Abstract
Inclusion of entropy is important and challenging for protein-protein binding prediction. Here, we present a statistical mechanics-based approach to empirically consider the effect of orientational entropy. Specifically, we globally sample the possible binding orientations based on a simple shape-complementarity scoring function using an FFT-type docking method. Then, for each generated orientation we calculate the probability through the partition function of the ensemble of accessible states, which are assumed to be represented by the set of nearby binding modes. For each mode, the interaction energy is calculated from our ITScorePP scoring function that was developed in our laboratory based on principles of statistical mechanics. Using the above protocol, we present the results of our participation in Rounds 22–27 of the CAPRI (Critical Assessment of PRedicted Interactions) experiment for ten targets (T46-T58). Additional experimental information, such as low-resolution SAXS data, was used when available. In the prediction (or docking) experiments of the ten target complexes, we achieved correct binding modes for six targets: one with high accuracy (T47), two with medium accuracy (T48 and T57), and three with acceptable accuracy (T49, T50, and T58). In the scoring experiments of seven target complexes, we obtained correct binding modes for six targets: one with high accuracy (T47), two with medium accuracy (T49 and T50), and three with acceptable accuracy (T46, T51, and T53).
Keywords: protein-protein interaction, CAPRI experiments, scoring function, entropy, molecular docking
1 INTRODUCTION
Protein-protein interactions play an important role in many biological processes. Due to the high cost and technical difficulty in experimentally determining the structures of protein complexes, protein docking has played an increasingly important role in computationally predicting the binding mode and free energy between proteins with known three-dimensional (3D) structures. There have been a number of algorithms developed for protein-protein docking with different speed and accuracy in the past two decades.1–6 It is widely accepted that bound docking, which attempts to reconstruct the complex assuming that the native conformations of the bound binding partners are known, has not been a problem, and many of the existing scoring functions can yield a relatively high success rate in binding mode predictions for this type of docking. However, the more realistic unbound docking, in which the complex is predicted from unbound structures, has been much more challenging because of the conformational changes and structural uncertainties of individual protein structures upon binding. Due to the difficulties of explicitly considering protein flexibility, different docking algorithms and scoring methods have been developed to improve the success rate in predicting correct binding modes. Furthermore, other biological information such as mutation data, residue contacts from experiments, and evolutionary analysis has also been used to assist in the traditional docking and scoring process. All of the developed docking and scoring approaches call for an objective benchmark for evaluation and further improvement. Meeting this demand, the CAPRI (Critical Assessment of PRedicted Interaction) challenge7–10 has been providing a valuable platform for evaluating docking and scoring protocols on realistic applications since it was launched in 2001, and is playing a significant role in promoting the development of new docking and scoring algorithms, and the improvement of existing methods.
In the previous rounds from 2007–2009, we have developed and evaluated a hierarchical protocol MDockPP11 using the CAPRI targets. In MDockPP, the possible binding modes of one protein are first globally sampled relative to the other using a Fast-Fourier Transform (FFT)-based sampling method,12 in which only the shape complementarity is used as the scoring function. Then the limited number of filtered binding orientations from the previous step are further scored and refined by our atomic scoring function for protein-protein interactions ITScorePP.13 The final predicted models were selected based on manual inspection and any additional biological information that was available. Recently, we have further improved our docking and scoring protocol for protein-protein interactions by empirically considering the orientational entropic effect. Although the improvement seems to be limited by the conformational changes of the proteins upon binding, the results indicate a possible direction for improving existing docking and scoring algorithms.
2 MATERIALS AND METHODS
2.1 Docking and Scoring Protocol
We have used a similar hierarchical protocol for the CAPRI docking (or prediction) and scoring experiments in the latest rounds from 2010–2013 to that in the previous rounds from 2007–2009, except that here we considered the effect of the orientational entropy at the scoring step in the latest rounds.
For the CAPRI docking experiments, our typical protocol was as follows. The putative binding modes were first generated using an FFT-based docking algorithm like ZDOCK 2.114 or a modified 3D-DOCK15 that is based on shape complementarity. During docking calculations, the default parameters were used unless otherwise specified. Namely, the grid spacing was set to 1.2 Å, and the interval of the Euler angles was set to 15°. For each rotation, according to the FFT calculation, one relative translation of the ligand with the best shape complementarity to the receptor was kept for further refinement. Then, the filtered binding modes were scored and optimized at the atomic level. If available, biological information about the binding site was also applied at this refinement step to choose binding modes that satisfy the biological information. The ranked binding modes were then clustered. For any two binding modes with an Lrmsd (root-mean-square deviation) less than Rclu, only the ligand orientation with lower score was kept. Here, Å Rclu is calculated based on the backbone atoms of the ligand protein and was set to 8 unless otherwise specified. The top 100 binding modes after clustering were kept for manual inspection to assure that the biological information could be properly considered. Ten binding modes were selected and submitted to CAPRI.
For the CAPRI scoring experiments, the protocol is similar except that the putative binding modes, that were constructed and kindly uploaded by the CAPRI predictors, were directly downloaded from the CAPRI site. Ten binding modes were finally submitted to CAPRI.
2.2 Ranking by considering the orientational entropic effect
In the present study, we restricted to rigid docking. Based on statistical mechanics, the free energy can be calculated according to the partition function theory:16
| (1) |
Here, Z is the canonical partition function and is approximated by 16,17
| (2) |
where β = 1/kBT and is the protein-protein interaction energy in the ith observed thermodynamic macrostate corresponding to a local minimum on the protein-protein binding energy landscape. In computational experiments, a microstate in a local minimum was approximated by a generated orientation in a cluster of its neighboring modes whose interaction potential energies can be calculated using our scoring function ITScorePP. The cluster size was set to 5 Å for Lrmsd. The parameter kBT was empirically set to 10 in the present study so that the loss of the entropy would not be overestimated or underestimated compared to the interaction potential energy URL. Namely, the calculated free energy G would still be comparable to the interaction potential energy URL.
