Key Points
AZD1208 is a selective pan-Pim kinase inhibitor with efficacy in AML cells, xenografts, and Flt3-internal tandem duplication or Flt3 wild-type patient samples.
AML cell growth inhibition is associated with suppression of p70S6K, 4EBP1 phosphorylation, and messenger RNA translation.
Abstract
Upregulation of Pim kinases is observed in several types of leukemias and lymphomas. Pim-1, -2, and -3 promote cell proliferation and survival downstream of cytokine and growth factor signaling pathways. AZD1208 is a potent, highly selective, and orally available Pim kinase inhibitor that effectively inhibits all three isoforms at <5 nM or <150 nM in enzyme and cell assays, respectively. AZD1208 inhibited the growth of 5 of 14 acute myeloid leukemia (AML) cell lines tested, and sensitivity correlates with Pim-1 expression and STAT5 activation. AZD1208 causes cell cycle arrest and apoptosis in MOLM-16 cells, accompanied by a dose-dependent reduction in phosphorylation of Bcl-2 antagonist of cell death, 4EBP1, p70S6K, and S6, as well as increases in cleaved caspase 3 and p27. Inhibition of p4EBP1 and p-p70S6K and suppression of translation are the most representative effects of Pim inhibition in sensitive AML cell lines. AZD1208 inhibits the growth of MOLM-16 and KG-1a xenograft tumors in vivo with a clear pharmacodynamic-pharmacokinetic relationship. AZD1208 also potently inhibits colony growth and Pim signaling substrates in primary AML cells from bone marrow that are Flt3 wild-type or Flt3 internal tandem duplication mutant. These results underscore the therapeutic potential of Pim kinase inhibition for the treatment of AML.
Introduction
The Pim serine/threonine kinase family is composed of three highly homologous members, Pim-1, -2, and -3, identified as proviral insertion sites of the Moloney murine leukemia virus associated with the development of T-cell lymphomas.1,2 Pim-1 and Pim-2 are upregulated in several hematologic malignancies, including acute myeloid leukemia (AML), chronic lymphocytic leukemia, acute lymphoblastic leukemia, multiple myeloma, and non-Hodgkin lymphoma.3-6 In AML, this is driven at least in part by activation of receptor tyrosine kinases such as the Flt3-internal tandem duplication (Flt3-ITD) mutation5,7,8 found in approximately one third of AML patients. The JAK/STAT pathway, a key mediator of cytokine and growth factor signaling, plays an important role in regulating Pim expression.9 Other pathways and transcription factors, such as nuclear factor κB10 and HOXA9,11 may also play a role, depending on cellular context. The Pims are constitutively active kinases whose expression is controlled primarily at the transcriptional and translational level.12,13
Pim kinases modulate the activity of a variety of substrates involved in the control of transcription, translation, cell proliferation, and survival.14 They have been shown to play a role in promoting survival of AML cells via phosphorylation of Bcl-2 antagonist of cell death (BAD), abrogating its inhibitory association with antiapoptotic protein Bcl-xL.12,15-17 In addition to BAD, Pims share other substrates with the AKT pathway such as PRAS40, which negatively regulates mTORC1, thereby modulating protein translation through the mTORC1 substrates p70S6K and 4EBP1.18 Evidence is accumulating for a convergent but independent role for Pim kinases and the AKT/mTOR pathway in the regulation of messenger RNA (mRNA) translation. Tamburini et al19 identified Pim-2 as an essential regulator of 4EBP1 and cap-dependent translation in AML, capable of maintaining translation in the presence of the mTORC1 inhibitor RAD001. Similar observations have been made in lymphoma models.20
Inhibition of Pim kinase activity provides a novel therapeutic approach to the treatment of cancer. The attribution of oncogenic activity to all three Pim isoforms, and the potential for redundancy, argues for the development of inhibitors capable of targeting all family members. Furthermore, gene knockout studies have demonstrated that mice deficient for all three Pims are viable and fertile,21 which supports the tolerability of pan-Pim kinase inhibition. AZD1208 is a highly selective and orally available inhibitor of all three Pim isoforms. In this article, the efficacy of AZD1208 in cultured AML cell lines, tumor xenograft models, and ex vivo cultures of primary tumor cells from Flt3-ITD and Flt3 wild-type patients is demonstrated, as well as accompanying modulation of Pim signaling substrates that can contribute to the inhibition of tumor growth. The results have supported the initiation of phase 1 clinical trials of AZD1208 in AML.
Materials and methods
Reagents
AZD1208 was synthesized by AstraZeneca R&D (Waltham, MA)22 and diluted in dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO). Cytarabine was purchased from Bedford Laboratories (Bedford, OH). Antibodies used are described in the supplemental Methods, available on the Blood Web site.
Enzyme assays
The activity of purified human Pim-1, -2, and -3 enzymes on a BAD peptide substrate was determined as previously described.22 To determine inhibition constants (Ki), 50% inhibition concentration (IC50) values were acquired at a series of adenosine triphosphate (ATP) concentrations and compound doses with 1 nM enzyme and 1.5 µM full-length BAD substrate in 50 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 1 mM dithiothreitol, 0.01% Tween 20, 50 μg/mL bovine serum albumin, and 10 mM MgCl2. The Ki values were calculated by global data fitting using the Cheng-Prusoff equation or the Morrison equation for tight-binding inhibitors.
To assess selectivity, 442 kinases were screened by using DiscoveRx KINOMEscan technology at a single concentration of 1 μM. Kinases inhibited by more than 50% were retested at DiscoveRx with a full dose-response to determine binding affinity.
ELISA for determination of cellular pBAD IC50
Details of enzyme-lined immunosorbent assay (ELISA) are available in the supplemental Methods.
