and quantify competition for different forms of 15N in situ within the rhizosphere of wheat.
Our specific aims were to determine: the relative uptake rate of organic and inorganic N supplied to roots in situ; subsequent N partitioning within root tissues and within the whole plant; and the amount of competition for the different N forms from bacteria living on the rhizoplane and in the ecto- and endo-rhizosphere.
Materials and Methods
Microcosm preparation and 15N labelling
A coarse-textured agricultural soil routinely used for wheat production in rotation with legumes and dominated by quartz sand grains (92% sand) was obtained from a freely draining soil located in Meckering, Western Australia (31°40′N, 117°00′E). Soil samples were collected from the Ap horizon (0–15 cm; 10 ± 1 mg organic C kg−1 and 0.45 mg total N kg−1) using a stainless steel corer and stored in CO2-permeable polypropylene bags for transport back to the laboratory where they were sieved (< 5 mm) and stored field-moist at 3°C. Seeds of wheat (Triticum aestivum L.) were soaked for 24 h in water and then allowed to germinate on moistened filter paper at 20°C. After 3 d, each plant had one main root axis c. 1 cm in length and two lateral roots 0.5 cm in length; the seedlings were then placed into individual soil-filled microcosms (Owen & Jones, 2001). The plant–soil microcosms were constructed from polyethylene tubing (30 cm long; 0.6 cm internal diameter) and filled with soil to a bulk density of 1.25 g cm−3 and wetted to 70% of their water-holding capacity with distilled water. To half the microcosms, 3-d-old seedlings were added to the top and covered with a further 1 cm of soil. Planted and unplanted microcosms were then maintained at 20°C, with a light intensity of 260 μmol m−2 s−1 photosynthetically active radiation and a 12-h photoperiod. Soil within the microcosms was kept moist by the daily addition of deionized water. A large number of microcosms were set up and the ones with poor plant growth (< 10% total) were discarded before the microcosms were randomly pre-allocated to treatments and labelled. When the roots and associated root hairs had completely occupied the microcosm, making it essentially all rhizosphere soil (15 d after transplantation; shoots 12.4 ± 0.5 cm long; mean ± SE, n = 12), 500 μl of 45 mg N l−1 (3 mM) 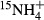 or 15N-labelled glutamate (0.99 15N : 14N ratio; Isotec, Miamisburg, OH, USA) was injected at three locations (3, 5 and 7 cm below the soil surface) through pre-made holes, giving a total injection volume of 1500 μl. The volume of solution used was sufficient to occupy soil pores to c. 1 cm either side of the injection point. All microcosms were injected by the same person to avoid bias. The tubes were then randomized for microcosm harvesting and maintained at 20°C as described above. As controls, microcosms with and without plants were injected with 18 MΏ deionized water instead of 15N using the same injection procedures as described above. Glutamate was chosen as a model amino acid as it is central to amino acid metabolism, is present at high concentrations in plant tissues and soil organic matter (Friedel & Scheller, 2002; Gattolin et al., 2008), can be the dominant free amino acid in soil solution (Jones et al., 2005a,2005b) and is known to be taken up by plant roots (Jones & Darrah, 1994; Forsum et al., 2008).
or 15N-labelled glutamate (0.99 15N : 14N ratio; Isotec, Miamisburg, OH, USA) was injected at three locations (3, 5 and 7 cm below the soil surface) through pre-made holes, giving a total injection volume of 1500 μl. The volume of solution used was sufficient to occupy soil pores to c. 1 cm either side of the injection point. All microcosms were injected by the same person to avoid bias. The tubes were then randomized for microcosm harvesting and maintained at 20°C as described above. As controls, microcosms with and without plants were injected with 18 MΏ deionized water instead of 15N using the same injection procedures as described above. Glutamate was chosen as a model amino acid as it is central to amino acid metabolism, is present at high concentrations in plant tissues and soil organic matter (Friedel & Scheller, 2002; Gattolin et al., 2008), can be the dominant free amino acid in soil solution (Jones et al., 2005a,2005b) and is known to be taken up by plant roots (Jones & Darrah, 1994; Forsum et al., 2008).
Microcosm harvesting
After 5, 30, 90, 360 or 1440 min of exposure to 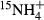 or 15N-glutamate, the microcosms were destructively harvested using three people to ensure rapid sample processing. The control, non-15N-labelled microcosms were harvested after 5 and 1440 min. Four independent microcosms were used for each harvest time for both the 15N-labelled substrates and the control treatment (i.e. n = 4 replications). At each harvest time, the shoots were excised and dried at 80°C, weighed and stored at 20°C for isotopic analysis. Immediately following recovery of the shoots, 2-cm-long microcosm soil sections were cut from around each of the three injection points using a razor blade to allow rapid cutting with minimal disturbance. The three sections were randomly allocated for NanoSIMS analysis, soil solution recovery, or KCl extraction and root recovery. Immediately after excision, the 2-cm-long microcosm sections destined for NanoSIMS analysis (microcosm harvest times = 5, 30 or 1440 min) and containing intact plant roots and soil were covered across the bottom with a cotton membrane, porous to water and solvents, which prevented soil movement during handling. These intact cores were then immersed in 2.5% glutaraldehyde at 4°C to enable extremely rapid fixation and then stored in 2.5% glutaraldehyde. The cross-linking nature of glutaraldehyde results in this chemical fixative being suitable for both the preservation and immobilization of 15N-labelled compounds in cellular material before NanoSIMS analysis, as demonstrated in Mays et al. (1984), Peteranderl & Lechene (2004), Herrmann et al. (2007), Clode et al. (2009) and Pernice et al. (2012). The time from excision to fixation was < 30 s and samples were stored in the glutaraldehyde for up to 24 h to ensure complete fixation of the soil core. An additional experiment performed on replicate 7-d-old wheat plants (n = 4) grown in hydroponic culture (200 μM KCl and 1 mM CaCl2) and then supplied with 15N-labelled glutamate for 30 min or 24 h also showed no significant differences in 15N content between roots that had undergone fixation and dehydration (as above) and roots that had not (30 min, P = 0.913; 24 h, P = 0.09).
or 15N-glutamate, the microcosms were destructively harvested using three people to ensure rapid sample processing. The control, non-15N-labelled microcosms were harvested after 5 and 1440 min. Four independent microcosms were used for each harvest time for both the 15N-labelled substrates and the control treatment (i.e. n = 4 replications). At each harvest time, the shoots were excised and dried at 80°C, weighed and stored at 20°C for isotopic analysis. Immediately following recovery of the shoots, 2-cm-long microcosm soil sections were cut from around each of the three injection points using a razor blade to allow rapid cutting with minimal disturbance. The three sections were randomly allocated for NanoSIMS analysis, soil solution recovery, or KCl extraction and root recovery. Immediately after excision, the 2-cm-long microcosm sections destined for NanoSIMS analysis (microcosm harvest times = 5, 30 or 1440 min) and containing intact plant roots and soil were covered across the bottom with a cotton membrane, porous to water and solvents, which prevented soil movement during handling. These intact cores were then immersed in 2.5% glutaraldehyde at 4°C to enable extremely rapid fixation and then stored in 2.5% glutaraldehyde. The cross-linking nature of glutaraldehyde results in this chemical fixative being suitable for both the preservation and immobilization of 15N-labelled compounds in cellular material before NanoSIMS analysis, as demonstrated in Mays et al. (1984), Peteranderl & Lechene (2004), Herrmann et al. (2007), Clode et al. (2009) and Pernice et al. (2012). The time from excision to fixation was < 30 s and samples were stored in the glutaraldehyde for up to 24 h to ensure complete fixation of the soil core. An additional experiment performed on replicate 7-d-old wheat plants (n = 4) grown in hydroponic culture (200 μM KCl and 1 mM CaCl2) and then supplied with 15N-labelled glutamate for 30 min or 24 h also showed no significant differences in 15N content between roots that had undergone fixation and dehydration (as above) and roots that had not (30 min, P = 0.913; 24 h, P = 0.09).
For soil solution recovery, the 2-cm-long microcosm section was weighed and then placed into a 1.5-ml Eppendorf tube in which a hole had been drilled in the bottom. This was then placed in the top of another 1.5-ml Eppendorf tube and the device was spun at 18 000 g for 5 min. The soil solution recovered in the lower Eppendorf tube was removed and respun at 18 000 g for 5 min, and the supernatant was stored at −20°C to await chemical analysis. The soil remaining was reweighed and dried overnight at 80°C and its dry weight determined. The soil solution recovered accounted for 83 ± 2% (mean ± SE) of the initial soil water present (264 ± 6 g H2O kg−1, mean ± SE).
The final 2-cm section was weighed and placed in 10 ml of 2 M KCl and shaken for 1 h on a rotary shaker at 60 rev min−1. After shaking, the excised roots were recovered and any loosely adhered soil was removed using forceps under a dissecting microscope and the samples were dried at 80°C, weighed and stored at 20°C for subsequent isotopic analysis. The 2 M KCl extract was then filtered through a GF-A glass fibre filter paper (vacuum-assisted), the residual soil was washed with 10 ml of 18 MΏ deionized water and the wash water was added to the 2 M KCl extract. The extracts were subsequently stored at −20°C to await analysis. The residual soil on the filter paper was recovered, using a small amount of 18 MΏ deionized water, into a Petri dish and the soil suspension was dried at 40°C (16 h) before weighing and storage at 20°C.
The amount of 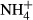 in soil solution and in the KCl extracts was determined using the hypochlorite-salicylic acid procedure of Mulvaney (1996) on a Cary Microplate Reader (Perkin-Elmer Corp., Beverly, MA, USA). Free amino acids were determined fluorometrially on a Cary Eclipse fluorimeter according to the o-phthialdehyde-β-mercaptoethanol procedure (Jones et al., 2002). Nitrate was determined colorimetrically on a San+ segmented flow autoanalyzer equipped with a Cd-Cu reduction column (Skalar Ltd, Breda, the Netherlands).
in soil solution and in the KCl extracts was determined using the hypochlorite-salicylic acid procedure of Mulvaney (1996) on a Cary Microplate Reader (Perkin-Elmer Corp., Beverly, MA, USA). Free amino acids were determined fluorometrially on a Cary Eclipse fluorimeter according to the o-phthialdehyde-β-mercaptoethanol procedure (Jones et al., 2002). Nitrate was determined colorimetrically on a San+ segmented flow autoanalyzer equipped with a Cd-Cu reduction column (Skalar Ltd, Breda, the Netherlands).
Quantification of 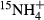 and
and 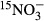 in the KCl extracts was performed using a two-stage microdiffusion over 7-d periods at 20°C. This was achieved by first diffusing 5 ml of the extracts with MgO, and subsequently with Devarda’s alloy (Brooks et al., 1989), and capturing separate 15N : 14N ratios in diffusion discs which were analysed by Tracer MS (Europa 20 : 20; Europa Scientific, Crewe, Cheshire, UK). Standards of 15N-enriched (NH4)2SO4 at 2 and 5 atom% excess were also diffused. Control (spiked) samples revealed no interference of 15N-glutamate in
in the KCl extracts was performed using a two-stage microdiffusion over 7-d periods at 20°C. This was achieved by first diffusing 5 ml of the extracts with MgO, and subsequently with Devarda’s alloy (Brooks et al., 1989), and capturing separate 15N : 14N ratios in diffusion discs which were analysed by Tracer MS (Europa 20 : 20; Europa Scientific, Crewe, Cheshire, UK). Standards of 15N-enriched (NH4)2SO4 at 2 and 5 atom% excess were also diffused. Control (spiked) samples revealed no interference of 15N-glutamate in 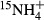 or
or 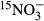 determination.
determination.