To calculate the partition function, putative protein-protein binding modes were generated by ZDOCK 2.1.14 During docking calculations, the default parameters were used except the number of output binding modes which was set to the number of maximum rotations (i.e. 3600 for the default interval of the Euler angles) in our preliminary test. Then, the generated binding modes were scored by ITScorePP and the ligand binding modes were optimized using a simplex algorithm.18 Those poses within a certain RMSD Rcut from a given binding mode serve as an approximation of the number of accessible microstates near the mode.19 Here, Rcut was set to be 5.0 Å. The RMSD was calculated based on the C α atoms to speed up the grouping calculations. Thus, under the assumption that the energies of the binding modes in each cluster are similar,20 one can obtain a partition function corresponding to the i-th mode as
| (3) |
where stands for the interaction energy of the k-th mode/microstate in the i-th cluster/group. The number of modes (“k”) in each cluster (“i”) depends on the sampling method and the scoring function in use. Then, the probability of finding the system around the i-th mode can be expressed as
| (4) |
In the present study we focused on relative ranking of the binding modes. Therefore, for mode ranking, instead of using the absolute values of Pi, we empirically chose to use lnPi and discarded the constant lnZ, which would not affect the ranking order but would help ranking by magnifying the difference in mode probability:
| (5) |
As a preliminary test, we have applied the above method to the 35 unbound inhibitor-enzyme pairs (or substrate-enzyme pairs) in the protein-protein docking benchmark 3.0.21 The inhibitor-enzyme complexes are relatively rigid during binding and often have a high degree of shape complementarity because of a main-chain-main-chain interacting mechanism.3,22 Other complexes in the protein-protein docking benchmark 3.0 are more complicated. For example, antigen-antibody binding involves both the main-chain and sidechain atoms and does not necessarily have the best-fit binding interface, a case that requires special training.3,22–24 Protein flexibility is another complication. Therefore, given the rigid-body assumption in our scoring method, the inhibitor-enzyme complexes will be more appropriate test cases to start with and the rest complexes will be left for future detailed study.
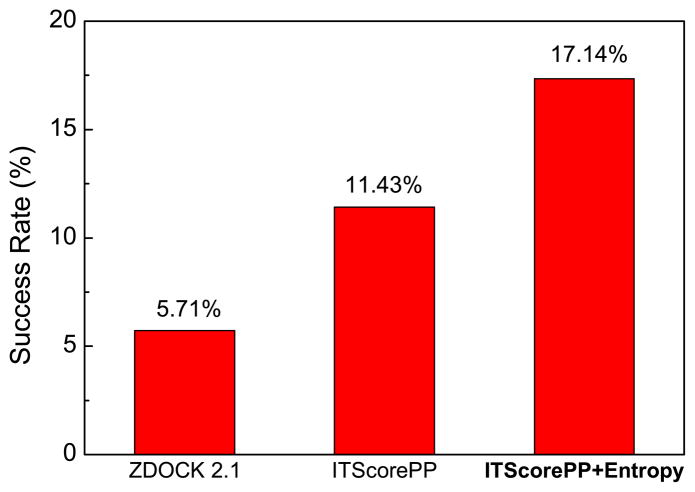
In this test study on the inhibitor-enzyme complexes, the final binding modes were clustered with an RMSD cutoff of 5.0 Å. Namely, if two binding modes have an RMSD within 5.0 Å, the one with the lower score was kept. The result is shown in Figure 1, which gives the binding mode prediction success rate when only the top binding mode is considered. Here, a successful prediction is defined as the lowest-scored binding mode having an Lrmsd less than 10 Å from the native structure. 25 For the purpose of references only, two other scoring methods, ZDOCK2.1 and ITScorePP, were also shown in the figure. ZDOCK2.114 is the simple shape complementarity-based scoring function that was used to generate putative binding modes in the present study.
Figure 1.
Success rates of three scoring functions (shape complementarity in ZDOCK 2.1, ITScorePP, and ITScore+Entropy) on the 35 enzyme/inhibitor test cases when the top binding mode was considered.
It can be seen from Figure 1 that the scoring function in ZDOCK 2.1, which is based on shape complementarity, had the lowest success rate of 5.71% when the top binding mode was considered. Compared to ITScorePP, with a success rate of 11.43%, the inclusion of entropy has substantially improved the performance of binding mode predictions to a success rate of 17.14%, supporting the efficacy of this statistical mechanics-based entropy method.
We have further examined the improvement for each case and found that the five medium and two difficult cases do not seem to contribute to the higher success rate where the definition of difficult groups is based on the degree of conformational change of the unbound structures at the interface compared to the native complex.21 In other words, the number of the cases with improvement from entropy would be the same after removing those seven medium/difficult cases, indicating that the conformational changes of individual structures may strictly limit the accuracy of our method. Further improvements may be possible if protein flexibility were properly considered.
3 RESULTS AND DISCUSSION
3.1 Overall Performance
From 2010 to 2013, we have participated in rounds 22–27 of the CAPRI experiment. We tested our method on ten targets for binding mode predictions. Table I summarizes our CAPRI results. For the docking (or prediction) experiments, we predicted at least one acceptable binding mode for six targets, including one high-accuracy prediction (Target 47), two medium-accuracy predictions (Targets 48 and 57), and three acceptable predictions (Targets 49, 50, and 58).
Table I.