Cell culture and proliferation assay
Cell lines were purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen, American Type Culture Collection, or European Collection of Cell Cultures and were cultured and assayed for the effect of AZD1208 on proliferation in the recommended media (supplemental Data).
Cell cycle, cell size, and apoptosis
Cells were plated at 0.13 to 0.5 × 106 cells per well in 6-well plates overnight and treated for 72 hours. For cell cycle analysis, cells were fixed in 70% ethanol before staining and treated with propidium iodide (20 μg/mL) and ribonuclease A (100 μg/mL). Samples were analyzed by using a BD FACSCalibur flow cytometer and ModFit software. For cell size measurements, the mean forward scatter parameter-height was determined from the G1 population by using FlowJo software (Tree Star, Inc.). For apoptosis, cells were stained with Annexin V (BD Bioscience) and 7-amino-actinomycin D (BD Bioscience). Samples were analyzed on a FACSCanto flow cytometer using FlowJo software.
Immunoblotting
Cells were plated at 0.8 × 106 cells per well in 6-well plates overnight and then treated for the indicated times. Cell lysates were prepared by using the following lysis buffers: 50 mM tris(hydroxymethyl)aminomethane (pH 7.5), 150 mM NaCl, 1% NP-40 or radioimmunoprecipitation assay buffer with phosphatase inhibitor and protease inhibitor cocktails (Thermo Scientific). Protein concentration was measured by using bicinchoninic acid reagent (Thermo Scientific), and 20 to 40 μg proteins were separated on bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane gels (Invitrogen). Immunoblotting was done with primary antibodies listed (supplemental Data). After incubation with appropriate secondary antibodies, signal was detected by using chemiluminescence and film or a FujiFilm Imager LAS-4000.
Cap-dependent translation assay
Cells were plated at 0.6 to 0.7 × 106 cells per milliliter in 15 mL of media overnight before drug treatment of 3 hours and lysis in 350 μL ice-cold radioimmunoprecipitation assay buffer lysis buffer containing protease and phosphatase inhibitors. Lysates were diluted to 1 μg/μL, and 450 μL was added to 55 μL methyl-7-guanosine-5′-triphosphate sepharose beads (GE Healthcare) and rotated overnight at 4°C. The beads were washed, and bound proteins were eluted with 1× sodium dodecyl sulfate buffer, heated to 95°C, and separated on bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane 4% to 12% gradient gels.
Polysome profiling
OCI-M1 or EOL-1 cell lines were treated with dimethylsulfoxide or AZD1208 (1 μM) for 9 hours, and polysomal fractionation was performed as described in previous studies.23-25
In vivo studies
Female CB17 SCID mice (5-6 weeks old) purchased from Charles River Laboratories were maintained under specific pathogen-free conditions and used in compliance with protocols approved by the Institutional Animal Care and Use Committees of AstraZeneca, which conform to institutional and national regulatory standards on experimental animal usage.
MOLM-16 cells (5 × 106) or KG-1a cells (6 × 106) were implanted with matrigel subcutaneously into the left flank of mice. When tumor size reached ∼150 to 200 mm3, mice were randomly assigned and treated once daily with either vehicle (0.5% hydroxypropyl methylcellulose) or AZD1208 by oral gavage with 10 to 15 mice per group. Cytarabine was diluted in 0.45% NaCl and dosed at 30 mg/kg twice per week on consecutive days by intraperitoneal injection. Tumor volume was measured twice per week with calipers and calculated as tumor volume = (length × width2) × 0.5.
For pharmacodynamic (PD) studies, mice with tumors of 200 to 400 mm3 were treated with either vehicle or AZD1208, with 3 mice per dose and time point, and tumors were harvested and lysed for immunoblot analysis. pBAD protein levels were quantified by using the phospho/total-BAD MULTI-SPOT Assay System from Meso Scale Discovery.
For pharmacokinetic analysis, blood samples were collected from the same mice via cardiac puncture. Total plasma concentrations of AZD1208 were determined by liquid chromatography/tandem mass spectrometry method.
Patient samples
Bone marrow aspirates were obtained from newly diagnosed AML patients (Dana-Farber Cancer Institute, Boston, MA) with patient consent and approval from the Dana-Farber Cancer Institute Institutional Review Board. This study was conducted in accordance with the Declaration of Helsinki. Mononuclear cells were isolated immediately by standard Ficoll-Paque gradient separation (Ficoll-Paque PLUS; GE Healthcare 17-1440-02) from samples delivered within 2 hours of collection. For the colony assay, cells were diluted in media and added to 3 mL of Methocult (STEMCELL Technologies #04435). AZD1208 (0.1, 0.3, 1, or 3 μM) or cytarabine (10 or 30 nM) was added, and cell suspensions were mixed and plated. After 14 days of incubation at 37°C, 5% CO2, and 95% humidity, colonies were enumerated by light microscopy. Data are shown for all 15 patient samples that yielded colony growth. For biomarker analysis, cells were plated in RPMI-1640 with 2% fetal bovine serum/1% l-glutamine and treated with vehicle or 0.3, 1, or 3 μM AZD1208 for 3 hours. Cell lysates were prepared in Cell Lysis Buffer (Cell Signaling Technology) supplemented with protease and phosphatase inhibitors.