Adsorption isotherms and amino acid turnover
To determine the soil-to-solid phase partitioning of 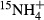 and 15N-glutamate after addition to the soil, concentration-dependent adsorption isotherms were performed. Briefly, solutions of 14C-glutamate (0.168 kBq ml−1) or
and 15N-glutamate after addition to the soil, concentration-dependent adsorption isotherms were performed. Briefly, solutions of 14C-glutamate (0.168 kBq ml−1) or 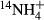 (0–5 mM; 10 ml) in a background of 0.01 M KCl were mixed with replicate (n = 3) batches of soil (5 g) for 15 min (200 rev min−1), the soil suspensions were centrifuged (18 000 g for 5 min) and the amount of
(0–5 mM; 10 ml) in a background of 0.01 M KCl were mixed with replicate (n = 3) batches of soil (5 g) for 15 min (200 rev min−1), the soil suspensions were centrifuged (18 000 g for 5 min) and the amount of 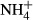 in the supernatant solution was determined as described in the previous section. The concentration of 14C-glutamate was determined using HiSafe 3® liquid scintillation fluid (Perkin-Elmer Inc., Waltham, MA, USA) and a Wallac 1408 liquid scintillation counter (Perkin-Elmer). To prevent microbial degradation of the added N sources, the soils were pre-sterilized at 80°C (for 30 min) before performing the assays (Kuzyakov & Jones, 2006). Our previous studies have shown that without this mild heat sterilization both N forms would be rapidly consumed by the soil microbial community, severely biasing the results (Rousk & Jones, 2010; Abaas et al., 2012). While heating caused no observable change in soil structure, we acknowledge that heating may cause the release of potentially competing amino acids from within microbial cells; however, we expect this to be minimal based on the large concentration of our added N in comparison to that present in the microbial community (Jörgensen, 1996). No significant effect of heating on the cation exchange properties of the soil was expected to occur (Forgeard & Frenot, 1996).
in the supernatant solution was determined as described in the previous section. The concentration of 14C-glutamate was determined using HiSafe 3® liquid scintillation fluid (Perkin-Elmer Inc., Waltham, MA, USA) and a Wallac 1408 liquid scintillation counter (Perkin-Elmer). To prevent microbial degradation of the added N sources, the soils were pre-sterilized at 80°C (for 30 min) before performing the assays (Kuzyakov & Jones, 2006). Our previous studies have shown that without this mild heat sterilization both N forms would be rapidly consumed by the soil microbial community, severely biasing the results (Rousk & Jones, 2010; Abaas et al., 2012). While heating caused no observable change in soil structure, we acknowledge that heating may cause the release of potentially competing amino acids from within microbial cells; however, we expect this to be minimal based on the large concentration of our added N in comparison to that present in the microbial community (Jörgensen, 1996). No significant effect of heating on the cation exchange properties of the soil was expected to occur (Forgeard & Frenot, 1996).
Glutamate mineralization in soil was determined as described in Hill et al. (2012). Briefly, 14C-U-glutamate (3 mM) was added to soil obtained from planted and unplanted microcosms (50 μl g−1) and 14CO2 captured over a 24-h period using 1 M NaOH traps. 14C in the traps was quantified by liquid scintillation counting using HiSafe 3® scintillation fluid and a Wallac 1408 scintillation counter. A double first-order kinetic model was fitted to the mineralization data and the first kinetic phase used to estimate the half-life of the 14C-glutamate added to the soil (see Boddy et al. (2008) and Hill et al. (2012) for further details of the modelling protocols). As the concentration of the introduced 14C-glutamate was much higher than the intrinsic 12C-glutamate concentration, the half-life estimates were not subject to major problems of isotopic pool dilution (Cobelli et al., 2001).
Preparation and imaging of samples
Preparation and imaging of samples before NanoSIMS analyses were performed as described in detail by Herrmann et al. (2007) and Clode et al. (2009). Briefly, microcosm sections were fixed and stored in 2.5% glutaraldehyde, before being dehydrated with acetone and infiltrated with araldite. Resin-embedded soil plus plant root cores were cut into slices and re-embedded into 10-mm NanoSIMS mounts. These mounts were polished and gold-coated before being imaged at 15 kV using scanning electron microscopy (SEM; Zeiss 55 field emission) to identify regions of interest (ROIs) for NanoSIMS analysis. For correlative structural and NanoSIMS imaging of bacteria associated with root cells, fixed roots from the microcosms were extracted from the soil, dehydrated and resin-embedded. Thin (120-nm) sections were cut on a diamond knife and mounted on C-filmed copper grids, and ROIs were identified and imaged at 120 kV in a transmission electron microscope (TEM; JEOL 2100) fitted with a digital camera (Gatan, ORIUS1000; Gatan Inc., Pleasanton, CA, USA).
NanoSIMS analyses
In situ isotopic mapping was performed using a NanoSIMS 50 (Cameca, Gennevilliers, France), with a 16-kV Cs+ primary ion beam. The analyses were performed in multi-collection mode, with the trolleys positioned to detect the negative secondary ions 12C−, 16O−, 12C14N−, 12C15N−, and 28Si−, simultaneously. The mass spectrometer was tuned to a high mass resolution of 9000 (CAMECA definition) to separate the 12C15N− from the 13C14N− peak on mass 27 using an entrance slit of 30 μm, an aperture slit of 200 μm, and a 10% reduction in the signal at the energy slit.
For standard resolution images, the primary current was set to c. 5 pA using a 300-μm primary aperture (D1), giving a spot size of c. 120 nm. Images were acquired by rastering the beam over an area 28 × 28 μm square, at a resolution of 256 × 256 pixels, giving a pixel size of 110 nm, slightly smaller than the beam diameter. For all samples, a systematic approach was taken to obtain comparable images from discrete distances extending from the root centre out into the rhizosphere – up to 500 μm. Poor preservation of root hairs and epidermal cells prevented their study in detail. High-resolution images of the sample prepared for TEM analysis were acquired with a c. 0.1-pA primary beam with a spot size of < 70 nm, using a smaller primary aperture (150 μm) and high voltage on the L1 lens. Initially, the same thin section samples were imaged in both TEM and NanoSIMS, but the samples were found to be extremely delicate under the high-energy ion beam, which would often ‘burn though’ after only one or two images had been acquired. Most of the isotope data for the bacteria were obtained from 1-μm-thick serial sections mounted on gold-coated aluminium stubs, with the adjacent thin sections prepared for TEM analysis, allowing the same bacteria and cortical cell walls to be imaged by both techniques. Image data consist of the total number of counts for a given secondary ion species recorded on each pixel, with count times kept constant at 40 ms pixel−1. All areas were implanted to the same ion dose by the primary beam before each acquisition to remove surface contamination and to enhance the generation of secondary ions.
Isotope data were extracted from the images using the openmims data analysis software (NIH/NIBIB National Resource for Imaging Mass Spectrometry, Cambridge, MA, USA) for the freeware package imagej (National Institute of Mental Health, Bethesda, MD, USA). Images were corrected for detector dead time (44 ns) on the individual pixels before any other data processing. Maps representing the 15N : 14N ratio were obtained by dividing the 12C15N counts by 12C14N counts on each pixel. Using the Hue-Saturation-Intensity function of the openmims data analysis software, it is possible to express the level of isotopic turnover in different components within a single image on a colour scale, thus highlighting the small, subtle differences that are not so apparent in a typical greyscale ratio image. Numerical 15N : 14N ratio data were extracted directly from the images by drawing ROI, discrete groups of pixels that define a particular feature, and extracting the total number of counts for the given ROIs. The ratio of the ROIs and the associated uncertainty (Poisson counting error) is then determined. The relative abundance of 15N : 14N from ROIs was then statistically compared with natural 15N abundance using Wilcoxon statistics. Counting errors for any given ROI were of the order of a few per cent. Quoted uncertainties are the standard error of the mean of the measured ROIs for any given bacterial/plant cell component, and were also of the order of a few per cent. This indicates that the reproducibility is tracking the counting statistics.
15N analysis of shoots, roots and soil
The entire shoots or roots for each replicate were weighed (5 decimal places; d.p.) into individual tin capsules for 15N determination. As the root samples were very small, a reference material (ANCA 56 Jarrah litter %N = 0.693; δ15N = −2.71; D.V. Murphy, University of Western Australia, WA, USA) was also weighed (5 d.p.) into the tin cups (c. 20 mg) to increase the N content and/or reduce the enrichment to within the range suitable for mass spectrometry. Tin cups receiving only the reference material were included to allow for back calculation of the reference material 15N : 14N ratio from the sample 15N : 14N ratio. Soil samples were ground using a mill adapted to the complete recovery of very small samples and subsequently weighed (c. 180 mg) into tin capsules for analysis. 15N in the prepared samples was determined using isotope ratio mass spectrometery (ANCA-NT system 20/20; Europa Scientific).
Statistical analysis
Linear regression analysis of uptake rates and first-order kinetic analysis of the mineralization data were undertaken with sigmaplot v12 (SPSS Inc., Chicago, IL, USA). t-tests and ANOVA (with Tukey pair-wise comparison) were undertaken with minitab v16 (Minitab Inc., State College, PA, USA). P < 0.05 was used as the upper limit for statistical significance.
Results
Nitrogen dynamics in the rhizosphere
The time-dependent uptake of 15N within the wheat shoots and roots (including any microbial cells still attached) derived from the added 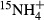 or 15N-glutamate is shown in Fig. 1. As expected, the rate of 15N uptake was fast and led to greater incorporation into root tissues in comparison to shoot tissues (P < 0.05 for
or 15N-glutamate is shown in Fig. 1. As expected, the rate of 15N uptake was fast and led to greater incorporation into root tissues in comparison to shoot tissues (P < 0.05 for 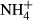 and glutamate). The first time-point at which significant 15N uptake could be detected in the roots relative to the background (microcosms not labelled with 15N) was 30 min for
and glutamate). The first time-point at which significant 15N uptake could be detected in the roots relative to the background (microcosms not labelled with 15N) was 30 min for 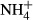 , while for glutamate it was significantly later at 90 min. With respect to the shoots, significant 15N enrichment was seen in both N addition treatments after 90 min, relative to the unamended control (P < 0.01). The rate of 15N uptake was linear in both treatments (r2 > 0.998), although the rate of 15N incorporation in the shoots was much greater from
, while for glutamate it was significantly later at 90 min. With respect to the shoots, significant 15N enrichment was seen in both N addition treatments after 90 min, relative to the unamended control (P < 0.01). The rate of 15N uptake was linear in both treatments (r2 > 0.998), although the rate of 15N incorporation in the shoots was much greater from 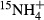 than from 15N-glutamate.
than from 15N-glutamate.
Figure 1.
