Abstract
FAS/FASL gene promoter polymorphisms are associated with cervical cancer risk, however, results from previous studies have been conflicting. To obtain a more precise estimation of the association between these polymorphisms and cancer risk, a meta-analysis was performed. All eligible studies up to November 1st, 2012, concerning FAS-670 A/G, FAS-1377 G/A and FASL-844 T/C polymorphisms and cervical cancer risk, were collected from the following electronic databases: PubMed, Excerpta Medica Database and Chinese Biomedical Literature Database. The odds ratio (OR) and 95% confidence interval (95% CI) were used to assess the strength of the association via the additive, codominant, dominant and recessive models. In total, 10 publications with 11 case-control studies (10 on FAS-670 A/G, 5 on FAS-1377 G/A and 6 on FASL-844 T/C polymorphisms) were included in this meta-analysis. No association between FAS-670 A/G, FAS-1377 G/A and FASL-844 T/C polymorphisms and cervical cancer susceptibility for all the genetic models was identified. Following stratification of the studies by ethnicity or source of controls, similar results were obtained. In conclusion, our findings showed that the FAS-670 A/G, FAS-1377 G/A and FASL-844 T/C polymorphisms are not associated with cervical cancer risk. Future studies with larger sample sizes are required to further evaluate these associations.
Keywords: cervical cancer, FAS, FASL, polymorphism, meta-analysis
Introduction
Cervical cancer was the third most commonly diagnosed cancer and the fourth leading cause of cancer death in females worldwide in 2008 (1). Risk factors for cervical cancer include early onset of sexual activity, multiple sexual partners, cigarette smoking, high parity, low socioeconomic status, immuno suppression, high-risk sexual partners (2) and particularly, human papillomavirus (HPV) infection (3). HPV infections are widespread in the general population. However, only a small proportion of infected women progress to cervical cancer (4), suggesting that the development of cervical cancer may be influenced by genetic factors.
FAS (also known as Apo-1 or CD95), a cell-surface receptor, plays a key role in apoptotic signaling in many cell types (5). FAS ligand (FASL) (also known as CD95L), a member of the tumor necrosis factor super family, can trigger an apoptotic cascade by cross-linking with its receptor, FAS (6,7). Single nucleotide polymorphisms in the promoter regions of FAS and FASL have been associated with the differential expression of these two genes. It was reported that a G→A transition at position −1377 and an A→G transition at position −670 in the promoter region of the FAS gene destroys stimulatory protein (Sp) 1 and signal transducer and activator of transcription (STAT) 1 protein-binding element, reducing promoter activity and decreasing FAS expression (8,9). In regards to the FASL gene, it has been shown that a T→C transition at position −844 in a binding motif for CAAT/enhancer-binding protein β induces a significantly higher basal expression of FASL (10).
Although evidence of an association between FAS/FASL polymorphisms and cervical cancer risk has been reported, the findings remain controversial (11–22). Therefore, we performed a meta-analysis of all published studies on the association between the FAS/FASL polymorphisms and cervical cancer.
Materials and methods
Publication search
A search of the literature was performed using PubMed, Excerpta Medica Database and the Chinese Biomedical Literature Database to identify articles that evaluated the associations between polymorphisms in FAS/FASL and cervical cancer risk (last search was updated on November 1st, 2012). The search terms used were: ‘FAS or CD95 or FASL or CD95L’, ‘cervical carcinoma or cervical cancer or cervix cancer’ and ‘polymorphism or polymorphisms’. In addition, we checked all the references of relevant reviews and eligible articles that our search retrieved. No language restrictions were applied. If more than one article was published by the same author using the same case series, we selected the research with the largest sample size.
Inclusion and exclusion criteria
All the studies included in the meta-analysis were required to meet the following criteria: i) case-control studies; ii) evaluation of the association between FAS/FASL polymorphisms and cervical cancer; iii) sufficient data for the estimation of an odds ratio (OR) with 95% confidence interval (CI). The exclusion criteria were: i) duplicate data; ii) no controls; iii) no sufficient data were reported and; iv) abstract, comment, review and editorial.
Data extraction
Two investigators independently extracted the data and reached consensus on all items. For each eligible study, the following were extracted: the first authors’ name, the year of publication, ethnicity (including Caucasian, Asian, patients of African descent and Mixed) of the study population, sources of controls (population- or hospital-based), sample size of cases and controls, genotyping method, distributions of every genotype and evidence of Hardy-Weinberg equilibrium (HWE).
Statistical analysis
ORs with 95% CIs were calculated to assess the strength of the association between FAS/FASL polymorphisms and cervical cancer risk. To estimate associations with cervical cancer risk, various genotypic models were explored, including the following models: i) additive (minor allele vs. major allele); ii) codominant (heterozygous vs. common homozygous carriers and rare homozygous vs. common homozygous carriers); iii) dominant (rare allele carriers vs. common homozygous carriers); iv) recessive (rare homozygous carriers vs. common allele carriers). Subgroup analyses were performed by ethnicity and source of controls.
Heterogeneity among studies was assessed by the χ2-based Q-statistic (23). Heterogeneity was considered significant for P<0.10. When significant heterogeneity was detected, the random-effects model (DerSimonian and Laird method) (23) was used, otherwise the fixed effects model (Mantel-Haenszel method) (24) was selected. The goodness-of-fit χ2 test was used to assess the deviation from HWE in controls, with statistical significance defined as P<0.05 (25).
Publication bias was evaluated using Begg’s funnel plot (26) and Egger’s linear regression test (27). Analyses were performed using the software Stata version 12.0 (Stata Corporation, College Station, TX, USA). P<0.05 was considered to indicate a statistically significant difference and all the P-values were two sided.
Results
Eligible studies
Based on a literature search and selection, a total of 10 publications (11–19,22) comparing the FAS/FASL polymorphism and cervical cancer susceptibility were identified. Among these studies, 1 study was in Chinese (22) and 1 study contained data on two different ethnicities (18). As a result, 11 case-control studies were included in this meta-analysis. Ten studies on FAS-670A/G polymorphism, five on FAS-1377G/A and six studies on FASL-844T/C polymorphisms met the inclusion criteria (Table I). The genotype distribution in the controls of the studies was consistent with HWE, with the exception of 2 studies for FAS-670 A/G polymorphism (17,18) and 1 study for FASL-844 T/C polymorphism (11) (Table I).
Table I.
Characteristics of studies considered in the meta-analysis.
| First author | Year | Ethnicity | Source of controls | Polymorphism | Genotype method | Sample size | Cases/controls | HWE | Refs. | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| Cases | Controls | AA | GA | GG | ||||||||
| Sun | 2005 | Asian | Population-based | FAS-670 A/G | PCR-RFLP | 314 | 615 | 138/268 | 144/272 | 32/75 | 0.641 | (11) |
| Lai | 2005 | Asian | