Abstract
An acid-degradable polymer-caged lipoplex (PCL) platform consisting of a cationic lipoplex core and a biocompatible, pH-responsive polymer shell, has been developed for the effective delivery of small interfering RNA (siRNA) through a combination of facile loading, rapid acid-triggered release, enhanced cellular internalization, and effective endosomal escape. In vitro testing of this degradable PCL delivery platform reveals ~45 and ~2.5 fold enhancement of EGFP knockdown in cancer cells in comparison to either free siRNA or siRNA-loaded nonacid- degradable lipoplex formulations, respectively. Together with the broad applicability of the PCL platform to many lipid formulations, the facile cellular release of siRNA through acid degradation of the PCL nanocarrier offer major advantages over other pH-responsive cationic lipid nanocarriers for effective release and endosomal escape of siRNA.
Since its first application in mammalian cell gene knockdown,1 small interfering RNA (siRNA) has garnered considerable interest as a versatile biological probe and as a highly potent therapeutic agent.2–3 The administration of siRNAs can induce enzyme-catalyzed degradation of their complementary mRNAs in diseased cells, thus disrupting the propagation of various diseases such as cancers, diabetes, and virus infections.2,4 However, siRNAs are not ideal drugs from the conventional pharmacokinetic standpoint: they are highly susceptible to renal clearance4 and nuclease- mediated degradation in vivo.4 In addition, they also have poor cellular membrane penetration5 due to their large molecular sizes and high negative charges. Hence, for siRNAs to be successful as a therapy, they must be coupled to an effective delivery system that overcomes these limitations.
One of the most popular siRNA delivery platforms is based on cationic lipid formulations (lipoplexes)4 that allow for direct siRNA encapsulation without a need for modification of the fragile siRNA payload and/or conjugation to the delivery vehicle and their facile cellular uptake. However, their positive surface charges often cause unwanted toxicity, poor serum stability, and rapid uptake by reticuloendothelial system in vivo,2,4,6–7 limiting their therapeutic efficacy.
Coating cationic lipid-derived carriers with hydrophilic polymers such as poly(ethylene glycol) (PEG) can reduce cytotoxicity caused by the stray binding of cationic lipids to cellular membrane 8 and prevent their unstable aggregation with serum proteins. 2 Unfortunately, the PEG coating layer also hinders the interactions between the carrier and the endosomal membranes that is necessary for the former’s escape from the endosome.9 As such, effective siRNA delivery into the cytosol by these platforms often requires a shedding of the PEG coating layer after cellular internalization.4,10 Remediating strategies based on electrostatic interaction10 and acid-cleavable linkers11–12 have been reported for this task, allowing for enhanced siRNA transfection efficacy. However, these often require specially designed pH-responsive cationic lipids and/or their covalent modification for effective release and endosomal escape of the encapsulated siRNA.
Herein, we report a lipoplex-based, core-shell siRNA delivery platform (Figure 1a) that can be triggered to degrade in acidic cellular endosomes, leading to selective siRNA release into the cytosol. Unmodified siRNA can be directly loaded into a lipoplex core composed of either a pH-responsive cationic lipid (1,2-dioleoyl-3-dimethylammonium-propane (DODAP), whose tertiary ammonium group has a 6.7 pKa13), or a non-pH-responsive cationic lipid (1,2-dioleoyl-3-trimethylammonium-propane (DOTAP), with a quaternary amino group), without the need for cumbersome covalent conjugation of siRNAs to the nanocarrier as necessary in other platforms.14–15 A biocompatible polymer shell comprising cholesterol-terminated poly(acrylic acid) (Chol-PAA) and acid-cleavable linkers was then constructed around the resulting lipoplex template to create a siRNA-loaded polymer-caged lipoplex (PCL). In acidic environments such as cellular endosomes, 16 this polymer network can shrink and induce the degradation of the PCL nanocarrier to accelerate siRNA release. This combined shrinking-degradation process is applied herein for the first time to acid-trigger siRNA-release from cationic lipoplexes (vide infra), a strategy that significantly differs from previously reported acid-degradable polymer-based nanoparticles17–18 in its integration of a polymer shell around lipid-based nanoscale containers. The Chol-PAA anionic polymer component can additionally facilitate endosomal escape of the released siRNA into the cytosol, presumably by destabilizing the endosomal membrane.19–20 That both A-cross-linked DODAP and DOTAP formulations show similar in vitro release properties (cf. Figure 1d and Figure S2a in the Supporting Information (SI)), suggests that the pH-responsive lipid DODAP does not play a major role in facilitating the acid-triggered release of encapsulated nucleic acids in our acid-degradable PCLs. As such our [Chol-PAA + acid-cleavable linkers] core-shell formulation offers a major fabrication advantage over other pH-responsive cationic lipid nanocarriers:11–12,21 it sidesteps the need to incorporate specially designed pH-responsive cationic lipids necessary for the release and endosomal escape of siRNA from these other platforms (vide infra).