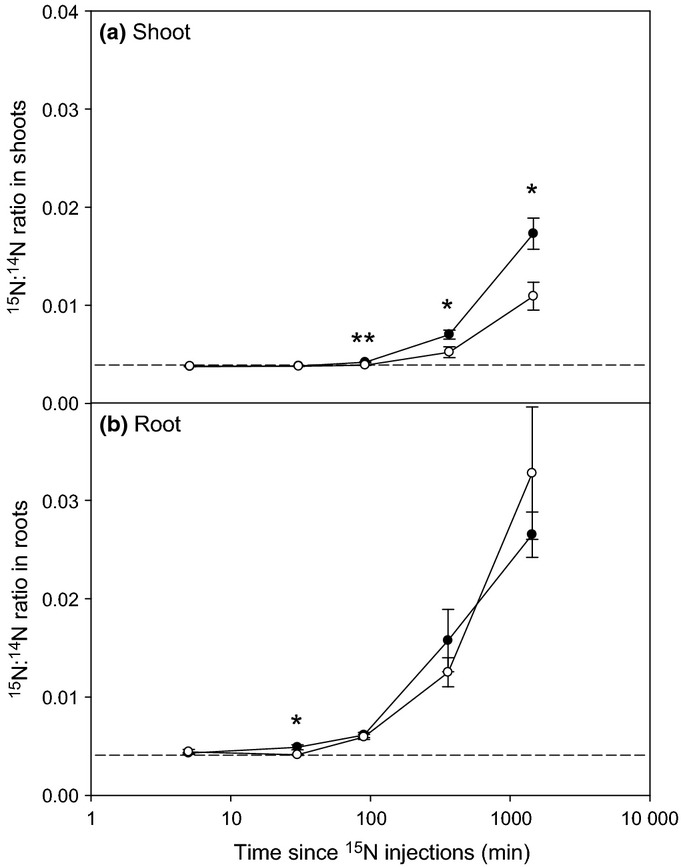
15N accumulation in the (a) shoots and (b) roots of wheat (Triticum aestivum) after the addition of 15NH4 (closed circles) or 15N-glutamate (open circles) to the rhizosphere. Dotted lines represent 15N natural abundance values in nonisotopically labelled plants. Values represent mean ± SE (n = 4 replicate microcosms). Significant differences between the 15NH4 and 15N-glutamate treatments: *, P < 0.05; **, P < 0.01.
Overall, there were no significant differences in root biomass (mean ± SE across treatments was 22.7 ± 1.1 mg DW per plant), shoot biomass (mean ± SE across treatments was 21.5 ± 0.3 mg DW per plant) or root-to-shoot ratio over the 1440-min (24-h) experimental period or between the two N treatments and the water amended control microcosms (P > 0.05). Similarly, there were no differences in root and shoot N contents between treatments or harvest times (root N, 1.29 ± 0.03% DW; shoot N, 5.6 ± 0.1% DW; mean ± SE, P > 0.05).
Nitrogen isotopic behaviour in the soil
Adsorption isotherms with sterile soil revealed that added 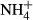 became preferentially bound to the soil’s solid phase (c. 85% of the total; Supporting Information Fig. S1). In relation to the initial concentration of
became preferentially bound to the soil’s solid phase (c. 85% of the total; Supporting Information Fig. S1). In relation to the initial concentration of 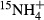 added to the microcosms (45 mg N l−1), and consistent with the adsorption isotherm data, there was a very rapid depletion of
added to the microcosms (45 mg N l−1), and consistent with the adsorption isotherm data, there was a very rapid depletion of 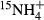 within the first 5 min to 3.5 mg N l−1 (Fig. 2a). The
within the first 5 min to 3.5 mg N l−1 (Fig. 2a). The 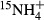 soil solution concentration then continued to decline, albeit at a much slower rate, over the subsequent 1440-min experimental period (Fig. 2a). By 1440 min, significantly greater amounts of
soil solution concentration then continued to decline, albeit at a much slower rate, over the subsequent 1440-min experimental period (Fig. 2a). By 1440 min, significantly greater amounts of 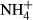 had been removed from soil solution in the planted microcosms in comparison to the unplanted microcosms (t-test P < 0.05 at 24 h; Fig. 2a). Extraction of the soil over time with KCl (to quantify the solution and exchangeable phase, that is, the plant-available 15N pool) revealed that most
had been removed from soil solution in the planted microcosms in comparison to the unplanted microcosms (t-test P < 0.05 at 24 h; Fig. 2a). Extraction of the soil over time with KCl (to quantify the solution and exchangeable phase, that is, the plant-available 15N pool) revealed that most 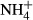 was not taken up by the plant in the first 360 min but remained bound to the soil’s solid phase (Fig. S2). By 1440 min, however, significant depletion of
was not taken up by the plant in the first 360 min but remained bound to the soil’s solid phase (Fig. S2). By 1440 min, however, significant depletion of 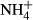 from the soil exchange phase had occurred in the planted microcosms.
from the soil exchange phase had occurred in the planted microcosms.
Figure 2.
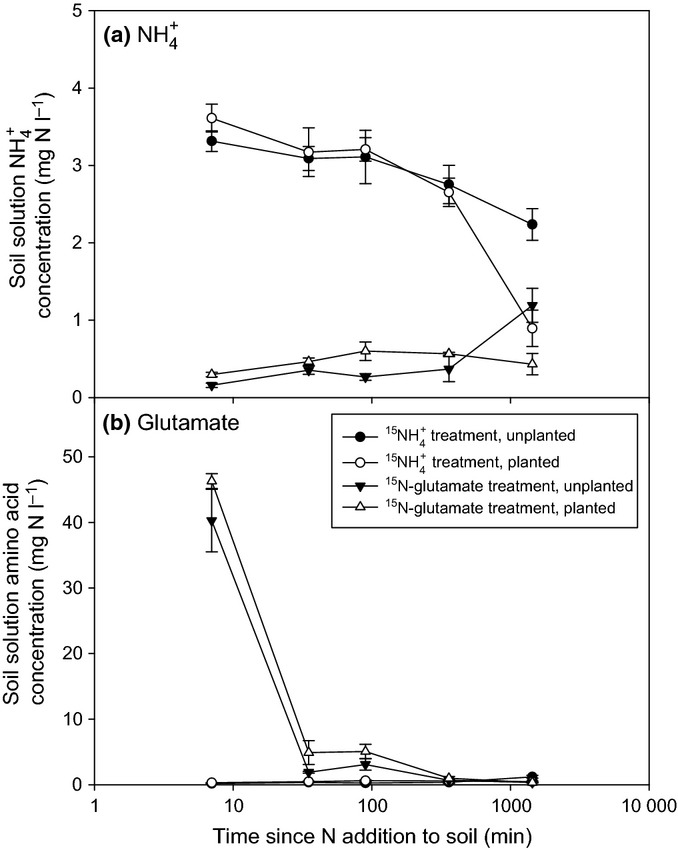
Concentration of nitrogen (N) in soil solution after the pulse-labelled addition of (a) 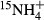 or (b) 15N-glutamate to soil in the presence and absence of wheat (Triticum aestivum) plants. The initial concentration was 45 mg N l−1. Because of the potential inter-conversion of N forms in soil,
or (b) 15N-glutamate to soil in the presence and absence of wheat (Triticum aestivum) plants. The initial concentration was 45 mg N l−1. Because of the potential inter-conversion of N forms in soil, 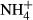 was also measured in the glutamate treatment and vice versa. Values represent mean ± SE (n = 4 replicate microcosms). The legend is the same for both panels (a) and (b).
was also measured in the glutamate treatment and vice versa. Values represent mean ± SE (n = 4 replicate microcosms). The legend is the same for both panels (a) and (b).
In the amino acid treatment, 15N-glutamate was also rapidly depleted (within 30 min) from soil solution but at a similar rate in the planted and unplanted microcosms (Fig. 2b). At the pH of the soil (pH 4.8 ± 0.1, mean ± SE), the net charge on glutamate was predicted to be −0.66. Adsorption isotherms revealed no detectable sorption of glutamate to the soil’s solid phase (data not presented). Some mineralization of the glutamate-N also occurred, as evidenced by a net accumulation of 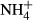 in the 15N-glutamate treatment, particularly in the absence of plants (Fig. 2a). The 14C-labelled glutamate was also shown to rapidly mineralize to 14CO2 after addition to both the planted and unplanted microcosms (Fig. S3). A double first-order kinetic model fitted well to the experimental mineralization data (r2 = 0.990 ± 0.002, mean ± SE) from which the half-life of the amino acid in soil solution was estimated to be 342 ± 36 min in planted and 708 ± 192 min in unplanted treatments (mean ± SE).
in the 15N-glutamate treatment, particularly in the absence of plants (Fig. 2a). The 14C-labelled glutamate was also shown to rapidly mineralize to 14CO2 after addition to both the planted and unplanted microcosms (Fig. S3). A double first-order kinetic model fitted well to the experimental mineralization data (r2 = 0.990 ± 0.002, mean ± SE) from which the half-life of the amino acid in soil solution was estimated to be 342 ± 36 min in planted and 708 ± 192 min in unplanted treatments (mean ± SE).
After extraction of the 15N-amended soils with KCl and removal of roots, the 15N content of the residual soil was determined. We ascribe this pool to 15N immobilized in the soil microbial biomass (Jörgensen & Brookes, 1990; Murphy et al., 2003), the size of which was the same in planted and unplanted microcosms (220 ± 18 mg microbial biomass-C kg−1 soil; mean ± SE, P > 0.05). The results presented in Fig. 3 indicate that both 15N forms became progressively immobilized by the microbial community over the 1440-min experimental period, although the rate of immobilization was much higher for 15N-glutamate than 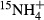 (P < 0.001, unplanted; P < 0.01, planted). The presence of plants had no influence on the rate of 15N-glutamate immobilization, whereas significantly greater immobilization occurred in the planted
(P < 0.001, unplanted; P < 0.01, planted). The presence of plants had no influence on the rate of 15N-glutamate immobilization, whereas significantly greater immobilization occurred in the planted  treatment (P < 0.001).
treatment (P < 0.001).
Figure 3.
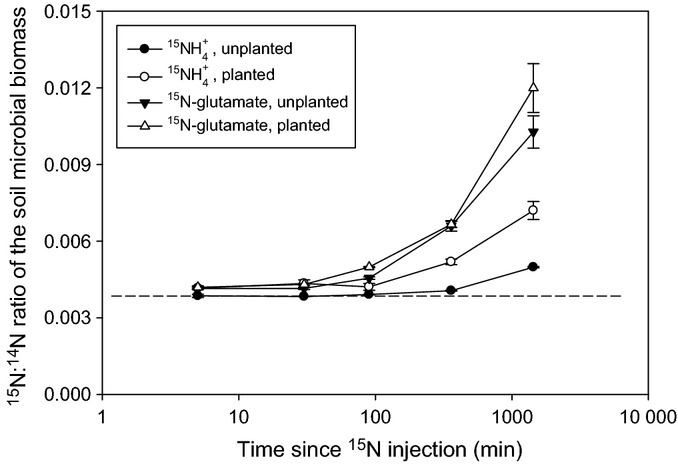
15N enrichment in the soil microbial biomass after the pulse-labelled addition of  or 15N-glutamate to soil in the presence and absence of wheat (Triticum aestivum) plants. The dotted line represents 15N natural abundance in the unlabelled microcosms. Values represent mean ± SE (n = 4 replicate microcosms).
or 15N-glutamate to soil in the presence and absence of wheat (Triticum aestivum) plants. The dotted line represents 15N natural abundance in the unlabelled microcosms. Values represent mean ± SE (n = 4 replicate microcosms).