Performance of our docking and scoring method in CAPRI rounds 22–28.
| Targeta | Complexb | Typec | Bio.d Info. | Predicting
|
Scoring
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| fnat (%) | Lrmsd (Å) | Irmsd (Å) | accuracye | fnat (%) | Lrmsd (Å) | Irmsd (Å) | accuracyf | ||||
| 46 | Mtq2/Trm112 | H/H | – | 0.040 | 19.646 | 10.356 | 0 | 0.406 | 7.621 | 3.395 | 1* |
| 47 | E2/IM2 | U/U | Y | 0.821 | 0.865 | 0.554 | 10*** | 0.821 | 0.904 | 0.535 | 10*** |
| 48 | T4moH/T4moC | U/U | Y | 0.375 | 3.921 | 1.653 | 8/1** | – | – | – | – |
| 49 | T4moH/T4moC | U/U | Y | 0.312 | 5.678 | 2.177 | 4* | 0.500 | 5.593 | 1.790 | 7/2** |
| 50 | HA/HB36.3C | U/H | Y | 0.673 | 9.618 | 2.239 | 1* | 0.571 | 5.650 | 1.912 | 2/1** |
| 51 | GH5-CBM6/CBM13/Fn3 | U/H/U | – | 0.000 | 17.052 | 11.562 | 0 | 0.241 | 20.501 | 3.483 | 1* |
| 53 | REP-4/REP-2 | U/H | – | 0.096 | 13.504 | 3.935 | 0 | 0.288 | 5.321 | 2.517 | 1* |
| 54 | NCS/REP16 | U/H | – | 0.040 | 18.315 | 7.831 | 0 | 0.040 | 12.231 | 5.397 | 0 |
| 57 | BT-4661/heparin | U/U | Y | 0.706 | 3.216 | 1.369 | 2/1** | – | – | – | – |
| 58 | SalG/PliG-Ec | U/U | Y | 0.275 | 3.602 | 1.807 | 3* | – | – | – | – |
T52 was cancelled and Targets 55 and 56 are for binding affinity prediction which are not listed in this table.
The first one is assigned as the receptor, and the second one as the ligand.
The symbol “B” stands for the bound experimental structure, “U” for the unbound experimental structure, and “H” for the homology-modeled structure.
“Y” represents that valid biological information was available for the binding site, “–” represents that no or little useful biological information was available.
The accuracy is categorized by three parameters following the CAPRI criteria:7,8 The percentage of the native residue-residue contacts (fnat), the ligand RMSD (Lrmsd), and the interface RMSD (Irmsd). “***” stands for high-accuracy, “**” for medium-accuracy, “*” for acceptable accuracy, and “0” for no correct prediction, respectively. For example, “8/1***” means that among 10 submitted binding modes for CAPRI, eight modes have at least acceptable accuracy, and one of these eight has high accuracy (***).
There were no scoring experiments for Targets 48, 57, and 58.
For the scoring experiments, we had a higher success rate than for the docking experiments, based on CAPRI’s criteria of acceptability. For six of the seven targets, we made at least one acceptable prediction, giving an overall success rate of 85.7%. For targets 46, 51, and 53, our scoring function gave an acceptable accuracy, even though our docking protocol failed to make a successful prediction in the ten submitted modes on these targets. Our predictions in the scoring experiments are also more correct with a medium accuracy for Targets 49 and 50, compared to the acceptable accuracy of the predictions by our docking protocol on these two targets, suggesting the relative accuracy and robustness of our scoring protocol.
3.2 Target 46 (Mtq2-Trm112)
Target 46 is a protein-protein complex between Mtq2 and Trm112 (PDB ID: 3Q87).26 There were no crystal structures available for either protein. The Mtq2 was modeled based on the crystal structure 1T4327 in the Protein Data Bank (PDB)28 using MODELLER,29 and the Trm112 was modeled based on the crystal structure 2J6A.30. No reliable binding site information was available from the literature. We did not make a correct prediction in the docking experiments for this target in which our best model gave an interface RMSD of 10.356 Å, although our scoring function gave one prediction of acceptable accuracy with fnat = 40.6%, Lrmsd = 7.621 Å and Irmsd = 3.395 Å in the scoring experiment (Figure 2).
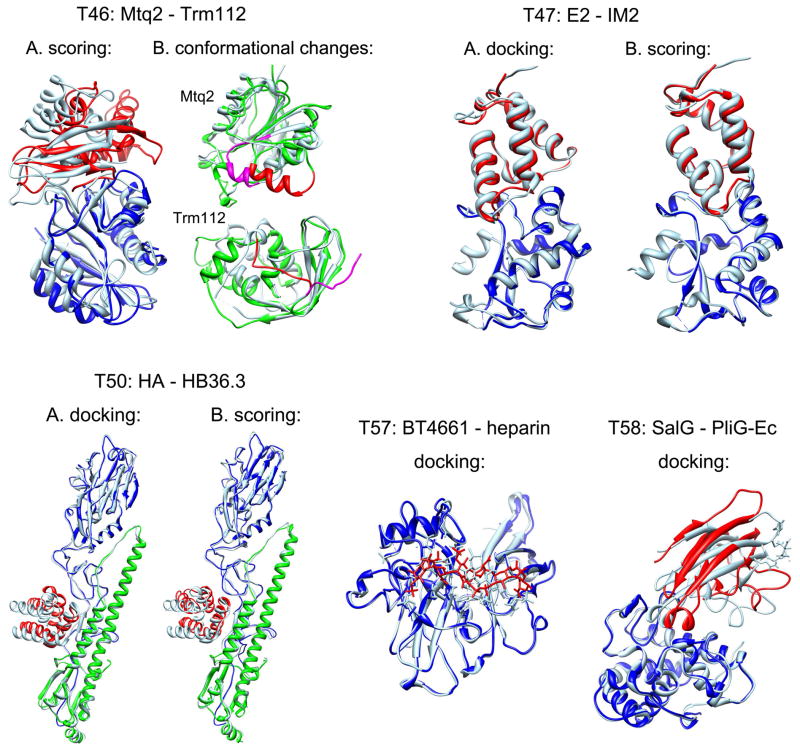
Figure 2. Target 46.