Figure 1.
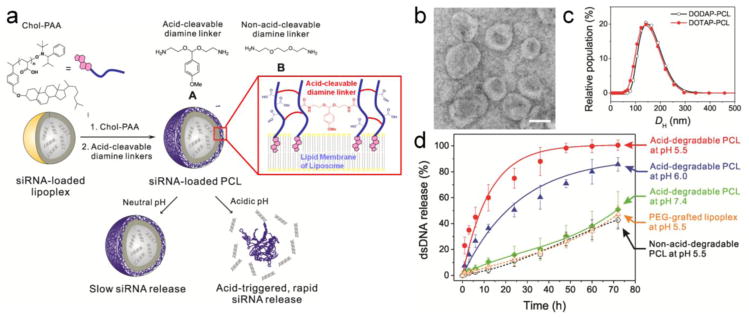
(a) A schematic presentation of the preparation of an acid-degradable siRNA-loaded PCL nanocarrier and its acid-triggered release of siRNAs. (b) Transmission electron microscopy (TEM) image of A-cross-linked DODAP-PCL (the scale bar is 100 nm) and (c) Plots of the hydrodynamic diameters (DH) of A-cross-linked DODAP-PCL (black open circle) and A-cross-linked DOTAP-PCL (red filled circle). (d) Time-dependent nucleic-acid-release profiles of the acid-degradable A-cross-linked DODAP PCL at different pHs. Also included for comparison are the time-dependent nucleic-acid-release profiles of the PEG-grafted DODAP lipoplex and the B-cross-linked DODAP-PCL, a non-acid-degradable analog. All experiments were carried out at 37 °C.
To evaluate the effectiveness of our concept in siRNA transfection, we employed an eGFP-siRNA (see the sequence in the SI), one of the siRNAs designed to knockdown the mRNAs that express enhanced green fluorescent protein (EGFP).10 This unmodified siRNA was simply loaded into the cationic lipoplex by hydrating the appropriate lipid film with a bufferred solution of eGFP-siRNA.10 The final loading of eGFP-siRNA in the cationic lipoplex is ~0.8 wt% siRNA/lipid (46% encapsulation efficiency), which is up to 3 fold higher than that shown for comparable formulations. 10 Chol-PAA (Mn = 5.4 kDa and polydispersity index = 1.10) was then incorporated into the siRNA-loaded lipoplex (hydrodynamic diameter (DH) = 140 ± 40 nm, polydispersity index (PDI) = 0.1 ± 0.05, zeta potential (ZP) = 7 ± 4 mV) at pH 7.4–8.0 using a “drop-in” method (Figure 1a, first step).22 As expected, the resulting polymer-grafted lipoplex (PGL) has a slightly increased diameter (DH = 160 ± 40 nm, PDI = 0.1 ± 0.05) and a more negative ZP (−32 ± 5 mV) as a result of Chol-PAA incorporation.23
The grafted polymer shell of the siRNA-loaded PGL was then cross-linked with the synthesized acid-cleavable diamine linker A (see section S2 in the SI)24 to ~50% of the total number of the Chol-PAA’s carboxyl groups. The unchanged diameter of the resulting PCL (DH = 155 ± 40 nm; Figures 1b and 1c) and its homogeneous distribution (PDI = 0.1 ± 0.05) indicate that these particles remains constant in size and do not aggregate in buffered saline solutions. The cross-linking of the PAA polymers increases the ZP of the PCL to a less negative value (−15 ± 5 mV), consistent with the ~50% conversion of the total carboxylic acid groups to amides.