The background concentrations of free amino acids,  and
and 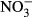 in both the planted and unplanted soils were very low (free amino acids 0.17 ± 0.01 mg N l−1;
in both the planted and unplanted soils were very low (free amino acids 0.17 ± 0.01 mg N l−1; 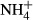 0.07 ± 0.01 mg N l−1;
0.07 ± 0.01 mg N l−1; 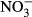 < 0.05 mg N l−1, mean ± SE), particularly in relation to the microcosms injected with 15N. The concentration of
< 0.05 mg N l−1, mean ± SE), particularly in relation to the microcosms injected with 15N. The concentration of 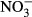 in soil solution remained low in both 15N treatments with no apparent conversion of 15N-glutamate or
in soil solution remained low in both 15N treatments with no apparent conversion of 15N-glutamate or 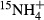 to
to 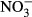 in any treatment at any harvest time (data not presented). Assuming perfect mixing of the added isotope with the intrinsic soil solution, we estimate that the initial isotopic pool dilution (i.e. by the 14N into the 15N pool) was very low, being 1.0% for
in any treatment at any harvest time (data not presented). Assuming perfect mixing of the added isotope with the intrinsic soil solution, we estimate that the initial isotopic pool dilution (i.e. by the 14N into the 15N pool) was very low, being 1.0% for 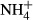 and 0.35% for glutamate. As a consequence of uncertainty over the native rates of 14N turnover and N gradients in the rhizosphere, it was not possible to estimate subsequent rates of isotopic pool dilution during the course of the experiment.
and 0.35% for glutamate. As a consequence of uncertainty over the native rates of 14N turnover and N gradients in the rhizosphere, it was not possible to estimate subsequent rates of isotopic pool dilution during the course of the experiment.
NanoSIMS imaging and quantification
Representative examples of roots within the experimental microcosms are presented in Fig. 4. The light micrograph shows the dense proliferation of root hairs, while the SEM images show cross-sections of the resin-embedded plant roots with the soil matrix. SEM or TEM images were used to aid identification of the specific location of cell components within the plant roots; these ROIs were subsequently analysed by NanoSIMS (Clode et al., 2009). The soil organic matter and mineral particles surrounding the root could also be easily distinguished using the 12C14N− and 28Si− (or 16O−) signals, respectively (Fig. 5). Areas of low signal strength correspond to resin-filled void spaces in the root (e.g. cell vacuoles) and soil (e.g. pore spaces); analyses of resin-only areas revealed that 15N abundance was at natural levels and had not been enriched. The cellular structure of the root, and particularly the stele with its thickened cell walls, could be readily distinguished in NanoSIMS images from transverse sections through a plant root embedded within the soil matrix (Figs 6, S4).
Figure 4.
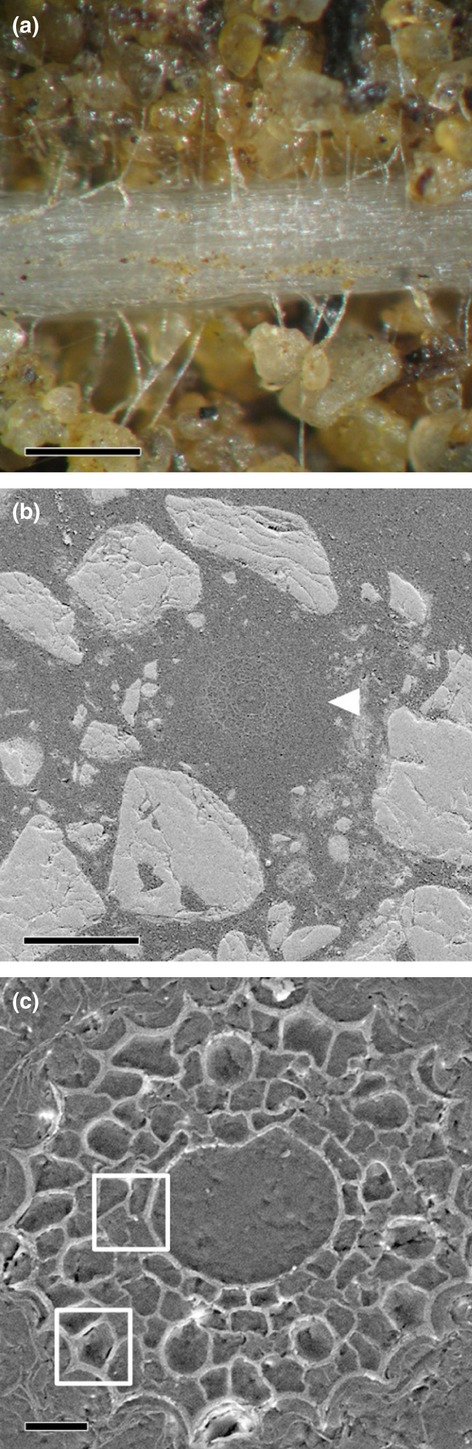
Imaging of plant roots in situ. (a) Optical micrograph of a wheat (Triticum aestivum) root growing within the microcosm. (b) Scanning electron micrograph (SEM) of a polished transverse section showing a plant root (arrow) within the embedded soil core. (c) SEM of a plant root within the embedded soil core at higher magnification, revealing the cellular root structure of inner cortical cells. Root hairs and epidermal cells cannot easily be seen in this image. Boxes indicate cellular regions typically analysed by nano-scale secondary ion mass spectrometry (NanoSIMS) (as seen in Fig. 6 and Supporting Information Fig. S4). Bars: (a) 400 μm; (b) 200 μm; (c) 20 μm.
Figure 5.
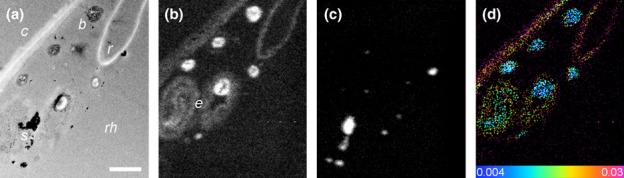
Correlative transmission electron micrograph (TEM) and nano-scale secondary ion mass spectrometry (NanoSIMS) imaging after 5 min of exposure of wheat (Triticum aestivum) to 15N-labelled 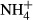 . (a) TEM of the rhizosphere (rh), showing bacteria (b) adjacent to a root cell (c), root hair (r) and soil particles (s). (b) 12C14N− NanoSIMS image of the same region, highlighting the bacteria and organic materials including extracellular mucilage (e) associated with soil particles and bacteria. (c) 16O− NanoSIMS image of the same region highlighting soil particles containing oxides. (d) Hue-Saturation-Intensity (HSI) image generated from the NanoSIMS 15N : 14N ratio of the same region, showing 15N : 14N ratio intensities from natural abundance (blue, 0.0037) to enriched (pink, 0.30). Bar, 1 μm for all images.
. (a) TEM of the rhizosphere (rh), showing bacteria (b) adjacent to a root cell (c), root hair (r) and soil particles (s). (b) 12C14N− NanoSIMS image of the same region, highlighting the bacteria and organic materials including extracellular mucilage (e) associated with soil particles and bacteria. (c) 16O− NanoSIMS image of the same region highlighting soil particles containing oxides. (d) Hue-Saturation-Intensity (HSI) image generated from the NanoSIMS 15N : 14N ratio of the same region, showing 15N : 14N ratio intensities from natural abundance (blue, 0.0037) to enriched (pink, 0.30). Bar, 1 μm for all images.
Figure 6.
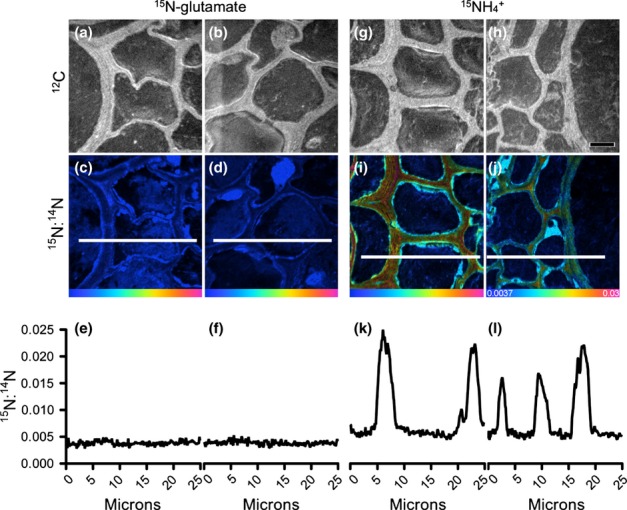
Nano-scale secondary ion mass spectrometry (NanoSIMS) imaging and analysis of wheat (Triticum aestivum) roots in situ within the embedded soil core after 5 min of exposure to 15N-labelled glutamate or 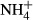 . Examples of the location of cellular regions analysed within a root are depicted in Fig. 4(c). Root and cell structure is visualized in 12C images (a, b; g, h). Enrichment levels of 15N within these regions are shown as 15N : 14N Hue-Saturation-Intensity (HSI) images (c, d; i, j). The HSI colour scale (j; 0.0037–0.03; 15N : 14N natural abundance = 0.0037) applies to all HSI images. Line scans show numerical levels of 15N enrichment across the subcellular regions (e, f; k, l), with data acquired from the lines indicated on each of the respective 15N : 14N HSI images. High levels of 15N enrichment can be seen in the
. Examples of the location of cellular regions analysed within a root are depicted in Fig. 4(c). Root and cell structure is visualized in 12C images (a, b; g, h). Enrichment levels of 15N within these regions are shown as 15N : 14N Hue-Saturation-Intensity (HSI) images (c, d; i, j). The HSI colour scale (j; 0.0037–0.03; 15N : 14N natural abundance = 0.0037) applies to all HSI images. Line scans show numerical levels of 15N enrichment across the subcellular regions (e, f; k, l), with data acquired from the lines indicated on each of the respective 15N : 14N HSI images. High levels of 15N enrichment can be seen in the 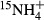 treatment within distinct cellular regions, whereas the 15N-glutamate treatment shows cells with very low levels of 15N enrichment just above natural abundance. Bar, 5 μm for all images.
treatment within distinct cellular regions, whereas the 15N-glutamate treatment shows cells with very low levels of 15N enrichment just above natural abundance. Bar, 5 μm for all images.