Results
AZD1208 is a potent and selective pan-Pim kinase inhibitor
The structure of AZD1208, a benzylidene-1,3-thiazolidine-2,4-dione, is shown in Figure 1. AZD1208 is a potent ATP-competitive inhibitor of all three Pim kinase isoforms (Table 1). The Ki values were determined to be 0.1 nM for Pim-1, 1.92 nM for Pim-2, and 0.4 nM for Pim-3. In enzymatic assays carried out at a concentration of ATP that leads to half-maximal reaction velocity, AZD1208 inhibited kinase activity with an IC50 of 0.4 nM for Pim-1, 5.0 nM for Pim-2, and 1.9 nM for Pim-3. In enzyme assays using 5 mM ATP, the high end of physiologic ATP concentration in human cells, the IC50 values were 2.6 nM for Pim-1, 164 nM for Pim-2, and 17 nM for Pim-3.
Figure 1.
Chemical structure of AZD1208.
Table 1.
Ki and enzyme assay IC50 values of AZD1208
| Assay value (nM) | Pim-1 | Pim-2 | Pim-3 |
|---|---|---|---|
| Ki | 0.10 | 1.92 | 0.40 |
| Enzyme IC50 at Km [ATP] | 0.4 | 5.0 | 1.9 |
| Enzyme IC50 at 5 mM [ATP] | 2.6 | 164 | 17 |
| Cell pBAD IC50 | 10 | 151 | 102 |
Km, half-maximal reaction velocity.
To measure selectivity for Pims relative to other kinases, AZD1208 was evaluated against a panel of 442 kinases using the DiscoveRx KINOMEscan competition binding assay. The three Pim kinases showed the highest percentage of inhibition. In addition, 13 other kinases were found to be inhibited by 50% or more. No inhibition of Flt3 or Flt3-ITD was observed. A dose-response follow-up conducted against the 16 kinases inhibited >50% showed AZD1208 to bind tightly to Pim-1, -2, and -3 with binding constants (Kd) of 0.2 nM, 0.88 nM, and 0.76 nM, respectively (Table 2), comparable to the Ki determined in functional enzyme assays. AZD1208 bound six other kinases with potencies ranging from 38 nM to 930 nM (Table 2) but did not show significant binding to the remaining seven kinases (Kd >10 μM). The potency difference between affinity of AZD1208 for Pim kinases and the next most potent “hit” in the panel (CDK7) was at least 43-fold.
Table 2.
Kd for top kinase hits in the KINOMEscan competition binding assay
| Entrez gene symbol | Kd (nM) |
|---|---|
| PIM1 | 0.20 |
| PIM3 | 0.76 |
| PIM2 | 0.88 |
| CDK7 | 38.0 |
| MAPK15 | 53.0 |
| CAMK4 | 360 |
| DAPK1 | 420 |
| HIPK3 | 480 |
| STK17B | 930 |
To assess the potency of AZD1208 on each of the three Pim isoforms in a cell-based assay, a substrate-enzyme tethered system was engineered. Constructs were created with full-length Pim-1, -2, and -3 kinases individually fused to a BAD peptide substrate via a flexible (G4S)4 linker and a c-Myc tag at the N terminus to allow capture in a sandwich ELISA. The fusion constructs were transiently transfected into U2-OS cells and treated with AZD1208 for 3 hours; the level of pBAD inhibition was determined by ELISA. The IC50 values were 10 nM for Pim-1, 151 nM for Pim-2, and 102 nM for Pim-3, consistent with enzyme inhibition potencies at 5 mM ATP concentration (Table 1).
AZD1208 inhibits AML cell-line growth and can induce cell-cycle arrest and apoptosis
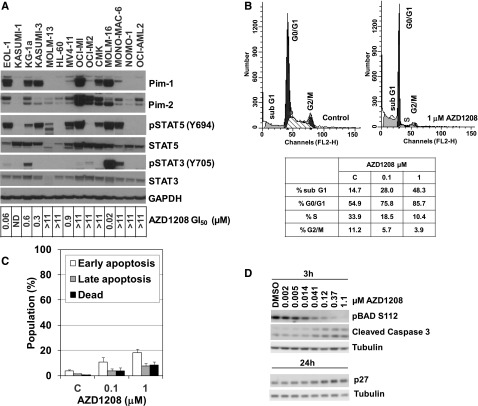
AZD1208 was tested for antiproliferative activity in a panel of AML cell lines (Figure 2A). Five of the cell lines tested, EOL-1, KG-1a, Kasumi-3, MV4-11, and MOLM-16, were sensitive to Pim inhibition with concentration for 50% of inhibition of cell proliferation (GI50) values <1 μM. Sensitivity in the AML panel generally correlated with high Pim-1 expression and phosphorylated STAT5 (pSTAT5). Three of the five sensitive cell lines, EOL-1, KG-1a, and MV4-11, carry known activated receptor tyrosine kinases (supplemental Table 1), and MOLM-16 cells carry a Tyk2 fusion gene (K.M., E.K.K., K.S.D., M.S., Asim Khwaja, Zhongwu Lai, and D.H., manuscript in preparation), which could account for Pim-1 upregulation downstream of STAT5 activation. MOLM-13 cells harbor the Flt3-ITD mutation and were not sensitive, but Pim-1 protein was not detected. Resistance to AZD1208 did not appear to be driven by drug efflux. Analysis of expression of 13 efflux pumps across the cell panel showed no correlation with AZD1208 GI50 (R2 < 0.1 for all pumps; data not shown).
Figure 2.