A. Comparison between the predicted complex between Mtq2 (blue) and Trm112 (red) by our scoring experiment and the experimentally determined crystal structure (light blue), where the complexes were aligned according to the receptor protein Mtq2. The predicted mode has an acceptable accuracy prediction with fnat = 40.6%, Lrmsd = 7.621 Å and Irmsd = 3.395 Å; B. Comparison between the modeled (green) and crystal (light blue) structures of Mtq2 (upper panel) and Trm112 (lower panel), in which the conformational changes are highlighted in red for the modeled structure and magenta for the crystal structure, respectively. Target 47: Comparison between the predicted complex of Colicin E2 (blue) and IM2 (red) and the corresponding crystal structure (light blue) for the docking (A) and scoring (B) experiments, where the complexes are aligned according to the receptor protein E2. Target 50: Comparison between the predicted complex of Colicin HA chains (green and blue) and HB36.3 (red) and the corresponding crystal structure (light blue) for the docking (A) and scoring (B) experiments, where the complexes are aligned according to the receptor protein HA chains. Target 57: Comparison between the predicted complex of BT4661 (blue) and heparin (red) and the corresponding crystal structure (light blue), where the complexes are aligned according to the receptor BT4661. Target 58: Comparison between the predicted complex of SalG (blue) and PliG-Ec (red) and the corresponding crystal structure (light blue), where the complexes are aligned according to the receptor SalG.
Further examination of the crystal structures and our modeled structures reveals that both Mtq2 and Trm112 undergo substantial conformational changes along the binding interface when binding, which have made it one of the most difficult targets; only two groups gave correct predictions of this target in the docking experiment. Comparison between the crystal structure and our modeled structures shows that there is a shift for the region around the helix 49–59 on Mtq2. There is also a significant conformational change on the C-terminus of Trm112 where the C-terminus blocks the binding site on the modeled Trm112 but moves away from the binding site on the crystal structure (Figure 2). These changes may account for the failure of our docking experiment prediction on this target. However, these conformational changes were adequately addressed in the binding modes constructed by Dr. Bonvin’s research group who built their homologous models based on the correct templates, which allowed our scoring function to identify one acceptable prediction in the scoring experiment (Figure 2).
3.3 Target 47 (E2-IM2)
Targets 47 is a protein-protein complex between the DNase domain of Colicin-E2 and the Colicin-E2 cognate immunity protein (IM2) (PDB ID: 3U43).31 We built the structures of the Colicin-E2 and Colicin-E2 immunity protein from its homologous complex E9-IM9 (1EMV)32 using MODELLER.29 Since there were several protein complexes in the PDB homologous to the complex of Colicin-E2 with Colicin-E2 immunity protein, the binding site of this target was clear, making it a relatively easy target. Due to our confidence in the biological information about the binding interface, we set the RMSD cutoff for clustering to 2.0 Å for this target. All of our ten predicted models had a high accuracy for both the docking experiments and scoring experiments, in which the best model has a very low Irmsd of about 0.55 Å (Figure 2). The challenging part about this target may have been the prediction of water molecule positions at the binding interface. We used a knowledge-based approach to predict the positions of molecules at the binding interface, which has been detailed elsewhere. Due to the high-accuracy models we predicted for this protein-protein complex, our method also performed well in predicting the positions of water molecules.
3.4 Targets 48 and 49 (two conformations of T4moH-T4moC)
Targets 48 and 49 both are complexes between the toluene 4-monooxygenase hydroxylase (T4moH) and the Rieske-type ferredoxin toluene-4-monooxygenase system protein C (T4moC) (PDB ID: 4I57).33 The structure of T4moC is the same between Targets 48 and 49. Both use the crystal structure of unbound T4moC from PDB ID: 1VM9.34 The difference between the two targets is the hexamer conformation of T4moH. Structures for both of these conformations were provided by CAPRI and were built from the crystal structure of T4moH hydroxylase as a hetero-hexamer (PDB ID: 3DHH).35 The T4moH hydrox-ylase has two binding sites for ferredoxin, but the ferrodoxin binding modes involving the first ABC trimer was required to be predicted. There was some biological information available about the binding sites of both proteins. According to the literature, the active site of T4moH hydroxylase for effector protein binding was expected to be the binding site of T4moC.35 The mutagenesis analysis of the ferre-doxin T4moC also indicated that the residues R65, W69, D82, and D83 may be involved in binding.36 In addition, since we used the T4moH trimer for our docking calculations and the T4moH hydroxylase is a hetero-hexamer, we excluded those T4moC binding modes that had severe atomic clashes with the constructed hexamer. Having been filtered based on all the above biological considerations, eight of our top ten selections were rated as acceptable or better binding modes for target 48. For our best model, which was of medium accuracy, fnat = 37.5%, Lrmsd = 3.921 Å and Irmsd = 1.653 Å. No scoring experiment was held for target 48. For target 49, we predicted four acceptable modes for the docking experiments with the best model having an acceptable accuracy of fnat = 31.2%, Lrmsd = 5.678 Å and Irmsd = 2.177. For the target 49 scoring experiment, we gave seven correct binding modes of which the best had a medium accuracy of fnat = 50.0%, Lrmsd = 5.593 Å and Irmsd = 1.790.
3.5 Targets 50 (HA-HB36.3)
Target 50 is a protein-protein complex between the influenza hemagglutinin (HA) and a designed protein HB36.3 (PDB ID: 3R2X).37 The unbound structure of the HA chains was taken from the crystal structure 3GBN,38 and the structure of HB36.3 was modeled based on the template 1U84 (not yet published) using MODELLER. The HB36.3 protein is a mutant designed to bind the HA chains. Therefore, we expected the mutated residues of HB36.3 to be located at the binding interface, to increase the binding affinity of the complex. With this in mind, we tried to select those HB36.3 binding modes in which more mutated residues are in contact with the protein HA chains. For this target, we made one acceptable prediction with an accuracy of fnat = 67.3%, Lrmsd = 9.618 Å and Irmsd = 2.239 in the docking experiments, and two correct predictions in the scoring experiments in which the best model had a medium accuracy of fnat = 57.1%, Lrmsd = 5.650 Å and Irmsd = 1.912 (Figure 2).