The isotopic enrichment of rhizosphere bacteria and root tissues normalized to unlabelled samples was determined for each treatment at the 5, 30 and 1440 min harvest times. Measurements of 15N : 14N ratios from corresponding regions of organic matter in unlabelled rhizosphere samples were equivalent to natural abundance, with a mean 15N : 14N ratio value of 0.003675 and standard error of 0.000015 (n = 15), thus giving a precision of 1.6% (standard deviation = 0.000059). Overall, the temporal pattern of 15N uptake within the bacterial cells was similar between the two 15N treatments (Fig. 7). Despite this similar trend, initially (0–5 min) the amount of 15N uptake into the bacterial cells was significantly greater (P < 0.05) in the 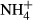 treatment (15N : 14N ratio data: mean and standard error = 0.009030 ± 0.000196; range = 0.003896–0.102855; n = 64 bacterial cells) compared with the glutamate treatment (15N : 14N ratio data: mean and standard error = 0.004657 ± 0.000269; range = 0.003807–0.01273; n = 36 bacterial cells), with both enriched relative to natural abundance (Fig. 7). It should be noted that a few bacterial cells (as identified by TEM) that did not take up 15N were also present in the rhizosphere (Fig. 5); the mean 15N : 14N ratio values for data presented in Fig. 7 only include data from the active (i.e. 15N enriched) populations. No difference was apparent between the two N treatments after 30 min; however, by 1440 min, a very high proportion of 15N had accumulated in the bacterial cells relative to natural abundance, and this proportion was significantly higher in the glutamate treatment than in the
treatment (15N : 14N ratio data: mean and standard error = 0.009030 ± 0.000196; range = 0.003896–0.102855; n = 64 bacterial cells) compared with the glutamate treatment (15N : 14N ratio data: mean and standard error = 0.004657 ± 0.000269; range = 0.003807–0.01273; n = 36 bacterial cells), with both enriched relative to natural abundance (Fig. 7). It should be noted that a few bacterial cells (as identified by TEM) that did not take up 15N were also present in the rhizosphere (Fig. 5); the mean 15N : 14N ratio values for data presented in Fig. 7 only include data from the active (i.e. 15N enriched) populations. No difference was apparent between the two N treatments after 30 min; however, by 1440 min, a very high proportion of 15N had accumulated in the bacterial cells relative to natural abundance, and this proportion was significantly higher in the glutamate treatment than in the 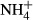 treatment.
treatment.
Figure 7.
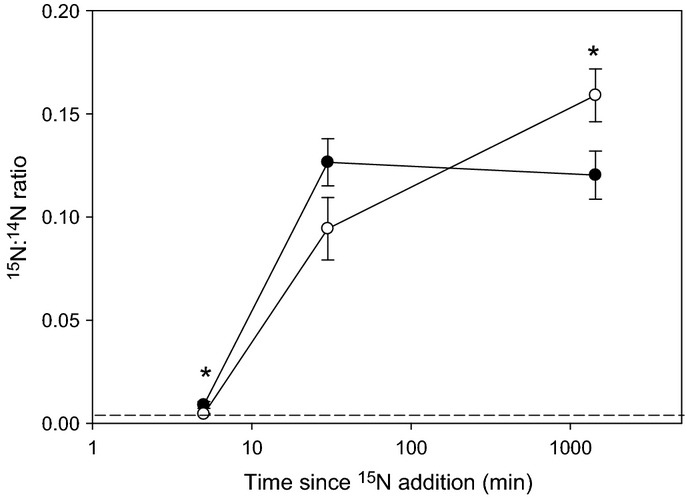
Nano-scale secondary ion mass spectrometry (NanoSIMS) derived 15N : 14N isotopic ratios of metabolically active (i.e. 15N-enriched) individual bacteria located in the rhizosphere of wheat (Triticum aestivum) plants after injection of 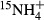 (closed circles) or 15N-glutamate (open circles) into the soil. The dotted line represents 15N natural abundance. Values represent mean ± SE across all four microcosms (n = 11–50 regions of interest containing bacterial cells per time period for
(closed circles) or 15N-glutamate (open circles) into the soil. The dotted line represents 15N natural abundance. Values represent mean ± SE across all four microcosms (n = 11–50 regions of interest containing bacterial cells per time period for 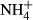 and n = 29–36 for 15N-glutamate treatments). The dotted line represents 15N natural abundance in the non-15N-labelled treatment. Significant differences between the 15NH4 and 15N-glutamate treatments: *, P < 0.05.
and n = 29–36 for 15N-glutamate treatments). The dotted line represents 15N natural abundance in the non-15N-labelled treatment. Significant differences between the 15NH4 and 15N-glutamate treatments: *, P < 0.05.
Enrichment of 15N within subcellular regions of plant root cells was easily distinguishable by NanoSIMS (Fig. 6) and could be traced over time (Fig. S4). After 5 min there were significant differences in plant tissue 15N enrichment between treatments; 15N enrichment in the 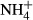 treatment was present across the entire root structure (Fig. 6), having reached the endodermis and vascular bundles (Fig. 8), whereas 15N enrichment in the glutamate treatment was only present in the outer cortical cells (Fig. 8). The levels of 15N enrichment in the plasma membrane of the plant tissues (Fig. 8) were generally 10-fold lower than observed in the rhizosphere bacteria (Fig. 7). In addition, the overall temporal pattern of 15N uptake was slightly different between the different tissue types (P < 0.05 for both N types) and unlike that observed for the microbial cells. The level of 15N enrichment was significantly lower in the cortex after 1440 min relative to that present at 30 min in the
treatment was present across the entire root structure (Fig. 6), having reached the endodermis and vascular bundles (Fig. 8), whereas 15N enrichment in the glutamate treatment was only present in the outer cortical cells (Fig. 8). The levels of 15N enrichment in the plasma membrane of the plant tissues (Fig. 8) were generally 10-fold lower than observed in the rhizosphere bacteria (Fig. 7). In addition, the overall temporal pattern of 15N uptake was slightly different between the different tissue types (P < 0.05 for both N types) and unlike that observed for the microbial cells. The level of 15N enrichment was significantly lower in the cortex after 1440 min relative to that present at 30 min in the 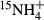 treatment (P < 0.01). 15N enrichment in root tissues treated with 15N-glutamate was significantly lower than that of the
treatment (P < 0.01). 15N enrichment in root tissues treated with 15N-glutamate was significantly lower than that of the 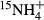 roots at all time-points (P < 0.001; Fig. 8). Although cell nuclei (e.g. see Fig. S4c) could not be seen in all replicates and at all time-points, when sufficient nuclei could be enumerated they were also similarly 15N enriched in both the
roots at all time-points (P < 0.001; Fig. 8). Although cell nuclei (e.g. see Fig. S4c) could not be seen in all replicates and at all time-points, when sufficient nuclei could be enumerated they were also similarly 15N enriched in both the 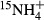 and 15N-glutamate treatments (15N : 14N ratio, 0.016 ± 0.001 at 1440 min, mean ± SE).
and 15N-glutamate treatments (15N : 14N ratio, 0.016 ± 0.001 at 1440 min, mean ± SE).
Figure 8.
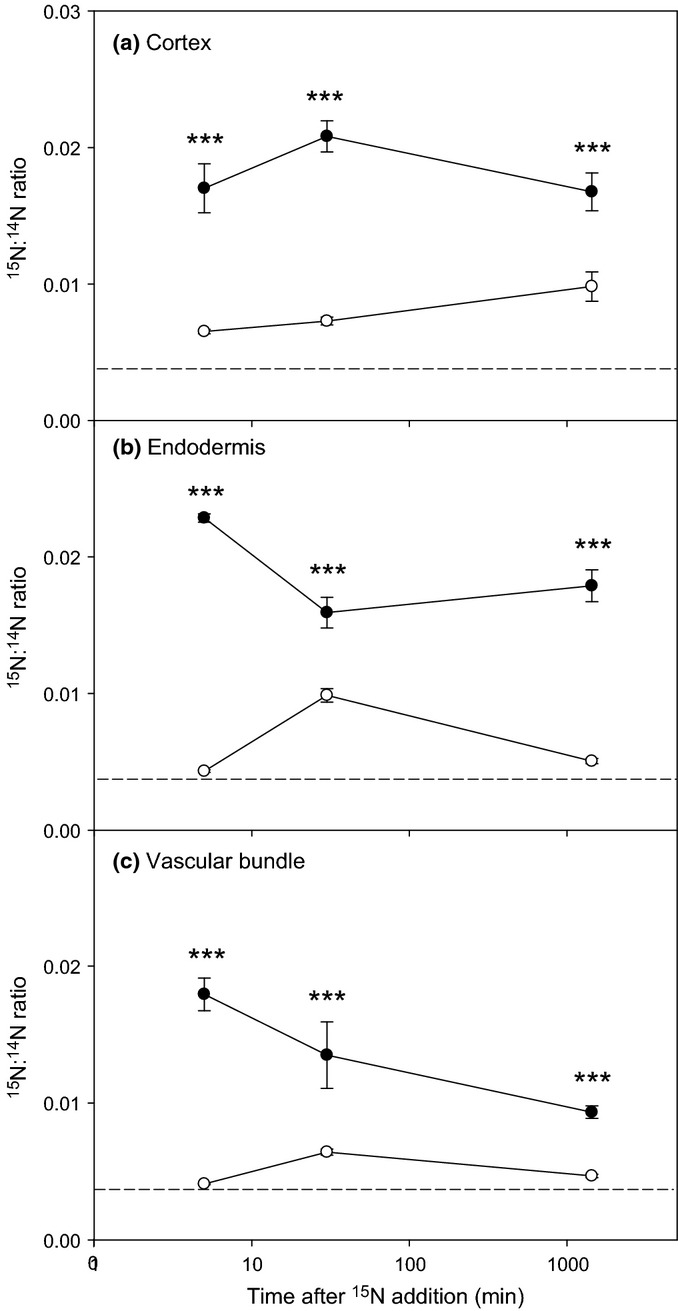
Nano-scale secondary ion mass spectrometry (NanoSIMS) derived 15N : 14N isotopic ratios for different wheat (Triticum aestivum) root tissues (intercellular cortex, endodermis and vascular bundle) after injection of 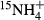 (closed circle) or 15N-glutamate (open circle) into the rhizosphere. Dotted lines represent 15N natural abundance. Values represent mean ± SE (n = 13 and n = 10 for roots in the
(closed circle) or 15N-glutamate (open circle) into the rhizosphere. Dotted lines represent 15N natural abundance. Values represent mean ± SE (n = 13 and n = 10 for roots in the 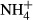 and glutamate treatments, respectively). The dotted line represents 15N natural abundance. Significant differences between the 15NH4 and 15N-glutamate treatments: ***, P < 0.001.
and glutamate treatments, respectively). The dotted line represents 15N natural abundance. Significant differences between the 15NH4 and 15N-glutamate treatments: ***, P < 0.001.
Discussion
Dynamics of 15N from a plant perspective
It has been demonstrated under field conditions that wheat has the potential to take up amino acids intact from soil, although its quantitative significance in terms of plant N nutrition remains highly uncertain (Näsholm et al., 2001, 2009). To assess this uncertainty, we took a microcosm approach to critically evaluate the temporal and spatial partitioning of 15N in different plant, microbial and soil pools. Our results showed an almost linear uptake of 15N by the wheat roots over the 1440-min labelling period and that, overall, plants competed better for 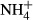 as a source of N in comparison to glutamate. Further, proportionally more N derived from
as a source of N in comparison to glutamate. Further, proportionally more N derived from 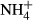 was allocated to the shoots in comparison to glutamate-derived N. This is in agreement with findings for wheat seedlings grown in sterile hydroponic culture, where a linear rate of N uptake was observed over a 360-min period and
was allocated to the shoots in comparison to glutamate-derived N. This is in agreement with findings for wheat seedlings grown in sterile hydroponic culture, where a linear rate of N uptake was observed over a 360-min period and 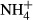 was also taken up preferentially in comparison to alanine, trialanine or
was also taken up preferentially in comparison to alanine, trialanine or 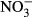 (Hill et al., 2011). We note that the plant–microbial competition scenario presented here is not reflective of severely N-limited ecosystems where N supply is regulated more by inputs of N from N2 fixation, atmospheric deposition and slow rates of organic matter turnover (e.g. Inselsbacher & Näsholm, 2012).