Effect of AZD1208 on cell growth and survival in AML cell lines. (A) Pim-1 and Pim-2 expression and STAT3 and STAT5 expression and phosphorylation across a panel of AML cell lines with varying sensitivity to AZD1208, as shown by immunoblot analysis of cell lysates and GI50 values from a 72-hour proliferation assay. (B) AZD1208 effect on G1 cell cycle arrest and subG1 population in MOLM-16 cells, assessed by propidium iodide staining and flow cytometry for DNA content 72 hours after treatment. Data shown are representative of 3 independent experiments. (C) Effect of AZD1208 on apoptosis in MOLM-16 cells determined by staining with Annexin V (early apoptosis), Annexin V and 7-amino-actinomycin D (7-AAD) (late apoptosis), and 7-AAD (dead/necrotic cells). Data shown are representative of 3 independent experiments. (D) AZD1208 effect on p27, pBAD, and cleaved caspase 3, as shown by immunoblot anaylsis 3 and 24 hours after treatment. FL2-H, peak emission values in the FL2 channel; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ND, not determined.
Cell cycle analysis of sensitive MOLM-16 cells showed dose-dependent increases in the G0/G1 and subG1 populations, indicating both cell cycle arrest and cell death in response to AZD1208 treatment (Figure 2B). The percentage of cells in G0/G1 and subG1 increased markedly after 1 μM compound treatment for 72 hours, from 54.9% to 85.7% and 14.7% to 48.3%, respectively. Two other sensitive cell lines tested, KG-1a and EOL-1, did not manifest cell cycle arrest in response to AZD1208, and none of the sensitive lines showed a marked apoptotic response like that seen for MOLM-16 (data not shown).
Consistent with the increased subG1 cell population observed in AZD1208-treated MOLM-16 cells, a dose-dependent increase in the percentage of apoptotic and dead cells measured by Annexin V and 7-amino-actinomycin D staining was also seen (Figure 2C). Inhibition of Pims can prevent phosphorylation of proapoptotic BAD leading to sequestration of the prosurvival proteins Bcl-2/Bcl-xL.17 Indeed, dose-dependent inhibition of BAD phosphorylation on Ser 112 was seen 3 hours after treatment of MOLM-16 cells with AZD1208, correlated with an increase in cleaved caspase 3 (Figure 2D).
Pim kinases have also been shown to directly phosphorylate the CDK inhibitor p27, resulting in its proteosome-dependent degradation.26 Pim inhibition resulted in accumulation of p27 24 hours after treatment with AZD1208 (Figure 2D), consistent with the G1 cell cycle arrest observed.
Biomarker modulation and downstream effects of AZD1208 in AML cell lines
In addition to phosphorylation of BAD, Pims have been shown to play a role in phosphorylation of other proteins shared with the PI3K/AKT/mTOR signaling pathway, as shown in Figure 3A. Modulation of several of these proteins was measured after treatment with AZD1208 in sensitive and resistant cell lines. Assessment of BAD phosphorylation on Ser 112 demonstrated inhibition by AZD1208 not only in MOLM-16 cells, but also in sensitive KG-1a, Kasumi-3, and MV4-11 cells (Figure 3B and supplemental Figure 1). However, no inhibition was observed in sensitive EOL-1 cells, and varying degrees of inhibition were also seen in resistant cells, indicating that modulation of pBAD does not necessarily correlate with growth inhibition or apoptosis in these cell lines.
Figure 3.
Effect of AZD1208 on downstream signaling in AML cell lines. (A) Pim signaling and interaction with PI3K/AKT/mTOR pathway showing activating (green) and inactivating (red) phosphorylation events. Rheb, Ras homolog enriched in brain. (B) Modulation of various biomarkers seen in MOLM-16, EOL-1, KG-1a, and OCI-M1 cells after 3 hours of treatment with control (“C”) dimethylsulfoxide (DMSO) or 0.01, 0.1, or 1 μM AZD1208 by immunoblot analysis of cell lysates. For p70S6K, the band shown represents the ∼60 kDa S6K1 isoform. (C) Effect of AZD1208 on cap-dependent translation complex formation after 3 hours of treatment with AZD1208 at the indicated doses. eIF4E was immunoprecipitated from treated cell lysates by using methyl-7-guanosine-5′-triphosphate cap sepharose beads (the eluted immunoprecipitated material is shown in the third row) and immunoblotted with eIF4G (first row) and 4EBP1 (second row) to assess the extent of association of each protein with eIF4E before and after treatment with AZD1208. The fourth row shows effects of drug treatment on p4EBP1 S65 prior to immunoprecipitation (IP) of cell lysates. (D) Effects of Pim kinase inhibition on polysomal assembly. OCI-M1 or EOL-1 cell lines were treated with solvent control (DMSO), or 1 μM AZD1208 for 9 hours, and the lysates were layered on a 10% to 50% sucrose gradient. The gradients were subjected to ultracentrifugation, and fractions were collected by continuous monitoring of optical density (OD) at 254 nm. The OD 254 nm was plotted as a function of gradient depth for each treatment. A representative profile from 1 of 2 studies is shown and the polysomal (PS) and monosomal (MS) peaks are indicated. The area under the polysome and monosome peaks was quantified for each treatment by using ImageJ software. The ratio of area under the polysomal and polysomal plus monosomal peaks was calculated for each treatment, and the results of 2 independent studies for each cell line are presented as percentage of respective DMSO control. Expt, experiment.
A better correlation with sensitivity to cell growth inhibition was seen for inhibition of phosphorylation of p70S6K on Thr 389 and its substrate the S6 ribosomal protein on Ser 235/236 (Figure 3B and supplemental Figure 1). In addition, moderate suppression of p4EBP1 on Ser 65, most evident in samples treated with 1 μM AZD1208, was seen preferentially in sensitive cell lines. Varying degrees of inhibition of phosphorylation of the mTORC1 inhibitory protein PRAS40 on Thr 246 was seen in both sensitive (Figure 3B and supplemental Figure 1) and resistant (supplemental Figure 1) cell lines. Levels of c-Myc protein were slightly reduced in some of the cell lines (Figure 3B), which could have an impact on its transcriptional activity.27 In fact, modulation of the c-Myc transcriptome was observed in cells treated with AZD1208 in both sensitive and resistant cell lines (data not shown). No decreases were seen in cyclin D1 and Mcl-1 levels (supplemental Figure 2 and data not shown). The levels of total BAD, 4EBP1, p70S6K, and S6 proteins were not reduced, although there was a slight reduction in total PRAS40 levels in two of the cell lines (supplemental Figure 2). Increases in p27 were not seen in cell lines other than MOLM-16 (data not shown).