Because the acid-cleavable diamine linker A can be hydrolyzed readily under mildly acidic conditions (Figure S1 in the SI), we expect A-cross-linked PCLs to show enhanced nucleic acid-release profiles at acidic pHs. To quantify this enhanced release, we employed the more stable, and less expensive, double-stranded DNA (dsDNA) analogue (see the sequence in the SI) of eGFP-siRNA as the payload. As shown in Figure 1d, our acid-degradable PCLs exhibit significantly enhanced nucleic acid-release rate at acidic pHs below pH 7.4. At pH 5.5 and 37 °C, ~60% of the total amount of encapsulated dsDNA is released from the A-cross-linked PCLs within 12 h and near-quantitative release is observed within 48 h. In contrast, only small amounts of dsDNA are released at similar time points (~10% at 12 h and ~30% at 48 h) at pH 7.4 and 37 °C. Particle concentration data (Figure S3 in the SI) clearly show a faster degradation of PCLs at pH 5.5 than at pH 7.4, suggesting that the accelerated release of dsDNA in acidic pH must have resulted from the degradation of these nanocarriers. This excellent acid-triggered release could also be applicable to the non-pH-responsive cationic lipid (DOTAP)-based PCL (Figure S2a in the SI).
In our PCL formulation, the shell-localized network created by amide cross-linking the inserted PAA polymer chains is essentially a poly[(AAc)-co-(AAm)] copolymer material that is well known to “shrink” at acidic pH.25 This chemomechanical response has been utilized to acid-trigger the release of encapsulated chemotherapeutics in macroscopic gels17,26 and anionic-lipid-derived polymer-caged nanobins.22,27 It is also observed for our A-cross-linked DODAP and DOTAP formulations, in contrast to the DODAP and DOTAP controls without PAA polymer and PEG-grafted DODAP lipoplex (Figure S4 in the SI). Although our study employs cationic lipoplexes, which may interact with the remaining negatively charged carboxylate residues of the Chol-PAA additives and reduce their “native shrinking” effect, the combination of the PAA polymer and acid-cleavable linker A ensures that the membrane of the PCL can still be quickly destabilized and ruptured, eventually leading to degradation of the PCL nanoparticle. Such a combined “shrinking”-degradation process could allow researchers to choose from a wide range of available cationic lipids for efficient siRNA delivery. This is more advantageous than previously reported approaches11–12,21,28 where one often must incorporate specially designed pH-responsive cationic lipids into lipoplexes before effective release and endosomal escape of siRNA can be achieved.
As controls, we have also examined the release of encapsulated dsDNAs from PEG-grafted DODAP lipoplex, a traditional formulation with pH-responsive cationic lipids, and B-cross-linked DODAP PCLs, the non-acid-degradable analogue of our formulation, 29 both of which do not show as significant a release of dsDNA (Figure 1d) as A-cross-linked PCLs under the same conditions. That the first control does not release nucleic acids at pH 5.5 is in line with recently reported observations that lipoplexes composed of pH-responsive cationic lipids do not significantly release encapsulated nucleic acids under acidic conditions unless they can make contact with anionic lipid membranes.4 That the second control also does not show significant release of nucleic acids under acidic conditions suggests that destabilization of the lipoplex core by the shrinking of the Chol-PAA polymer cage alone22 is not enough to release encapsulated nucleic acids from the PCL. These control experiments, together with the aforementioned fast degradation profile of our acid-degradable A-cross-linked PCL, clearly support our hypothesis that a combination of acid-triggered “shrinking” and polymer shell degradation is critical for nucleic acid release from cationic-lipid-derived A-cross-linked PCL nanocarriers.