(Hill et al., 2011). We note that the plant–microbial competition scenario presented here is not reflective of severely N-limited ecosystems where N supply is regulated more by inputs of N from N2 fixation, atmospheric deposition and slow rates of organic matter turnover (e.g. Inselsbacher & Näsholm, 2012).
The type and amount of N taken up by plants are dependent on several factors, including the concentration of N in soil solution, its diffusion rate through soil, the activity and affinity of the root’s membrane transporters, the N status of the plant and the competition from microbes inhabiting the rhizosphere (Glass et al., 2002; Forsum et al., 2008; Jones et al., 2009). The soil used here was naturally low in available N and the amount of N added to the soil was chosen to reflect a pulse addition of N that would occur after the addition of an inorganic- or organic-based fertilizer or as a result of the lysis of plant cells in response to physical (e.g. abrasion or osmotic stress), chemical (e.g. metal-induced bursting of epidermal cells) or biological (e.g. toxin) stress (typically 2–30 mM 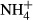 or free amino acids in root cell sap; Jones & Darrah, 1994; Miller et al., 2001). Measurements of soil N availability indicated a rapid depletion of the added glutamate and especially
or free amino acids in root cell sap; Jones & Darrah, 1994; Miller et al., 2001). Measurements of soil N availability indicated a rapid depletion of the added glutamate and especially 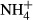 from the soil solution pool. However, the reasons for this disappearance were solute-specific, being mainly attributable to abiotic ion attraction to soil particles for
from the soil solution pool. However, the reasons for this disappearance were solute-specific, being mainly attributable to abiotic ion attraction to soil particles for 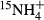 (i.e. cation exchange) and to biological immobilization for 15N-glutamate. This supports the findings of previous studies that have shown amino acids are rapidly mineralized by the soil microbial community, whose growth is more limited by C than N availability (Dempster et al., 2012). These findings also suggest that the timing of plant 15N uptake was significantly different between the two N treatments. Specifically, as
(i.e. cation exchange) and to biological immobilization for 15N-glutamate. This supports the findings of previous studies that have shown amino acids are rapidly mineralized by the soil microbial community, whose growth is more limited by C than N availability (Dempster et al., 2012). These findings also suggest that the timing of plant 15N uptake was significantly different between the two N treatments. Specifically, as 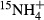 was not irreversibly bound to the soil’s exchange phase, it was capable of being readily released back into the available pool as the soil solution
was not irreversibly bound to the soil’s exchange phase, it was capable of being readily released back into the available pool as the soil solution 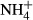 concentration fell as a result of progressive biotical removal. This indicates that a large reservoir of potentially available
concentration fell as a result of progressive biotical removal. This indicates that a large reservoir of potentially available 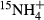 existed for the entire duration of our experiment. By contrast, after microbial uptake of almost all of the 15N-glutamate within 30 min, the plant-available 15N amino acid pool was effectively depleted (although roots may have taken up some unlabelled amino acids produced de novo from soil organic matter turnover). Thus, the period for root
existed for the entire duration of our experiment. By contrast, after microbial uptake of almost all of the 15N-glutamate within 30 min, the plant-available 15N amino acid pool was effectively depleted (although roots may have taken up some unlabelled amino acids produced de novo from soil organic matter turnover). Thus, the period for root 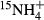 uptake was actually 50-fold longer than that for 15N-glutamate. Our results further indicate that the microbial community fed 15N-glutmate also effluxed a significant amount of N back into the soil as
uptake was actually 50-fold longer than that for 15N-glutamate. Our results further indicate that the microbial community fed 15N-glutmate also effluxed a significant amount of N back into the soil as 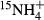 and that this process occurred within 5 min of glutamate addition in the planted microcosms. This
and that this process occurred within 5 min of glutamate addition in the planted microcosms. This 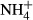 efflux process is induced by the internal catabolism of the added amino acids (in which 40% of the amino acid-C is typically respired) which causes a C : N ratio imbalance between the microbial biomass and the added glutamate, leading to the dumping of excess N (Jones et al., 2005a,2005b; Manzoni et al., 2008). Thus, our view is that plant 15N uptake in the glutamate treatment was actually a result of a small amount of intact amino acid uptake in the first 60 min followed by the uptake of microbially effluxed
efflux process is induced by the internal catabolism of the added amino acids (in which 40% of the amino acid-C is typically respired) which causes a C : N ratio imbalance between the microbial biomass and the added glutamate, leading to the dumping of excess N (Jones et al., 2005a,2005b; Manzoni et al., 2008). Thus, our view is that plant 15N uptake in the glutamate treatment was actually a result of a small amount of intact amino acid uptake in the first 60 min followed by the uptake of microbially effluxed 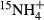 over the subsequent 1380-min (23-h) period. This is supported by Roberts et al. (2009) and the lack of reappearance of 15N-glutamate in soil solution after microbial uptake (Fig. 2). This may also explain the apparent lag phase in 15N uptake in the glutamate treatment in comparison to
over the subsequent 1380-min (23-h) period. This is supported by Roberts et al. (2009) and the lack of reappearance of 15N-glutamate in soil solution after microbial uptake (Fig. 2). This may also explain the apparent lag phase in 15N uptake in the glutamate treatment in comparison to 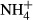 (as revealed by both NanoSIMS and conventional mass spectrometry in addition to the observed slower translocation of 15N to the shoots). This poor root capture of glutamate supports previous work showing that plants could only capture a very small amount of amino acid C supplied to nonsterile wheat roots (Owen & Jones, 2001).
(as revealed by both NanoSIMS and conventional mass spectrometry in addition to the observed slower translocation of 15N to the shoots). This poor root capture of glutamate supports previous work showing that plants could only capture a very small amount of amino acid C supplied to nonsterile wheat roots (Owen & Jones, 2001).
Although 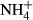 diffuses 50% faster in free solution than glutamate, we calculated that, because of its strong attraction to the solid phase, its effective linear rate of diffusion in soil over the 1440-min experimental period was fivefold slower than for glutamate (0.13 cm d−1 for
diffuses 50% faster in free solution than glutamate, we calculated that, because of its strong attraction to the solid phase, its effective linear rate of diffusion in soil over the 1440-min experimental period was fivefold slower than for glutamate (0.13 cm d−1 for 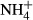 and 0.62 cm d−1 for glutamate). In our experimental set-up, the inter-root distance was c. 1–2 mm, implying that nearly all the
and 0.62 cm d−1 for glutamate). In our experimental set-up, the inter-root distance was c. 1–2 mm, implying that nearly all the 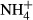 was available to the roots during the 1440-min experimental period. In other situations, if the inter-root distance were larger (c. 5 mm), then much of the
was available to the roots during the 1440-min experimental period. In other situations, if the inter-root distance were larger (c. 5 mm), then much of the 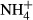 would have been incapable of diffusing towards the root within the experimental period. We hypothesize that this alternative scenario would additionally favour root capture of glutamate over
would have been incapable of diffusing towards the root within the experimental period. We hypothesize that this alternative scenario would additionally favour root capture of glutamate over 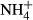 .
.
At high exogenous concentrations (> 1 mM) in axenic soil-less culture, glutamate has been shown to be inhibitory to root growth, particularly when applied to the root apex (Sivaguru et al., 2003; Forde & Walch-Liu, 2009). In this soil-based study, however, the localized addition of glutamate revealed no negative effect of glutamate on either root or shoot growth. We ascribe this lack of negative response to the very rapid lowering of the glutamate concentration in solution by the soil microbial community (and to some extent root consumption), which lowered the glutamate concentration to below the critical response threshold. In addition, inhibitory effects of exogenously applied glutamate on root growth were only observed in hydroponic culture at concentrations > 5 mM (Fig. S5).
Spatial distribution of 15N in plant tissues
Our results showed that the added 15N was quickly taken up and rapidly distributed throughout the root tissues (i.e. within 5 min of 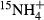 addition). Although we did not quantify 15N from root hair or epidermal cells (because of either poor preservation or lack of alignment in the field of view), our results suggest no preferential accumulation of 15N in any root cell type (there was never any accumulation of 15N within xylem tissue; e.g. Fig. S4). This supports the view that 15N is rapidly translocated away from the point of uptake to the active growing regions (e.g. root tips and shoots). In terms of root uptake, NanoSIMS did reveal the presence of 15N super-enriched bacteria in both the ecto-rhizosphere (Fig. 5) and endo-rhizosphere (Clode et al., 2009). It is likely that the typical root-washing procedure will not remove these bacteria and that these may have represented a significant contamination of the root 15N signal in many previous soil-based studies. As we injected 15N into root regions with fully expanded root hairs, this may favour the microbial capture of uptake of amino acids because of the higher microbial population in comparison to the low density of microorganisms surrounding root tips (Marschner et al., 2011). Further, the distribution and expression of root N transporters may be different between the actively and nonactively growing root regions (Yuan et al., 2007).
addition). Although we did not quantify 15N from root hair or epidermal cells (because of either poor preservation or lack of alignment in the field of view), our results suggest no preferential accumulation of 15N in any root cell type (there was never any accumulation of 15N within xylem tissue; e.g. Fig. S4). This supports the view that 15N is rapidly translocated away from the point of uptake to the active growing regions (e.g. root tips and shoots). In terms of root uptake, NanoSIMS did reveal the presence of 15N super-enriched bacteria in both the ecto-rhizosphere (Fig. 5) and endo-rhizosphere (Clode et al., 2009). It is likely that the typical root-washing procedure will not remove these bacteria and that these may have represented a significant contamination of the root 15N signal in many previous soil-based studies. As we injected 15N into root regions with fully expanded root hairs, this may favour the microbial capture of uptake of amino acids because of the higher microbial population in comparison to the low density of microorganisms surrounding root tips (Marschner et al., 2011). Further, the distribution and expression of root N transporters may be different between the actively and nonactively growing root regions (Yuan et al., 2007).
Another benefit of the NanoSIMS approach is that 15N in the apoplast can be readily distinguished from 15N present in the symplast (i.e. true uptake). At high external concentrations (> 1 mM), apoplastic diffusion of 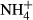 can readily take place until it reaches the endodermis, at which point the
can readily take place until it reaches the endodermis, at which point the 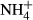 is taken up before translocation (Frensch et al., 1996; Yuan et al., 2007; Schreiber, 2010). Our NanoSIMS images indicated that some apoplastic 15N was present in the cortex at 5 min but that this extracellular pool was readily depleted to background levels by 30 min. Based on the equilibrium soil solution
is taken up before translocation (Frensch et al., 1996; Yuan et al., 2007; Schreiber, 2010). Our NanoSIMS images indicated that some apoplastic 15N was present in the cortex at 5 min but that this extracellular pool was readily depleted to background levels by 30 min. Based on the equilibrium soil solution 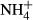 concentration (< 225 μM), we speculate that most
concentration (< 225 μM), we speculate that most 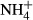 was taken up using high-affinity transporters in the root hairs and epidermis and that
was taken up using high-affinity transporters in the root hairs and epidermis and that 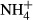 transport across the root to the stele occurred symplastically.
transport across the root to the stele occurred symplastically.