To assess the functional consequence of inhibition of 4EBP1 phosphorylation, the effect of AZD1208 on assembly of the cap-dependent translation complex was measured. Binding of unphosphorylated 4EBP1 to the cap binding protein eIF4E blocks its interaction with eIF4G, resulting in inhibition of cap-dependent translation, whereas phosphorylation of 4EBP1 inhibits binding to eIF4E and allows its interaction with eIF4G to promote translation.28 In response to AZD1208 treatment, three sensitive cell lines, MOLM-16, EOL-1, and KG-1a, showed reduced interaction of immunoprecipitated eIF4E with eIF4G and increased interaction with inhibitory 4EBP1 that correlated with reduced phosphorylation of 4EBP1 on Ser 65, most evident at the 1 μM dose (Figure 3C). The effects were much less marked in AZD1208 insensitive OCI-M1 cells.
To determine whether the observed inhibition of formation of translation initiation complexes manifests at the level of mRNA translation, polysome profiling was carried out. Treatment with 1 μM AZD1208 resulted in marked suppression of polysomal peaks with a reciprocal increase in the 80S monosome peak in sensitive EOL-1 cells, but not in insensitive OCI-M1 cells (Figure 3D), consistent with suppression of polysomal assembly preferentially in sensitive cells treated with AZD1208.
Functional deficiency of p70S6K or S6 has been linked to reduced cell size.29-32 Cell size analysis by flow cytometry demonstrated a 10% to 15% decrease in the size of MOLM-16 and KG-1a cells following treatment with AZD1208 (supplemental Figure 3), coincident with the decreases in p70S6K and S6 phosphorylation seen and comparable to that observed following treatment with rapamycin in other reports.29,30
AZD1208 efficacy and PK-PD in AML xenografts
The in vivo activity of AZD1208 was assessed in a MOLM-16 xenograft model treated daily for 2 weeks with drug or vehicle control. Dose-dependent inhibition of MOLM-16 tumor growth was observed (Figure 4A). Treatment with 10 mg/kg or 30 mg/kg of AZD1208 (the latter representing the typical maximum-tolerated dose on this treatment schedule) led to 89% tumor growth inhibition or slight regression, respectively. PD analyses following a single dose of 30 mg/kg showed strong suppression of pBAD, p4EBP1, and p-p70S6K for up to 12 hours after dosing (Figure 4B).
Figure 4.

AZD1208 efficacy and pharmacokinetic (PK)-PD analyses in AML xenograft models. (A) CB17 SCID mice implanted subcutaneously with MOLM-16 cells were treated once daily (QD) with either vehicle or AZD1208 by oral gavage for 14 days to assess effect on tumor growth. Data shown are the average ± standard error of the mean (SEM). **Significant difference in tumor size at day 25 between vehicle- and AZD1208-treated mice; Student t test P < .001. (B) Immunoblot analysis of MOLM-16 tumors from mice treated with 30 mg/kg AZD1208 at the times shown. (C) pBAD levels as percent of vehicle-treated controls measured at the indicated times and doses, as described in “Methods.” Data represent the average of triplicates + SEM. (D) Percent pBAD inhibition at multiple plasma drug levels was measured to determine the PK-PD relationship and concentration required for 50% inhibition. Data are from the study in (C) and an additional data set including 18-hour time points. The graph was generated by using Phoenix WinNonlin 6.3 software (Pharsight). (E) CB17 SCID mice implanted subcutaneously with KG-1a cells were treated once daily with vehicle or AZD1208 by oral gavage or twice per week, on consecutive days, with cytarabine by intraperitoneal injection from day 14 to day 39 postimplantation. Data shown are the average ± SEM. **Significant difference in tumor size at days 39 and 50 between AZD1208-treated or cytarabine-treated mice and mice treated with the combination of AZD1208 and cytarabine. Student t test P < .001.
To determine the PK-PD relationship, inhibition of pBAD was quantified by mesoscale ELISA in MOLM-16 tumor xenografts, and plasma drug concentrations were measured following acute doses of 3, 10, and 30 mg/kg AZD1208. Dose proportional inhibition of pBAD Ser 112 was observed (Figure 4C). Following a single dose of 30 mg/kg AZD1208, BAD phosphorylation was suppressed maximally by an average of 72% for 12 hours and by 46% at 24 hours after dosing. At 10 mg/kg, AZD1208 inhibited pBAD comparably to 30 mg/kg up to 12 hours after dosing, whereas administration of 3 mg/kg showed 40% to 60% inhibition up to 9 hours after dosing. Plasma drug concentrations correlated with increasing pBAD inhibition, establishing a clear PK-PD correlation (Figure 4D). The total plasma drug concentration required to reach 50% inhibition of pBAD was 241 ng/mL (0.63 μM) (Figure 4D).
AZD1208 was also tested in the KG-1a xenograft. Once daily treatment with 30 mg/kg resulted in 71% tumor growth inhibition at the end of the dosing period (day 39; Figure 4E). Treatment with the AML standard-of-care chemotherapeutic cytarabine (30 mg/kg twice per week on consecutive days) resulted in 79% inhibition of tumor growth. Combination of AZD1208 with cytarabine produced 96% inhibition of tumor growth and indication of a decreased rate of tumor regrowth following cessation of treatment (Figure 4E).