To evaluate the biocompatibility of PCLs, human breast cancer (MDA-MB-231) and human cervical cancer (HeLa) cells were treated for 24 h at 37 °C with eGFP-siRNA-loaded PCLs. The cells subjected to both lipid formulations of eGFP-siRNA-loaded PCLs (DODAP-PCL and DOTAP-PCL) exhibit ~100% viability over all tested siRNA concentrations (≤ 400 nM; ≤ 45.2 μg/mL of cationic lipids). This is in stark contrast to the significant cytotoxicity shown by Lipofectamine 2000, one of the most popularly used cationic lipid-based formulations for gene and siRNA transfection (Figure 2). In our hands, the cells treated with the eGFP-siRNA-loaded Lipofectamine 2000 formulation shows < 5% viability at the upper range of tested siRNA concentration (400 nM; 20 μg/mL of lipids). Thus, our acid-degradable PCL platform would be safer for clinical applications in comparison to currently used toxic cationic transfection agents.8
Figure 2.
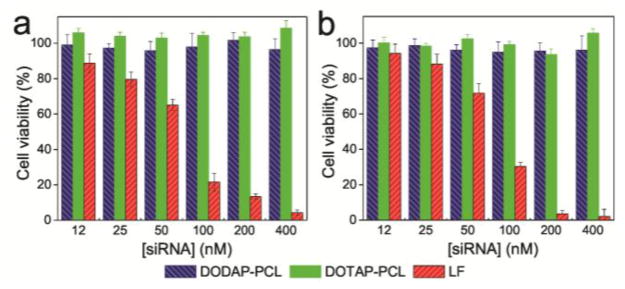
Dose-responsive cell viability of (a) MDA-MB-231 and (b) HeLa cells exposed to the eGFP-siRNA-loaded, A-cross-linked, acid-degradable DODAP-(left blue bar); DOTAP-PCLs (middle green bar); and eGFP-siRNA-loaded Lipofectamine 2000 (LF, right red bar) at 37 °C for 24 h. After rinsing off the excess siRNA-containing formulations, the cells were further incubated at 37 °C for 48 h before cell viability was measured using MTS assay. Error bars indicate the standard deviation of the mean with n = 3.
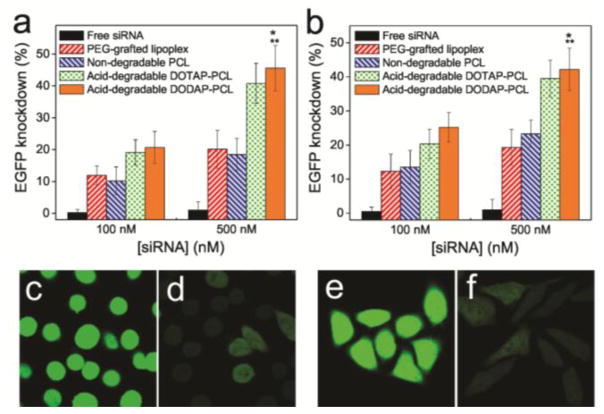
To evaluate EGFP knockdown efficiencies, MDA-MB-231 cells that have been engineered to stably express EGFPs were exposed to free eGFP-siRNA and four different nanocarriers (PEG-grafted lipoplex, non-acid-degradable B-cross-linked PCL, and acid-degradable DOTAP- and DODAP-PCLs that have been loaded with eGFP-siRNA ([siRNA] = 100 and 500 nM)). The acid-degradable PCLs exhibit ~45% EGFP knockdown at 500 nM of siRNA, ~45 fold and ~2.5 fold enhancement over the free eGFP-siRNA (~1% EGFP knockdown at 500 nM of eGFP-siRNA) and the non-acid-degradable nanocarriers (PEG-grafted lipoplexes and non-acid-degradable B-cross-linked PCLs; ~20% EGFP knockdown at 500 nM of siRNA), respectively (Figure 3a). Similar enhancement in EGFP knockdown by the acid-degradable PCLs was also observed for EGFP-expressing HeLa cells (Figure 3b). Indeed, confocal laser-scanning microscopy (CLSM) imaging (Figures 3c–3f) shows a visually clear decrease in the EGFP fluorescence signal intensity in both types of cells after treatment with the acid-degradable PCLs. These gene knockdown efficiencies are similar to those previously reported for PEG-coated lipoplexes10 and carbon nanotubes.15
Figure 3.