Dynamics of 15N from a microbial perspective
NanoSIMS data revealed that initial bacterial uptake of 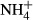 was faster than for glutamate but that the internal capacity for
was faster than for glutamate but that the internal capacity for 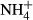 quickly became saturated (< 30 min), after which little
quickly became saturated (< 30 min), after which little 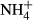 uptake occurred. By contrast, 15N-glutamate incorporation into the active bacterial community continued to rise throughout the experiment, suggesting that the added C was fuelling the formation of new cell biomass that necessitated the uptake of more N. These highly 15N-enriched bacteria were most often seen close to the root surface or in the endorhizosphere. We hypothesized that these rhizosphere bacteria would already be highly active and primed for glutamate uptake in response to the continual release of acidic amino acids from the roots into the soil via passive root exudation (Ayers & Thornton, 1968). Therefore, the lack of appreciable uptake of 15N derived from glutamate within the bacterial cells within 5 min was surprising. It is possible that wheat rhizodeposition contains low amounts of glutamate, as reported by Phillips et al. (2004), or that the bacterial transporters and internal metabolic capacity were initially saturated by the added glutamate pulse (Jones & Hodge, 1999). The latter is supported by the intrinsically low solution concentrations of total amino acids in this soil (12 ± 1 μM, mean ± SE), suggesting that the microbial community will have been operating high-affinity transport systems (Anraku, 1980). It should also be noted that we only investigated 15N uptake in rhizobacteria and not in the wider microbial community (i.e. fungi and actinomycetes). Although no distinct arbuscular mycorrhizal structures were observed in our roots when viewed by TEM, NanoSIMS or conventional light microscopy of trypan blue-stained roots, we cannot discount the potential for mycorrhizal transfer of 15N from the soil into the inner root tissues. The likelihood that this represents a major uptake N pathway in comparison to direct root uptake, however, is very low (Hawkins et al., 2000).
uptake occurred. By contrast, 15N-glutamate incorporation into the active bacterial community continued to rise throughout the experiment, suggesting that the added C was fuelling the formation of new cell biomass that necessitated the uptake of more N. These highly 15N-enriched bacteria were most often seen close to the root surface or in the endorhizosphere. We hypothesized that these rhizosphere bacteria would already be highly active and primed for glutamate uptake in response to the continual release of acidic amino acids from the roots into the soil via passive root exudation (Ayers & Thornton, 1968). Therefore, the lack of appreciable uptake of 15N derived from glutamate within the bacterial cells within 5 min was surprising. It is possible that wheat rhizodeposition contains low amounts of glutamate, as reported by Phillips et al. (2004), or that the bacterial transporters and internal metabolic capacity were initially saturated by the added glutamate pulse (Jones & Hodge, 1999). The latter is supported by the intrinsically low solution concentrations of total amino acids in this soil (12 ± 1 μM, mean ± SE), suggesting that the microbial community will have been operating high-affinity transport systems (Anraku, 1980). It should also be noted that we only investigated 15N uptake in rhizobacteria and not in the wider microbial community (i.e. fungi and actinomycetes). Although no distinct arbuscular mycorrhizal structures were observed in our roots when viewed by TEM, NanoSIMS or conventional light microscopy of trypan blue-stained roots, we cannot discount the potential for mycorrhizal transfer of 15N from the soil into the inner root tissues. The likelihood that this represents a major uptake N pathway in comparison to direct root uptake, however, is very low (Hawkins et al., 2000).
The C and N status of rhizosphere microorganisms remains an area of hot debate. Depending on the prevailing soil conditions, microbial growth could be C, N or phosphorus (P) limited (Denison et al., 2003; Marschner et al., 2011). Our 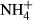 results obtained with NanoSIMS suggest that they were N limited in the short term and then became C limited. Previous work in sterile hydroponic culture has suggested that passive exudation is dominated by simple low-molecular-weight C compounds that are released in response to the large cytoplasmic–soil solution diffusion gradient (e.g. glucose and sucrose) or by compounds that are released to alleviate stress (e.g. organic acids). In both these cases we would expect this to lead to microbial N limitation in the rhizosphere, which would be exacerbated by root removal of available soil N. The rapid immobilization of
results obtained with NanoSIMS suggest that they were N limited in the short term and then became C limited. Previous work in sterile hydroponic culture has suggested that passive exudation is dominated by simple low-molecular-weight C compounds that are released in response to the large cytoplasmic–soil solution diffusion gradient (e.g. glucose and sucrose) or by compounds that are released to alleviate stress (e.g. organic acids). In both these cases we would expect this to lead to microbial N limitation in the rhizosphere, which would be exacerbated by root removal of available soil N. The rapid immobilization of 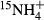 observed here would support this view.
observed here would support this view.
Conclusions
The rhizosphere represents a site of intense resource competition between plant roots and soil microorganisms. Here, we demonstrate that wheat roots represent poor competitors for glutamate in the soil in comparison to 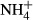 when both N forms are applied at the same rate. We ascribe this poor ability of the root to capture glutamate to the rapid immobilization and super-accumulation of amino acid-N in rhizobacteria, which prevents root uptake. This favouring of inorganic N by wheat roots may also have been exacerbated by the targeted breeding of wheat to favour the efficient acquisition of inorganic fertilizers (Reeve et al., 2009; Sadras & Lawson, 2013). Although the microbial community does immobilize
when both N forms are applied at the same rate. We ascribe this poor ability of the root to capture glutamate to the rapid immobilization and super-accumulation of amino acid-N in rhizobacteria, which prevents root uptake. This favouring of inorganic N by wheat roots may also have been exacerbated by the targeted breeding of wheat to favour the efficient acquisition of inorganic fertilizers (Reeve et al., 2009; Sadras & Lawson, 2013). Although the microbial community does immobilize 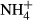 , this was low in comparison to glutamate. After amino acid assimilation, our results suggest that the rhizosphere bacteria rapidly excrete excess
, this was low in comparison to glutamate. After amino acid assimilation, our results suggest that the rhizosphere bacteria rapidly excrete excess 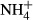 back into the soil, at which point it becomes available again for plant uptake. This study clearly demonstrates that N is rapidly transformed and cycled within the rhizosphere and highlights the extremely spatially localized nature of N dynamics at the plant–soil interface.
back into the soil, at which point it becomes available again for plant uptake. This study clearly demonstrates that N is rapidly transformed and cycled within the rhizosphere and highlights the extremely spatially localized nature of N dynamics at the plant–soil interface.
Acknowledgments
This research was funded through the Australian Research Council (FT110100246), Grains Research and Development Corporation’s Soil Biology Initiative II (D.V.M.) and a UK Natural Environment Research Council (NERC) grant (D.L.J.). The NanoSIMS and microscopy were carried out using facilities within the School of Earth and Environment and at the Centre for Microscopy, Characterisation and Analysis, at The University of Western Australia (UWA), which are supported by University, State and Federal Government funding. D.V.M. is the recipient of an Australian Research Council Future Fellowship (project number FT110100246). NanoSIMS data processing was performed using openmims software, whose development is funded by the NIH/NIBIB National Resource for Imaging Mass Spectrometry, NIH/NIBIB 5P41 EB001974-10. We thank Frank Nemeth for sample preparation.
Supporting Information
Additional supporting information may be found in the online version of this article.
Fig S1 Amount of 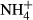 held on the soil’s solid phase as a function of solution
held on the soil’s solid phase as a function of solution 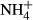 concentration.
concentration.
Fig S2 Total amount of 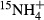 present in soil solution and held on the soil’s cation exchange phase in Triticum aestivum planted and unplanted microcosms.
present in soil solution and held on the soil’s cation exchange phase in Triticum aestivum planted and unplanted microcosms.
Fig S3 Time-dependent mineralization of 14C-labelled glutamate in soil in the presence and absence of wheat (Triticum aestivum) roots.
Fig S4 NanoSIMS imaging and analysis of wheat (Triticum aestivum) roots in situ within the embedded soil core after exposure to 15N-labelled glutamate uptake.
Fig S5 Influence of external solution glutamate concentration on the growth rate of wheat (Triticum aestivum) roots exposed to various concentrations of glutamate.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
References
- Abaas E, Hill PW, Roberts P, Murphy DV, Jones DL. Microbial activity differentially regulates the vertical mobility of nitrogen compounds in soil. Soil Biology & Biochemistry. 2012;53:120–123. [Google Scholar]
- Anraku Y, Payne JW. Microorganisms and nitrogen sources. New York, NY, USA: Wiley; 1980. Transport and utilization of amino acids by bacteria; pp. 9–33. [Google Scholar]
- Ayers WA, Thornton RH. Exudation of amino-acids by intact and damaged roots of wheat and peas. Plant and Soil. 1968;18:193–207. [Google Scholar]
- Boddy E, Roberts P, Hill PW, Farrar J, Jones DL. Turnover of low molecular weight dissolved organic C (DOC) and microbial C exhibit different temperature sensitivities in Arctic tundra soils. Soil Biology & Biochemistry. 2008;40:1557–1566. [Google Scholar]
- Brooks PD, Stark JM, McInteer BB, Preston T. Diffusion method to prepare soil extracts for automated 15N analysis. Soil Science Society of America Journal. 1989;53:1707–1711. [Google Scholar]
- Clode PL, Kilburn MR, Jones DL, Stockdale EA, Cliff JB, Herrmann AM, Murphy DV. In situ mapping of nutrient uptake in the rhizosphere using nanoscale secondary ion mass spectrometry. Plant Physiology. 2009;151:1751–1757. doi: 10.1104/pp.109.141499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobelli C, Foster D, Toffolo G. Tracer kinetics in biomedical research: from data to model. New York, NY, USA: Springer; 2001. [Google Scholar]
- Dempster DN, Jones DL, Murphy DV. Organic nitrogen mineralisation in two contrasting agro-ecosystems is unchanged by biochar addition. Soil Biology & Biochemistry. 2012;48:47–50. [Google Scholar]