AZD1208 activity in primary AML samples
Bone marrow aspirates obtained from newly diagnosed patients were used to evaluate the effects of AZD1208 on colony formation and Pim pathway biomarkers in primary AML cells. Mononuclear cells isolated from bone marrow aspirates were cultured in methylcellulose supplemented with cytokines and growth factors in the presence of increasing concentrations of AZD1208, cytarabine, or dimethylsulfoxide control for 14 days. Dose-dependent inhibition of colony growth was seen, as shown for examples in Figure 5B. Marked inhibition of colony formation (>60%) by 3 μM AZD1208 was seen in the majority of samples, including both FLT3 wild-type and FLT3-ITD mutant samples and was comparable to cytarabine in most samples (Figure 5A). Of the two samples that showed no growth inhibitory response to AZD1208, sample 020 was from a patient with acute promyelocytic leukemia, and sample 036 was uniquely resistant to colony growth inhibition by both AZD1208 and cytarabine.
Figure 5.
Inhibition of primary AML colony growth. (A) Summary of percent inhibition of colony growth of primary marrow mononuclear cells from AML patients treated with AZD1208 or cytarabine. Data shown are from cells continuously treated for 14 days with 3 μM AZD1208 or 30 nM cytarabine. AraC, cytarabine; nd, not determined. (B) Result of colony growth inhibition in Flt3 wild-type (WT or wt) and Flt3-ITD patient samples treated with vehicle control or 0.1, 0.3, 1, or 3 μM AZD1208 in a 14-day methylcellulose assay.
Biomarker responses were also assessed in mononuclear cells treated for 3 hours with up to 3 μM AZD1208. The extent of inhibition of phosphorylation of BAD, 4EBP1, and p70S6K was evaluated by immunoblot analysis (supplemental Figure 4). Inhibition of phosphorylation of 4EBP1 and p70S6K was much more prevalent than that for BAD and, as was the case in the cell lines tested, showed better correlation with growth inhibition by AZD1208. Consistent with the absence of Flt3 inhibition by AZD1208, suppression of pSTAT5, as seen for Pim inhibitors with Flt3 activity,33-35 was not observed in the Flt3-ITD patient samples (supplemental Figure 4 and data not shown).
Discussion
AZD1208 is an orally available Pim kinase inhibitor with an excellent selectivity profile, low nanomolar activity against all three Pim isoforms at the enzyme level, and potent activity in cells.22 AZD1208 inhibited growth in AML cell lines and xenograft tumor models, both as monotherapy and in combination with the standard-of-care cytarabine. AZD1208 also showed activity in ex vivo colony assays of progenitor cells from AML patients, in which inhibition of colony growth was seen in the majority of samples tested. This included three samples with the Flt3-ITD mutation, demonstrating activity of AZD1208 in Flt3-ITD leukemic cells. Notably, this was in the absence of the off-target inhibition of Flt3 activity characteristic of other Pim inhibitors reported to have activity in this setting.33-35 These results indicate that Pim inhibition may be beneficial for the treatment of both Flt3 wild-type and Flt3-ITD AML.
Treatment with AZD1208 was shown to suppress Ser 112 phosphorylation of proapoptotic BAD, a well-documented mediator of Pim kinase prosurvival activity. In MOLM-16 cells, significant but incomplete inhibition of pBAD was seen and correlated with induction of apoptosis. However, in AZD1208-sensitive KG-1a cells, suppression of pBAD did not correspond with increased apoptosis, and suppression of pBAD was also observed in cells insensitive to growth inhibition by AZD1208. This variability likely reflects the molecular heterogeneity of AML, the multiplicity of pathways that can contribute to BAD phosphorylation,36 and differing dependencies of the cell lines tested on BAD sequestration for cell survival.
Pim kinases have also been shown to exert more global cell growth effects by regulation of protein translation through modulation of 4EBP1 and p70S6K. Reduced phosphorylation of 4EBP1 and particularly p70S6K and its substrate S6 were the most consistent downstream effects of Pim inhibition observed in this study and were most closely correlated with growth inhibition in the cell lines and patient samples studied. Suppression of p4EBP1 was shown to correlate with inhibition of formation of the mRNA translation initiation complex and suppression of polysome assembly, consistent with inhibition of translation. In addition, AZD1208 treatment was also associated with decreased cell size, consistent with suppression of p-p70S6K and inhibition of translation.
Although oncogenic protein synthesis through 4EBP1 was reported to be mTORC1 independent and directly controlled by Pim-2 in AML, partly on the basis of Pim-2 small interfering RNA studies,19 an mTORC1-dependent mechanism of Pim activity may be at least in part responsible for the inhibition of both p4EBP1 and p-p70S6K. One potential mechanism is through PRAS40, a known Pim substrate, but in our studies, variable suppression of PRAS40 phosphorylation by AZD1208 was seen in both sensitive and insensitive cell lines. Alternatively, Pims could regulate mTORC1 activity in AML through upstream regulation of the mTORC1 negative regulator TSC2 (Figure 3A), as recently demonstrated in multiple myeloma models.37
Ongoing preclinical and clinical analyses will be required to better understand the mechanism of action and context dependency of Pim kinase inhibition. The pan-Pim potency and selectivity of AZD1208 render it a particularly useful pharmacologic tool for such studies. In addition, the activity of AZD1208 in multiple preclinical models of AML indicates its potential for therapeutic benefit in AML, and the ex vivo data demonstrate activity of AZD1208 in both Flt3 wild-type and Flt3-ITD AML primary samples. Beneficial combination with cytarabine in the tumor xenograft model supports exploration of combination of Pim inhibition with standard-of-care agents in AML. AZD1208 is currently in phase 1 clinical trials.