EGFP knockdown efficiency in (a) MDA-MB-231 and (b) HeLa cells by free eGFP-siRNA (black), siRNA-loaded PEG-grafted lipoplex (red), siRNA-loaded non-acid-degradable PCLs (blue), and siRNA-loaded acid-degradable DOTAP- (green) and DODAP-PCLs (orange) at 100 and 500 nM of eGFP-siRNA. The data were assessed by flow cytometry. CLSM images for MDA-MB-231 and HeLa cells were acquired before (c and e) and after (d and f) treatment with acid-degradable DODAP-PCLs ([eGFP-siRNA] = 500 nM), respectively. Error bars indicate the standard deviation of the mean with n = 3. * denotes p < 0.001 compared to free siRNAs. ** denotes p < 0.01 compared to siRNA-loaded PEG-grafted lipoplex and siRNA-loaded B-cross-linked PCL.
While the gene knockdown efficiency by our acid-degradable A-cross-linked PCL increase over time (Figure S6 in the SI), its behavior in 95% serum does not changed significantly from that in 10% serum-included media. This notable behavior is completely different from that of Lipofectamine 2000, whose gene knockdown efficiency dramatically dropped when being transitioned from 10% serum-included media into 95% serum (Figure S7 in the SI) and points to a serum-independent advantage that our PCL may have over Lipofectamine and similar formulations.
An additional advantage of our PCL design is the use of the polyanionic Chol-PAA polymer, which has been known to destabilize endosomal membranes and lead to endosomal escape of biomolecules.19 As such, we hypothesized that the Chol-PAAs component of our coated PCLs could further facilitate the escape of nucleic acids from endosomes into cytosol after acid degradation, which facilitate their desorption from the shell. To verify this, MDA-MB-231 and HeLa cells were treated at 37 °C for 24 h with PCL formulations that have been loaded with Cy5-labeled dsDNA (Cy5-dsDNA) analogue of eGFP-siRNA as the payload. These cells were then treated with Hoechst and LysoTraker Green dyes to stain nuclei and acidic endosomes/lysosomes, respectively, before analysis by CLSM. CLSM images of the cells that have been exposed to the dsDNA-loaded PCLs (Figure 4) show that Cy5-dsDNA (red-colored regions) have spread throughout the cells with minimal overlap with the acidic endosomes/lysosomes (green-colored regions), indicating successful endosomal escape of Cy5-dsDNA and its localization in the cytosol. In contrast, CLSM images of MDA-MB-231 and HeLa cells that have been exposed to a Cy5-dsDNA-loaded PEG-grafted lipoplex formulation (Figure S8 in the SI) show more localized Cy5-dsDNAs regions that significantly overlap (yellow-colored regions) with the acidic endosomes/lysosomes, indicating that there are more nucleic acids trapped inside the endosomes/lysosomes in these cells. Together, these results strongly support our hypothesis that the Chol-PAA components of the PCL could indeed facilitate nucleic acid release into the cytosol without them being trapped in the endosomal/lysosomal stage.
Figure 4.
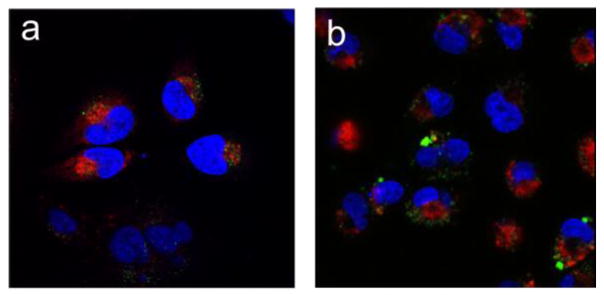
CLSM images illustrating the intracellular distribution of Cy5-dsDNA (red) and acidic endosomes/lysosomes stained with LysoTracker Green (green) in (a) HeLa and (b) MDA-MB-231 cells. The cells were treated with Cy5-dsDNA-encapsulated DODAP-PCLs (100 nM DNA) for 24 h at 37 °C. The nuclei were stained with Hoechst dye (blue)
In conclusion, we have demonstrated an acid-degradable cationic lipid-based PCL core-shell system that exhibit excellent potential as a delivery platform for siRNA transfection. This outstanding biocompatible PCL platform can be facily fabricated from easily accessible components and the desired nucleic acids via a drop-in method. It shows high efficiencies in the knockdown of EGFP in cancer cells through rapid acid-triggered release, enhanced cellular internalization, and effective endosomal escape of unmodified siRNA. As a platform, it is more advantageous than Lipofectamine 2000 by being much less cytotoxic and can maintain the same gene-knockdown efficiency over a broad range of serum-containing media.
Moreover, siRNA release by the acid-degradable PCLs, through their acid-triggered degradation, offers a major fabrication advantage over previously reported pH-responsive, PEG-coated cationic lipid-based nanocarriers11–12,21 in its broad applicability to many lipid formulations: the drop-in modification replaces the need for incorporating specially designed pH-responsive cationic lipids traditionally required for the effective release and endosomal escape of siRNA. In addition, the Chol-PAA-based polymer shell of the PCLs has carboxylic acid groups that can be used as chemical handles for further conjugation with cancer-targeting ligands,23 imaging probes,30 and chemotherapeutics, 31 all of which are not easily accessible in conventional PEG-grafted lipoplexes. Together, these multifunctional features of the acid-degradable PCL platform bring us closer to an effective siRNA delivery technology that can be utilized for clinical gene therapy. Our future work will focus on increasing the amount of loaded siRNAs in a single delivery particle,32 to achieve better gene knockdown efficiency.
Supplementary Material
Acknowledgments
This work is financially supported by the NIH (NCI CCNE Grant C54CA151880, CCNP Grant U01CA151461, and Core Grant P30CA060553 to the Lurie Cancer Center of Northwestern University (NU)). Instruments in the NU IMSERC, Keck Biophysics, and NUANCE facilities were purchased with grants from NSF-NSEC, NIH-CCNE, NSF-MRSEC, the Keck Foundation, the State of Illinois, and NU. We thank Prof. T. V. O’Halloran for access to the cell-culture facility, Mr. E. P. Swindell and Dr. K. W. MacRenaris for advices concerning the endosomal escape studies, and the reviewers of the initial version of this manuscript for suggestions that greatly strengthened the final work.
Footnotes
The authors declare no competing financial interest.
Descriptions of experimental procedures, the synthetic procedure for the acid-cleavable diamine linker A, nucleic acid-release profiles, gene-knockdown data, CLSM images, and the full citation for reference 28. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 2.Whitehead KA, Langer R, Anderson DG. Nat Rev Drug Discovery. 2009;8:516–516. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oh YK, Park TG. Adv Drug Delivery Rev. 2009;61:850–862. doi: 10.1016/j.addr.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 4.Huang L, Liu Y. Annu Rev Biomed Eng. 2011;13:507–530. doi: 10.1146/annurev-bioeng-071910-124709. [DOI] [PubMed] [Google Scholar]
- 5.Salcher EE, Wagner E. Top Curr Chem. 2010;296:227–249. doi: 10.1007/128_2010_69. [DOI] [PubMed] [Google Scholar]
- 6.Allen TM, Cullis PR. Adv Drug Delivery Rev. 2013;65:36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 7.Kulkarni A, DeFrees K, Hyun SH, Thompson DH. J Am Chem Soc. 2012;134:7596–7599. doi: 10.1021/ja300690j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lv H, Zhang S, Wang B, Cui S, Yan J. J Controlled Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 9.Although PEG coating is believed to also slow down endosomal uptake (see refs. 3 and 4), recent evidences have suggested that this hindrance does not play an important role in therapies. Rather, the endosomal escape is more critical for siRNA delivery. See: ref. 13 and Kostarelos K, Miller AD. Chem Soc Rev. 2005;34:970–994. doi: 10.1039/b307062j.