3.6 Targets 51 (GH5-CBM6/CBM13/Fn3)
Target 51 is a multidomain protein including three domains GH5-CBM6, XYLAN binding-domain CBM13, and Fn3-like domain (PDB ID: 4BXG).39 CAPRI participants were invited to predict the domain-domain interfaces between these three domains. Three domain-domain interfaces, i.e. CBM13/Fn3, GH5-CBM6/CBM13, and GH5-CBM6/CBM13-Fn3, were evaluated during the assessment. In Target 35, GH5-CBM6 was provided with an unpublished crystal structure by the authors of this target. The structure of CBM13 was built based on the crystal structure 1KNL40 using MODELLER, and the unbound structure of Fn3-like protein was taken from the crystal structure 3MPC.41 Due to lack of clear information about the interaction interface and uncertainties in domain-domain interactions, this target has been one of the most difficult targets and only three groups gave correct predictions for the docking experiments. We did not predict any correct modes for this target in the docking experiments, and our best model gave an Irmsd of 11.56 Å. However, our scoring function predicted one mode with an acceptable accuracy of fnat = 24.1%, Lrmsd = 20.501 Å and Irmsd = 3.483 for the GH5-CBM6/CBM13-Fn3 interface, indicating that sufficient sampling of the binding modes during docking may have played an important role in identifying the correct binding mode of this target.
3.7 Target 53 (Rep4-Rep2)
Target 53 is a protein-protein complex between an artificial α-repeat protein Rep4 and a short α-repeat Rep2 (PDB ID: 4JW2).42 The unbound structure of Rep4 was taken from the crystal structure 3LTJ,43 and Rep2 was built based on the template 3LTJ using MODELLER. Although the Rep4 forms a homodimer according to its crystal structure, it was not clear whether or not the Rep4 dimer interface is the binding interface between Rep4 and Rep2. Therefore, we did not apply this biological information during the filtering step for the final model selection. In addition, because Rep4 is a repeat protein, there were several possible sequence alignments between Rep4 and Rep2, which would result in significantly different conformations of the modeled Rep2. In this target, we did not predict any correct binding modes in the docking experiments, although our best model gave a low Irmsd of 3.935 Å. Our scoring function was able to predict one correct mode with an acceptable accuracy of fnat = 28.8%, Lrmsd = 5.321 Å and Irmsd = 2.517 in the scoring experiment.
3.8 Target 54 (NCS/Rep16)
Target 54 is a protein-protein complex between a designed protein neocarzinostatin (NCS) and an α-repeat Rep16 (PDB ID: 4JW3).42 The unbound structure of the neocarzinostatin was taken from the crystal structure 2CBO,44 and the structure of Rep16 was built based on the crystal structure 3LTJ43 using MODELLER. No biological information about the binding site was found in the literature. This target also turned out to be a difficult target with only four groups giving correct predictions in the docking experiment and no groups giving a successful prediction in the scoring experiment. In this target, we were not able to give any correct predictions for both the docking experiment, in which the best model had an Irmsd of 7.832 Å, and the scoring experiment, in which the best model had an Irmsd of 5.397 Å. Since the crystal structure of this complex is not available at the time of writing, we do not yet know the reason for the difficulty of this target. However, given that the repeat protein Rep16 is expected to have less conformational changes in its backbone due to its helical structure, significant conformational changes might have occurred in the neocarzinostatin.
3.9 Target 57 (BT4661/heparin)
Target 57 is a protein-oligosaccharide complex between heparin hexasaccharide and BT4661, a SusE-like polysaccharide binding protein from Bacteroides thetaiotaomicron (PDB ID: 4AK2).45 The unbound conformation of BT4661 and an extended conformation of heparin were provided by CAPRI. Heparin is highly negatively charged, so we manually inspected all of the crystal structures containing heparin oligosaccharides that are available in the PDB and found that in nearly every case heparin binds to a positively charged region of the protein. In addition, the binding mode of heparin is often more solvent-exposed than is typical for biological ligands. The BT4661 structure was found to have one promising site for binding: a positively charged region with a concentration of four arginine residues (R581, R582, R623, and R688). There was also a nearby positively-charged region that was sufficiently close to the first to permit the possibility of heparin interacting with both.
To account for the flexibility of heparin, we obtained most of the putative conformations from PDB ID: 3IRI, a set of solution structures of heparin octadecasaccharide that were built through constrained modeling with data from analytical ultracentrifugation and small-angle X-ray scattering.46 We divided each of these nine solution structures into six hexasaccharide-sized fragments consisting of saccharides 1–6, 3–8, 5–10, 9–14, 11–16, and 13–18, yielding a total of 54 putative conformations. We also included two other conformations in the ensemble: the heparin structure from PDB: 3OJV47 and the extended conformation of heparin provided by CAPRI. These 56 conformations were all docked to the presumed binding site of BT4661 using MDock, the protein-ligand docking software developed by our laboratory.48–51 For this target, we achieved one prediction of medium accuracy (Figure 2) and one other acceptable prediction. Our medium-accuracy prediction had the lowest Irmsd (1.369 Å) among all the predictions for this target.
3.10 Target 58 (SalG/PliG-Ec)
Target 58 is a protein-protein complex between SalG, the Atlantic salmon goose-type lysozyme, and PliG-Ec, a goose-type lysozyme inhibitor from E. coli (PDB ID: 4G9S).52 The unbound conformations of both SalG and PliG-Ec were provided by CAPRI. A small-angle X-ray scattering (SAXS) profile for the inhibitor-lysozyme complex was also provided. We used the program CRYSOL to fit this experimental profile to a theoretical SAXS profile generated for each of the docked poses.53 The default CRYSOL parameters were used, except for the parameter specifying the angular units of the input file, which was set to match the format of the experimental profile. The quality of the theoretical-experimental fit was evaluated by the chi-squared test, which is handled automatically by CRYSOL. We also evaluated the poses in terms of their consistency with the binding site information available in the literature. SalG residues E73, D86, and D97 are known to be part of its active site, so we presumed that the binding mode of PliG-Ec should block these residues.54 In addition, the mutations Y47A, R115A, and R119A on PliG-Ec significantly reduce its inhibitory activity and thus these residues were considered likely to be part of the inhibitor-lysozyme interface.55 Our final ten selections were those non-redundant poses that provided the best combinations of low docking scores, theoretical-experimental SAXS agreement, and consistency with the experimental binding site information. For this target, we achieved three acceptable predictions (Figure 2).