- Denison RF, Bledsoe C, Kahn M, O’Gara F, Simms EL, Thomashow LS. Cooperation in the rhizosphere and the “free rider” problem. Ecology. 2003;84:838–845. [Google Scholar]
- Forde BG, Walch-Liu P. Nitrate and glutamate as environmental cues for behavioural responses in plant roots. Plant, Cell & Environment. 2009;32:682–693. doi: 10.1111/j.1365-3040.2008.01927.x. [DOI] [PubMed] [Google Scholar]
- Forgeard F, Frenot Y. Effects of burning on heathland soil chemical properties: an experimental study on the effect of heating and ash deposits. Journal of Applied Ecology. 1996;33:803–811. [Google Scholar]
- Forsum O, Svennerstam H, Ganeteg U, Näsholm T. Capacities and constraints of amino acid utilization in Arabidopsis. New Phytologist. 2008;179:1058–1069. doi: 10.1111/j.1469-8137.2008.02546.x. [DOI] [PubMed] [Google Scholar]
- Frensch J, Hsiao TC, Steudle E. Water and solute transport along developing maize roots. Planta. 1996;198:348–355. [Google Scholar]
- Friedel JK, Scheller E. Composition of hydrolysable amino acids in soil organic matter and soil microbial biomass. Soil Biology & Biochemistry. 2002;34:315–325. [Google Scholar]
- Gattolin S, Newbury HJ, Bale JS, Tseng HM, Barrett DA, Pritchard J. A diurnal component to the variation in sieve tube amino acid content in wheat. Plant Physiology. 2008;147:912–921. doi: 10.1104/pp.108.116079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass ADM, Britto DT, Kaiser BN, Kinghorn JR, Kronzucker HJ, Kumar A, Okamoto M, Rawat S, Siddiqi MY, Unkles SE, et al. The regulation of nitrate and ammonium transport systems in plants. Journal of Experimental Botany. 2002;53:855–864. doi: 10.1093/jexbot/53.370.855. [DOI] [PubMed] [Google Scholar]
- Hawkins HJ, Johansen A, George E. Uptake and transport of organic and inorganic nitrogen by arbuscular mycorrhizal fungi. Plant and Soil. 2000;226:275–285. [Google Scholar]
- Herrmann AM, Clode PL, Fletcher IR, Nunan N, Stockdale EA, O’Donnel AG, Murphy DV. A novel method for the study of the biophysical interface in soils using nano-scale secondary ion mass spectrometry. Rapid Communications in Mass Spectrometry. 2007;21:29–34. doi: 10.1002/rcm.2811. [DOI] [PubMed] [Google Scholar]
- Hill PW, Farrell M, Jones DL. Bigger may be better in soil N cycling: does rapid acquisition of small L-peptides by soil microbes dominate fluxes of protein-derived N in soil? Soil Biology & Biochemistry. 2012;48:106–112. [Google Scholar]
- Hill PW, Quilliam RS, DeLuca TH, Farrar J, Farrell M, Roberts P, Newsham KK, Hopkins DW, Bardgett RD, Jones DL. Acquisition and assimilation of nitrogen as peptide-bound and D-enantiomers of amino acids by wheat. PLoS ONE. 2011;6:e19220. doi: 10.1371/journal.pone.0019220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inselsbacher E, Näsholm T. The below-ground perspective of forest plants: soil provides mainly organic nitrogen for plants and mycorrhizal fungi. New Phytologist. 2012;195:329–334. doi: 10.1111/j.1469-8137.2012.04169.x. [DOI] [PubMed] [Google Scholar]
- Jones DL, Darrah PR. Amino-acid influx at the soil–root interface of Zea mays L. and its implications in the rhizosphere. Plant and Soil. 1994;163:1–12. [Google Scholar]
- Jones DL, Healey JR, Willett VB, Farrar JF, Hodge A. Dissolved organic nitrogen uptake by plants – an important N uptake pathway? Soil Biology & Biochemistry. 2005a;37:413–423. [Google Scholar]
- Jones DL, Hodge A. Biodegradation kinetics and sorption reactions of three differently charged amino acids in soil and their effects on plant organic nitrogen availability. Soil Biology & Biochemistry. 1999;31:1331–1342. [Google Scholar]
- Jones DL, Nguyen C, Finlay RD. Carbon flow in the rhizosphere: carbon trading at the soil–root interface. Plant and Soil. 2009;321:5–33. [Google Scholar]
- Jones DL, Owen AG, Farrar JF. Simple method to enable the high resolution determination of total free amino acids in soil solutions and soil extracts. Soil Biology & Biochemistry. 2002;34:1893–1902. [Google Scholar]
- Jones DL, Shannon D, Junvee-Fortune T, Farrar JF. Plant capture of free amino acids is maximized under high soil amino acid concentrations. Soil Biology & Biochemistry. 2005b;37:179–181. [Google Scholar]
- Jörgensen RG. Quantification of the microbial biomass by determining ninhydrin-reactive N. Soil Biology and Biochemistry. 1996;28:301–306. [Google Scholar]
- Jörgensen RG, Brookes PC. Ninhydrin-reactive nitrogen measurements of microbial biomass in 0.5 M K2SO4 soil extracts. Soil Biology & Biochemistry. 1990;22:1023–1027. [Google Scholar]
- Kuzyakov Y, Jones DL. Glucose uptake by maize roots and its transformation in the rhizosphere. Soil Biology & Biochemistry. 2006;38:851–860. [Google Scholar]
- Loomis RS, Connor DJ. Crop ecology. Cambridge, UK: Cambridge University Press; 1992. [Google Scholar]
- Manzoni S, Porporato A, Schimel JP. Soil heterogeneity in lumped mineralization-immobilization models. Soil Biology & Biochemistry. 2008;40:1137–1148. [Google Scholar]
- Marschner P, Crowley D, Rengel Z. Rhizosphere interactions between microorganisms and plants govern iron and phosphorus acquisition along the root axis – model and research methods. Soil Biology & Biochemistry. 2011;43:883–894. [Google Scholar]
- Mays ET, Feldhoff RC, Nettleton GS. Determination of protein loss during aqueous and phase partition fixation using formalin and glutaraldehyde. Journal of Histochemistry & Cytochemistry. 1984;32:1107–1112. doi: 10.1177/32.10.6434629. [DOI] [PubMed] [Google Scholar]
- McKee HS. Nitrogen metabolism in plants. Oxford, UK: Clarendon Press; 1962. [Google Scholar]
- Miller AJ, Cookson SJ, Smith SJ, Wells DM. The use of microelectrodes to investigate compartmentation and the transport of metabolized inorganic ions in plants. Journal of Experimental Botany. 2001;52:541–549. [PubMed] [Google Scholar]
- Mosier AR, Syers JK, Freney JR. Agriculture and the nitrogen cycle: assessing the impacts of fertilizer use on food production and the environment. Washington, DC, USA: Island Press; 2004. [Google Scholar]
- Mulvaney RL, Sparks DL. Methods of soil analysis. Madison, WI, USA: Soil Science Society of America, American Society of Agronomy Inc; 1996. Nitrogen – inorganic forms; pp. 1123–1184. [Google Scholar]
- Murphy DV, Recous S, Stockdale EA, Fillery IRP, Jensen LS, Hatch DJ, Goulding KWT. Gross nitrogen fluxes in soil: theory measurement and application of 15N pool dilution techniques. Advances in Agronomy. 2003;79:69–118. [Google Scholar]
- Näsholm T, Huss-Danell K, Hogberg P. Uptake of glycine by field grown wheat. New Phytologist. 2001;150:59–63. [Google Scholar]
- Näsholm T, Kielland K, Ganeteg U. Uptake of organic nitrogen by plants. New Phytologist. 2009;182:31–48. doi: 10.1111/j.1469-8137.2008.02751.x. [DOI] [PubMed] [Google Scholar]
- Owen AG, Jones DL. Competition for amino acids between wheat roots and rhizosphere microorganisms and the role of amino acids in plant N acquisition. Soil Biology & Biochemistry. 2001;33:651–657. [Google Scholar]
- Pernice M, Meibom A, Van Den Heuvel A, Kopp C, Domart-Coulon I, Hoegh-Guldberg O, Dove S. A single-cell view of ammonium assimilation in coral–dinoflagellate symbiosis. ISME Journal. 2012;6:1314–1324. doi: 10.1038/ismej.2011.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson J, Näsholm T. A GC-MS method for determination of amino acid uptake by plants. Physiologia Plantarum. 2001;113:352–358. doi: 10.1034/j.1399-3054.2001.1130308.x. [DOI] [PubMed] [Google Scholar]
- Peteranderl R, Lechene C. Measure of carbon and nitrogen stable isotope ratios in cultured cells. Journal of the American Society for Mass Spectrometry. 2004;15:478–485. doi: 10.1016/j.jasms.2003.11.019. [DOI] [PubMed] [Google Scholar]
- Phillips DA, Fox TC, King MD, Bhuvaneswari TV, Teuber LR. Microbial products trigger amino acid exudation from plant roots. Plant Physiology. 2004;136:2887–2894. doi: 10.1104/pp.104.044222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen J, Sauheitl L, Eriksen J, Kuzyakov Y. Plant uptake of dual-labeled organic N biased by inorganic C uptake: results of a triple labeling study. Soil Biology & Biochemistry. 2010;42:524–527. [Google Scholar]
- Reeve JR, Smith JL, Carpenter-Boggs L, Reganold JP. Glycine, nitrate, and ammonium uptake by classic and modern wheat varieties in a short-term microcosm study. Biology and Fertility of Soils. 2009;45:723–732. [Google Scholar]
- Roberts P, Stockdale R, Khalid M, Iqbal Z, Jones DL. Carbon-to-nitrogen ratio is a poor predictor of low molecular weight organic nitrogen mineralization in soil. Soil Biology & Biochemistry. 2009;41:1750–1752. [Google Scholar]
- Rousk J, Jones DL. Loss of low molecular weight dissolved organic carbon (DOC) and nitrogen (DON) in H2O and 0.5 M K2SO4 soil extracts. Soil Biology & Biochemistry. 2010;42:2331–2335. [Google Scholar]
- Sadras VO, Lawson C. Nitrogen and water-use efficiency of Australian wheat varieties released between 1958 and 2007. European Journal of Agronomy. 2013;46:34–41. [Google Scholar]
- Sauheitl L, Glaser B, Weigelt A. Advantages of compound-specific stable isotope measurements over bulk measurements in studies on plant uptake of intact amino acids. Rapid Communications in Mass Spectrometry. 2009;23:3333–3342. doi: 10.1002/rcm.4255. [DOI] [PubMed] [Google Scholar]
- Schreiber L. Transport barriers made of cutin, suberin and associated waxes. Trends in Plant Science. 2010;15:546–553. doi: 10.1016/j.tplants.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Sivaguru M, Pike S, Gassmann W, Baskin TI. Aluminum rapidly depolymerizes cortical microtubules and depolarizes the plasma membrane: evidence that these responses are mediated by a glutamate receptor. Plant and Cell Physiology. 2003;44:667–675. doi: 10.1093/pcp/pcg094. [DOI] [PubMed] [Google Scholar]
- Soper FM, Paungfoo-Lonhienne C, Brackin R, Rentsch D, Schmidt S, Robinson N. Arabidopsis and Lobelia anceps access small peptides as a nitrogen source for growth. Functional Plant Biology. 2011;38:788–796. doi: 10.1071/FP11077. [DOI] [PubMed] [Google Scholar]
- Thornton B. Uptake of glycine by non-mycorrhizal Lolium perenne. Journal of Experimental Botany. 2001;52:1315–1322. [PubMed] [Google Scholar]
- Warren CR. Post-uptake metabolism affects quantification of amino acid uptake. New Phytologist. 2012;193:522–531. doi: 10.1111/j.1469-8137.2011.03933.x. [DOI] [PubMed] [Google Scholar]
- Yuan L, Loque D, Kojima S, Rauch S, Ishiyama K, Inoue E, Takahashi H, von Wiren N. The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell. 2007;19:2636–2652. doi: 10.1105/tpc.107.052134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1 Amount of 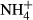 held on the soil’s solid phase as a function of solution
held on the soil’s solid phase as a function of solution 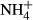 concentration.
concentration.
Fig S2 Total amount of 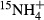 present in soil solution and held on the soil’s cation exchange phase in Triticum aestivum planted and unplanted microcosms.
present in soil solution and held on the soil’s cation exchange phase in Triticum aestivum planted and unplanted microcosms.
Fig S3 Time-dependent mineralization of 14C-labelled glutamate in soil in the presence and absence of wheat (Triticum aestivum) roots.
Fig S4 NanoSIMS imaging and analysis of wheat (Triticum aestivum) roots in situ within the embedded soil core after exposure to 15N-labelled glutamate uptake.
Fig S5 Influence of external solution glutamate concentration on the growth rate of wheat (Triticum aestivum) roots exposed to various concentrations of glutamate.