- 10.Auguste DT, Furman K, Wong A, Fuller J, Armes SP, Deming TJ, Langer R. J Controlled Release. 2008;130:266–274. doi: 10.1016/j.jconrel.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolli S, Wong SP, Harbottle R, Johnston B, Thanou M, Miller AD. Bioconjugate Chem. 2013;24:314–332. doi: 10.1021/bc3004099. [DOI] [PubMed] [Google Scholar]
- 12.Carmona S, Jorgensen MR, Kolli S, Crowther C, Salazar FH, Marion PL, Fujino M, Natori Y, Thanou M, Arbuthnot P, Miller AD. Mol Pharm. 2009;6:706–717. doi: 10.1021/mp800157x. [DOI] [PubMed] [Google Scholar]
- 13.Auguste DT, Armes SP, Brzezinska KR, Deming TJ, Kohn J, Prud’homme RK. Biomaterials. 2006;27:2599–2608. doi: 10.1016/j.biomaterials.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 14.Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. J Am Chem Soc. 2009;131:2072–2073. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kam NWS, Liu Z, Dai H. J Am Chem Soc. 2005;127:12492–12493. doi: 10.1021/ja053962k. [DOI] [PubMed] [Google Scholar]
- 16.Casey JR, Grinstein S, Orlowski J. Nat Rev Mol Cell Biol. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 17.Cohen JA, Beaudette TT, Cohen JL, Broaders KE, Bachelder EM, Fréchet JMJ. Adv Mater. 2010;22:3593–3597. doi: 10.1002/adma.201000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui L, Cohen JL, Chu CK, Wich PR, Kierstead PH, Fréchet JMJ. J Am Chem Soc. 2012;134:15840–15848. doi: 10.1021/ja305552u. [DOI] [PubMed] [Google Scholar]
- 19.Yessine MA, Leroux JC. Adv Drug Delivery Rev. 2004;56:999–1021. doi: 10.1016/j.addr.2003.10.039. [DOI] [PubMed] [Google Scholar]
- 20.Guo ST, Huang L. J Nanomater. 2011 [Google Scholar]
- 21.Sato Y, Hatakeyama H, Sakurai Y, Hyodo M, Akita H, Harashima H. J Controlled Release. 2012;163:267–276. doi: 10.1016/j.jconrel.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Lee SM, Chen H, Dettmer CM, O’Halloran TV, Nguyen ST. J Am Chem Soc. 2007;129:15096–15097. doi: 10.1021/ja070748i. [DOI] [PubMed] [Google Scholar]
- 23.Lee SM, Chen H, O’Halloran TV, Nguyen ST. J Am Chem Soc. 2009;131:9311–9320. doi: 10.1021/ja9017336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murthy N, Thng YX, Schuck S, Xu MC, Fréchet JMJ. J Am Chem Soc. 2002;124:12398–12399. doi: 10.1021/ja026925r. [DOI] [PubMed] [Google Scholar]
- 25.Nayak S, Lyon LA. Angew Chem, Int Ed. 2005;44:7686–7708. doi: 10.1002/anie.200501321. [DOI] [PubMed] [Google Scholar]
- 26.Hoffman AS. J Control Release. 1987;6:297–305. [Google Scholar]
- 27.Lee SM, Lee OS, O’Halloran TV, Schatz GC, Nguyen ST. ACS Nano. 2011;5:3961–3969. doi: 10.1021/nn200478m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Semple SC, et al. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 29.The use of B-cross-linked DODAP PCLs is a better control than the non-cross-linked Chol-PAA-inserted DODAP lipoplex because it mimics better the surface environment of the acid-cleavable A-cross-linked DODAP PCLs and directly addresses the question if it is necessary to have A being acid-cleavable.
- 30.Lee SM, Song Y, Hong BJ, MacRenaris KW, Mastarone DJ, O’Halloran TV, Meade TJ, Nguyen ST. Angew Chem, Int Ed. 2010;49:9960–9964. doi: 10.1002/anie.201004867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SM, O’Halloran TV, Nguyen ST. J Am Chem Soc. 2010;132:17130–17138. doi: 10.1021/ja107333g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ornelas-Megiatto C, Wich PR, Fréchet JMJ. J Am Chem Soc. 2012;134:1902–1905. doi: 10.1021/ja207366k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.