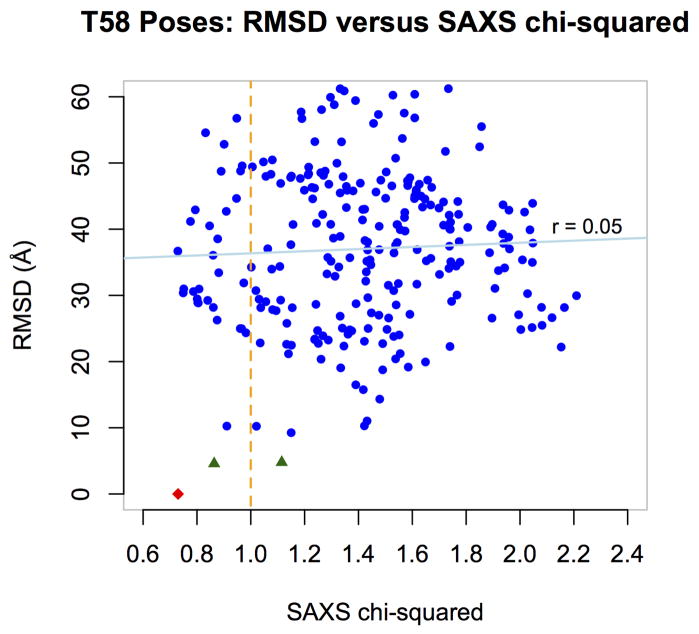
After the release of the native complex, we computed the Cα-RMSD between each of the docked complexes and the native complex, and compared these RMSDs to the chi-squared goodness of fit values between the theoretically generated SAXS profiles and the experimental SAXS profile. This comparison showed them to be uncorrelated (correlation coefficient r = 0.05, see Figure 3) and therefore not useful for ranking in this case. Nevertheless, our results are consistent with the usefulness of SAXS for filtering out the worst models. To show this in the figure, we labeled the docked complexes within 5.0 Å Cα-RMSD of the native complex as ‘distant’ and colored their data points blue, and we labeled the two closer docked complexes as ‘close’ and colored them black. The native complex is colored red in this figure. The orange vertical line indicates an example cutoff at a chi-squared goodness of fit of 1.0. Using this cutoff, the number of distant poses drops from 248 to 29, while one of the two close poses is preserved, as is the native complex. One challenge was the sphericity of the subunits, which can yield similar SAXS profiles for substantially different poses when either subunit rotates in situ.
Figure 3.
RMSD versus SAXS goodness of fit for the 250 docked poses of Target 58 and the native complex. The x-axis gives the chi-squared goodness of fit between the theoretically-predicted SAXS profile for each pose and the experimentally-determined SAXS profile of the native complex. The y-axis gives the Cα-RMSD between each pose and the native complex. The two close docked complexes are plotted as black triangles (see text). The native complex is plotted as a red diamond.
4 CONCLUSION AND DISCUSSIONS
We have further improved our hierarchical approach for protein-protein docking (MDockPP) by considering the orientational entropic effect using an empirical statistical mechanics-based method. This method was utilized by our group in the CAPRI experiments, a community-wide blind test for protein-protein interactions. For the docking experiments, we predicted correct models for six out of ten targets, including one high-accuracy, two medium-accuracy, and three acceptable predictions. For the scoring experiments, our scoring function correctly identified a model with at least acceptable accuracy for six out of seven target complexes, including one high-accuracy, two medium-accuracy, and three acceptable models. During our participation in CAPRI, we found that biological information about the binding site played a valuable role in the selection of correct modes (e.g. Targets 47 and 48). Protein flexibility was still a challenge and needs to be properly considered for protein-protein docking (e.g. Target 46). Multidomain protein docking is also a challenge due to the issue of enormous sampling. In the case of Target 58, we used SAXS data to aid in our predictions. Its use was limited to filtering out those models predicted to have highly different SAXS profiles from the experimentally-determined SAXS profile of the target.
Our current scoring scheme is based on the rigid-body assumption. Future studies include the development of more advanced methods to account for protein flexibility, validation of these methods on the protein-protein benchmark 3.0,21 and comparison with other existing docking algorithms.
Acknowledgments
This work was supported by the National Science Foundation CAREER Award DBI0953839, the National Institute of Health grant R21GM088517, and the American Heart Association Midwest Affiliate grant 13GRNT16990076 (XZ). Many of the computations were performed on the HPC resources at the University of Missouri Bioinformatics Consortium (UMBC). SZG is supported through the NLM Biomedical Informatics Research Training Program (T15 LM07089, PI: Simoes).