Supplementary Material
Acknowledgments
We thank Sylvie Guichard for helpful discussions, Dharmaraj Chinnappan and Kelly Jacques for technical assistance, Zhongming Yang for statistical analyses, and Shaun Grosskurth for help with cell line annotation.
This work was supported in part by National Institutes of Health, National Cancer Institute grant CA77816 and a Merit Review grant from the Department of Veterans Affairs (L.C.P.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: E.K.K., K.M., S.P., Y. Chen, Y. Cao, M.R.G., S.W., K.S.D., A.W., S.K., and M.S. performed research and interpreted data; R.M.S. and D.J.D. provided patient samples and annotation; M.L.L., P.D.L., X.Z., and L.A.D. designed and synthesized AZ1208; E.K.K., P.D.L., H.C., F.D.G., S.P., L.C.P., and D.H. designed and interpreted studies; and E.K.K. and D.H. wrote the paper.
Conflict-of-interest disclosure: E.K.K., K.M., K.S.D., S.P., Y. Cao, M.R.G., S.W., Y. Chen, A.W., M.S., F.D.G., M.L.L., X.Z., L.A.D., H.C., P.D.L., and D.H. are current or former employees of AstraZeneca. The remaining authors declare no competing financial interests.
Correspondence: Dennis Huszar, 35 Gatehouse Dr, Waltham, MA 02451; e-mail: dennis.huszar@astrazeneca.com.
References
- 1.Cuypers HT, Selten G, Quint W, et al. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell. 1984;37(1):141–150. doi: 10.1016/0092-8674(84)90309-x. [DOI] [PubMed] [Google Scholar]
- 2.Breuer ML, Cuypers HT, Berns A. Evidence for the involvement of pim-2, a new common proviral insertion site, in progression of lymphomas. EMBO J. 1989;8(3):743–748. doi: 10.1002/j.1460-2075.1989.tb03434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amson R, Sigaux F, Przedborski S, Flandrin G, Givol D, Telerman A. The human protooncogene product p33pim is expressed during fetal hematopoiesis and in diverse leukemias. Proc Natl Acad Sci USA. 1989;86(22):8857–8861. doi: 10.1073/pnas.86.22.8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen AM, Grinblat B, Bessler H, et al. Increased expression of the hPim-2 gene in human chronic lymphocytic leukemia and non-Hodgkin lymphoma. Leuk Lymphoma. 2004;45(5):951–955. doi: 10.1080/10428190310001641251. [DOI] [PubMed] [Google Scholar]
- 5.Mizuki M, Schwable J, Steur C, et al. Suppression of myeloid transcription factors and induction of STAT response genes by AML-specific Flt3 mutations. Blood. 2003;101(8):3164–3173. doi: 10.1182/blood-2002-06-1677. [DOI] [PubMed] [Google Scholar]
- 6.Asano J, Nakano A, Oda A, et al. The serine/threonine kinase Pim-2 is a novel anti-apoptotic mediator in myeloma cells. Leukemia. 2011;25(7):1182–1188. doi: 10.1038/leu.2011.60. [DOI] [PubMed] [Google Scholar]
- 7.Kim KT, Baird K, Davis S, et al. Constitutive Fms-like tyrosine kinase 3 activation results in specific changes in gene expression in myeloid leukaemic cells. Br J Haematol. 2007;138(5):603–615. doi: 10.1111/j.1365-2141.2007.06696.x. [DOI] [PubMed] [Google Scholar]
- 8.Kim KT, Baird K, Ahn JY, et al. Pim-1 is up-regulated by constitutively activated FLT3 and plays a role in FLT3-mediated cell survival. Blood. 2005;105(4):1759–1767. doi: 10.1182/blood-2004-05-2006. [DOI] [PubMed] [Google Scholar]
- 9.Bachmann M, Möröy T. The serine/threonine kinase Pim-1. Int J Biochem Cell Biol. 2005;37(4):726–730. doi: 10.1016/j.biocel.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Zhu N, Ramirez LM, Lee RL, Magnuson NS, Bishop GA, Gold MR. CD40 signaling in B cells regulates the expression of the Pim-1 kinase via the NF-kappa B pathway. J Immunol. 2002;168(2):744–754. doi: 10.4049/jimmunol.168.2.744. [DOI] [PubMed] [Google Scholar]
- 11.Hu YL, Passegué E, Fong S, Largman C, Lawrence HJ. Evidence that the Pim1 kinase gene is a direct target of HOXA9. Blood. 2007;109(11):4732–4738. doi: 10.1182/blood-2006-08-043356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox CJ, Hammerman PS, Cinalli RM, Master SR, Chodosh LA, Thompson CB. The serine/threonine kinase Pim-2 is a transcriptionally regulated apoptotic inhibitor. Genes Dev. 2003;17(15):1841–1854. doi: 10.1101/gad.1105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qian KC, Wang L, Hickey ER, et al. Structural basis of constitutive activity and a unique nucleotide binding mode of human Pim-1 kinase. J Biol Chem. 2005;280(7):6130–6137. doi: 10.1074/jbc.M409123200. [DOI] [PubMed] [Google Scholar]
- 14.Nawijn MC, Alendar A, Berns A. For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat Rev Cancer. 2011;11(1):23–34. doi: 10.1038/nrc2986. [DOI] [PubMed] [Google Scholar]
- 15.Aho TL, Sandholm J, Peltola KJ, Mankonen HP, Lilly M, Koskinen PJ. Pim-1 kinase promotes inactivation of the pro-apoptotic Bad protein by phosphorylating it on the Ser112 gatekeeper site. FEBS Lett. 2004;571(1-3):43–49. doi: 10.1016/j.febslet.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 16.Yan B, Zemskova M, Holder S, et al. The PIM-2 kinase phosphorylates BAD on serine 112 and reverses BAD-induced cell death. J Biol Chem. 2003;278(46):45358–45367. doi: 10.1074/jbc.M307933200. [DOI] [PubMed] [Google Scholar]