References
- 1.Wodak SJ, Janin J. Computer analysis of protein-protein interaction. J Mol Biol. 1978;124:323–342. doi: 10.1016/0022-2836(78)90302-9. [DOI] [PubMed] [Google Scholar]
- 2.Smith GR, Sternberg MJ. Prediction of protein-protein interactions by docking methods. Curr Opin Struct Biol. 2002;12:28–35. doi: 10.1016/s0959-440x(02)00285-3. [DOI] [PubMed] [Google Scholar]
- 3.Halperin I, Ma B, Wolfson H, Nussinov R. Principles of docking: an overview of search algorithms and a guide to scoring functions. Proteins. 2002;47:409–43. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- 4.Schneidman-Duhovny D, Nussinov R, Wolfson HJ. Predicting molecular interactions in silico: II. Protein-protein and protein-drug docking. Curr Med Chem. 2004;11:91–107. doi: 10.2174/0929867043456223. [DOI] [PubMed] [Google Scholar]
- 5.Gray JJ. High-resolution protein-protein docking. Curr Opin Struct Biol. 2006;16:183–193. doi: 10.1016/j.sbi.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Bonvin AM. Flexible protein-protein docking. Curr Opin Struct Biol. 2006;16:194–200. doi: 10.1016/j.sbi.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Janin J, Henrick K, Moult J, Ten Eyck L, Sternberg MJE, Vajda S, Vasker I, Wodak SJ. CAPRI: a critical assessment of predicted interactions. Proteins: Struct Funct Genet. 2003;52:2–9. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- 8.Méndez R, Leplae R, Lensink MF, Wodak SJ. Assessment of CAPRI predictions in rounds 3–5 shows progress in docking procedures. Proteins. 2005;60:150–169. doi: 10.1002/prot.20551. [DOI] [PubMed] [Google Scholar]
- 9.Lensink MF, Wodak SJ, Méndez R. Docking and scoring protein complexes: CAPRI 3rd edition. Proteins. 2007;69:704–718. doi: 10.1002/prot.21804. [DOI] [PubMed] [Google Scholar]
- 10.Lensink MF, Wodak SJ. Blind predictions of protein interfaces by docking calculations in CAPRI. Proteins. 2010;78:3085–3095. doi: 10.1002/prot.22850. [DOI] [PubMed] [Google Scholar]
- 11.Huang S-Y, Zou X. MDockPP: A hierarchical approach for protein-protein docking and its application to CAPRI rounds 15–19. Proteins. 2010;78:3096–3103. doi: 10.1002/prot.22797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katchalski-Katzir E, Shariv I, Eisenstein M, Friesem AA, Aflalo C, Vakser IA. Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques. Proc Natl Acad Sci USA. 1992;89:2195–9. doi: 10.1073/pnas.89.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang S-Y, Zou X. An iterative knowledge-based scoring function for protein-rotein recognition. Proteins. 2008;72:557–579. doi: 10.1002/prot.21949. [DOI] [PubMed] [Google Scholar]
- 14.Chen R, Weng ZP. A novel shape complementarity scoring function for protein-protein docking. Proteins. 2003;51:397–408. doi: 10.1002/prot.10334. [DOI] [PubMed] [Google Scholar]
- 15.Gabb HA, Jackson RM, Sternberg MJ. Modelling protein docking using shape complementarity, electrostatics and biochemical information. J Mol Biol. 1997;272:106–120. doi: 10.1006/jmbi.1997.1203. [DOI] [PubMed] [Google Scholar]
- 16.McQuarrie DA. Statistical Mechanics. University Science Books; 2000. [Google Scholar]
- 17.Gilson MK, Zhou H-X. Calculation of protein-ligand binding affinities. Annu Rev Biophys Biomol Struct. 2007;36:21–42. doi: 10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- 18.Nelder JA, Mead R. A simplex method for function minimization. Computer J. 1965;7:308–313. [Google Scholar]
- 19.Huang S-Y, Zou X. Inclusion of solvation and entropy in the knowledge-based scoring function for protein-ligand interactions. J Chem Inf Model. 2010;50:262–273. doi: 10.1021/ci9002987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruvinsky AM, Kozintsev AV. New and fast statistical-thermodynamic method for computation of protein-ligand binding entropy substantially improves docking accuracy. J Comput Chem. 2005;26:1089–1095. doi: 10.1002/jcc.20246. [DOI] [PubMed] [Google Scholar]
- 21.Hwang H, Pierce B, Mintseris J, Janin J, Weng Z. Protein-protein docking benchmark version 3. 0. Proteins. 2008;73:705–709. doi: 10.1002/prot.22106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 23.Chen R, Li L, Weng ZP. ZDOCK: An initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 24.Brenke R, Hall DR, Chuang GY, Comeau SR, Beglov D, Vajda S, Kozakov D. Application of asymmetric statistical potentials to antibody-antigen docking. Bioinformatics. 2012;28:2608–2614. doi: 10.1093/bioinformatics/bts493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozakov D, Brenke R, Comeau SR, Vajda S. PIPER: An FFT-based protein docking program with pairwise potentials. Proteins. 2006;65:392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 26.Liger D, Mora L, Lazar N, Figaro S, Henri J, Scrima N, Buckingham RH, van Tilbeurgh H, Heurgué-Hamard V, Graille M. Mechanism of activation of methyltransferases involved in translation by the Trm112 ‘hub’ protein. Nucleic Acids Res. 2011;14:6249–6259. doi: 10.1093/nar/gkr176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Z, Shipman L, Zhang M, Anton BP, Roberts RJ, Cheng X. Structural characterization and comparative phylogenetic analysis of Escherichia coli HemK, a protein (N5)-glutamine methyltransferase. J Mol Biol. 2004;340:695–706. doi: 10.1016/j.jmb.2004.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marti-Renom MA, Stuart A, Fiser A, Sánchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 30.Heurgue-Hamard V, Graille M, Scrima N, Ulryck N, Champ S, Van Tilbeurgh H, Buckingham RH. The zinc finger protein Ynr046w is plurifunctional and a component of the eRF1 methyltransferase in yeast. J Biol Chem. 2006;281:36140–36148. doi: 10.1074/jbc.M608571200. [DOI] [PubMed] [Google Scholar]
- 31.Wojdyla JA, Fleishman SJ, Baker D, Kleanthous C. Structure of the ultra-high-affinity colicin E2 DNase–Im2 complex. J Mol Biol. 2012;417:79–94. doi: 10.1016/j.jmb.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 32.Kuhlmann UC, Pommer AJ, Moore GR, James R, Kleanthous C. Specificity in protein-protein interactions: the structural basis for dual recognition in endonuclease colicin-immunity protein complexes. J Mol Biol. 2000;301:1163–1178. doi: 10.1006/jmbi.2000.3945. [DOI] [PubMed] [Google Scholar]
- 33.Acheson JF, Bailey LJ, Fox BG. Structure of a diiron hydroxylase ferredoxin electron transfer complex. In preparation. [Google Scholar]
- 34.Moe LA, Bingman CA, Wesenberg GE, Phillips GN, Fox BG. Structure of T4moC, the Rieske-type ferredoxin component of toluene 4-monooxygenase. Acta Crystallogr Sect D. 2006;62:476–482. doi: 10.1107/S0907444906006056. [DOI] [PubMed] [Google Scholar]
- 35.Bailey LJ, McCoy JG, Phillips GN, Jr, Fox BG. Structural consequences of effector protein complex formation in a diiron hydroxylase. Proc Natl Acad Sci USA. 2008;105:19194–19198. doi: 10.1073/pnas.0807948105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elsen NL, Moe LA, McMartin LA, Fox BG. Redox and functional analysis of the Rieske ferredoxin component of the toluene 4-monooxygenase. Biochemistry. 2007;46:976–986. doi: 10.1021/bi0616145. [DOI] [PubMed] [Google Scholar]
- 37.Fleishman SJ, Whitehead TA, Ekiert DC, Dreyfus C, Corn JE, Strauch EM, Wilson IA, Baker D. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bras JLA, Gilbert HJ, Ferreira LMA, Fontes CMGA, Najmudin S. The penta-modular cellulosomal arabi-noxylanase CtXy15A structure as revealed by X-ray crystallography. In preparation. [Google Scholar]
- 40.Notenboom V, Boraston AB, Williams SJ, Kilburn DG, Rose DR. High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding. Biochemistry. 2002;41:4246–4254. doi: 10.1021/bi015865j. [DOI] [PubMed] [Google Scholar]
- 41.Alahuhta M, Xu Q, Brunecky R, Adney WS, Ding SY, Himmel ME, Lunin VV. Structure of a fibronectin type III-like module from Clostridium thermocellum. Acta Crystallogr Sect F. 2010;66:878–880. doi: 10.1107/S1744309110022529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guellouz A, Valerio-Lepiniec M, Urvoas A, Chevrel A, Graille M, Fourati-Kammoun Z, Desmadril M, van Tilbeurgh H, Minard P. Selection of specific protein binders for pre-defined targets from an optimized library of artificial helicoidal repeat proteins (alphaRep) PLoS One. 2013;8:e71512. doi: 10.1371/journal.pone.0071512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urvoas A, Guellouz A, Valerio-Lepiniec M, Graille M, Durand D, Desravines DC, van Tilbeurgh H, Desmadril M, Minard P. Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins (αRep) based on thermostable HEAT-like repeats. J Mol Biol. 2010;404:307–327. doi: 10.1016/j.jmb.2010.09.048. [DOI] [PubMed] [Google Scholar]
- 44.Drevelle A, Graille M, Heyd B, Sorel I, Ulryck N, Pecorari F, Desmadril M, Van Tilbeurgh H, Minard P. Structures of in vitro evolved binding sites on neocarzinostatin scaffold reveal unanticipated evolutionary pathways. J Mol Biol. 2006;358:455–471. doi: 10.1016/j.jmb.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 45.Lowe EC, Basle A, Czjzek M, Thomas S, Murray H, Firbank SJ, Bolam DN. Structure of BT4661, a SusE-like surface located polysaccharide binding protein from the Bacteroides thetaiotaomicron heparin utilisation locus. In preparation. [Google Scholar]
- 46.Khan S, Gor J, Mulloy B, Perkins SJ. Semi-rigid solution structures of heparin by constrained X-ray scattering modelling: new insight into heparin-protein complexes. J Mol Biol. 2010;395:504–21. doi: 10.1016/j.jmb.2009.10.064. [DOI] [PubMed] [Google Scholar]
- 47.Beenken A, Eliseenkova AV, Ibrahimi OA, Olsen SK, Mohammadi M. Plasticity in interactions of fibrob-last growth factor 1 (FGF1) N terminus with FGF receptors underlies promiscuity of FGF1. J Biol Chem. 2012;287:3067–3078. doi: 10.1074/jbc.M111.275891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang S-Y, Zou X. An iterative knowledge-based scoring function to predict protein-ligand interactions: I. Derivation of interaction potentials. J Comput Chem. 2006;27:1865–1875. doi: 10.1002/jcc.20504. [DOI] [PubMed] [Google Scholar]
- 49.Huang S-Y, Zou X. An iterative knowledge-based scoring function to predict protein-ligand interactions: II. Derivation of interaction potentials. J Comput Chem. 2006;27:1876–1882. doi: 10.1002/jcc.20504. [DOI] [PubMed] [Google Scholar]
- 50.Huang S-Y, Zou X. Ensemble docking of multiple protein structures: considering protein structural variations in molecular docking. Proteins. 2007;66:399–421. doi: 10.1002/prot.21214. [DOI] [PubMed] [Google Scholar]
- 51.Huang S-Y, Zou X. Efficient molecular docking of NMR structures: application to HIV-1 protease. Protein Sci. 2007;16:43–51. doi: 10.1110/ps.062501507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leysen S, Vanderkelen L, Weeks SD, Michiels CW, Strelkov SV. Crystal structure of Escherichia coli PliG in complex with Atlantic salmon g-type lysozyme. Cell Mol Life Sci. 2013;70:1113–1122. doi: 10.1007/s00018-012-1184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Svergun DI, Barberato C, Koch MHJ. CRYSOL — a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J Appl Cryst. 1995;28:768–773. [Google Scholar]
- 54.Kyomuhendo P, Myrnes B, Brandsdal BO, Smalas AO, Nilsen IW, Helland R. Thermodynamics and structure of a salmon cold-active goose-type lysozyme. Comp Biochem Physiol B Biochem Mol Biol. 2010;156:254–263. doi: 10.1016/j.cbpb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 55.Leysen S, Vanderkelen L, Van Asten K, Vanheuverzwijn S, Theuwis V, Michiels CW, Strelkov SV. Crystal structure of Escherichia coli PliG, a periplasmic lysozyme inhibitor of g-type lysozyme. J Struct Biol. 2012;180:235–242. doi: 10.1016/j.jsb.2012.05.006. [DOI] [PubMed] [Google Scholar]