- 17.Macdonald A, Campbell DG, Toth R, McLauchlan H, Hastie CJ, Arthur JS. Pim kinases phosphorylate multiple sites on Bad and promote 14-3-3 binding and dissociation from Bcl-XL. BMC Cell Biol. 2006;7:1. doi: 10.1186/1471-2121-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang F, Beharry ZM, Harris TE, et al. PIM1 protein kinase regulates PRAS40 phosphorylation and mTOR activity in FDCP1 cells. Cancer Biol Ther. 2009;8(9):846–853. doi: 10.4161/cbt.8.9.8210. [DOI] [PubMed] [Google Scholar]
- 19.Tamburini J, Green AS, Bardet V, et al. Protein synthesis is resistant to rapamycin and constitutes a promising therapeutic target in acute myeloid leukemia. Blood. 2009;114(8):1618–1627. doi: 10.1182/blood-2008-10-184515. [DOI] [PubMed] [Google Scholar]
- 20.Schatz JH, Oricchio E, Wolfe AL, et al. Targeting cap-dependent translation blocks converging survival signals by AKT and PIM kinases in lymphoma. J Exp Med. 2011;208(9):1799–1807. doi: 10.1084/jem.20110846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikkers H, Nawijn M, Allen J, et al. Mice deficient for all PIM kinases display reduced body size and impaired responses to hematopoietic growth factors. Mol Cell Biol. 2004;24(13):6104–6115. doi: 10.1128/MCB.24.13.6104-6115.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dakin LA, Block MH, Chen H, et al. Discovery of novel benzylidene-1,3-thiazolidine-2,4-diones as potent and selective inhibitors of the PIM-1, PIM-2, and PIM-3 protein kinases. Bioorg Med Chem Lett. 2012;22(14):4599–4604. doi: 10.1016/j.bmcl.2012.05.098. [DOI] [PubMed] [Google Scholar]
- 23.Kaur S, Sassano A, Dolniak B, et al. Role of the Akt pathway in mRNA translation of interferon-stimulated genes. Proc Natl Acad Sci USA. 2008;105(12):4808–4813. doi: 10.1073/pnas.0710907105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carayol N, Vakana E, Sassano A, et al. Critical roles for mTORC2- and rapamycin-insensitive mTORC1-complexes in growth and survival of BCR-ABL-expressing leukemic cells. Proc Natl Acad Sci USA. 2010;107(28):12469–12474. doi: 10.1073/pnas.1005114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altman JK, Sassano A, Kaur S, et al. Dual mTORC2/mTORC1 targeting results in potent suppressive effects on acute myeloid leukemia (AML) progenitors. Clin Cancer Res. 2011;17(13):4378–4388. doi: 10.1158/1078-0432.CCR-10-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morishita D, Katayama R, Sekimizu K, Tsuruo T, Fujita N. Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res. 2008;68(13):5076–5085. doi: 10.1158/0008-5472.CAN-08-0634. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Wang Z, Li X, Magnuson NS. Pim kinase-dependent inhibition of c-Myc degradation. Oncogene. 2008;27(35):4809–4819. doi: 10.1038/onc.2008.123. [DOI] [PubMed] [Google Scholar]
- 28.Gingras AC, Raught B, Gygi SP, et al. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15(21):2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruvinsky I, Sharon N, Lerer T, et al. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19(18):2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16(12):1472–1487. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohanna M, Sobering AK, Lapointe T, et al. Atrophy of S6K1(-/-) skeletal muscle cells reveals distinct mTOR effectors for cell cycle and size control. Nat Cell Biol. 2005;7(3):286–294. doi: 10.1038/ncb1231. [DOI] [PubMed] [Google Scholar]
- 32.Dowling RJ, Topisirovic I, Alain T, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328(5982):1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fathi AT, Arowojolu O, Swinnen I, et al. A potential therapeutic target for FLT3-ITD AML: PIM1 kinase. Leuk Res. 2012;36(2):224–231. doi: 10.1016/j.leukres.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen LS, Redkar S, Taverna P, Cortes JE, Gandhi V. Mechanisms of cytotoxicity to Pim kinase inhibitor, SGI-1776, in acute myeloid leukemia. Blood. 2011;118(3):693–702. doi: 10.1182/blood-2010-12-323022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hospital MA, Green AS, Lacombe C, Mayeux P, Bouscary D, Tamburini J. The FLT3 and Pim kinases inhibitor SGI-1776 preferentially target FLT3-ITD AML cells. Blood. 2012;119(7):1791–1792. doi: 10.1182/blood-2011-11-393066. [DOI] [PubMed] [Google Scholar]
- 36.Danial NN. BAD: undertaker by night, candyman by day. Oncogene. 2008;27(Suppl 1):S53–S70. doi: 10.1038/onc.2009.44. [DOI] [PubMed] [Google Scholar]
- 37.Lu J, Zavorotinskaya T, Dai Y, et al. Pim2 is required for maintaining multiple myeloma cell growth through modulating TSC2 phosphorylation. Blood. 2013;122(9):1610-1620. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.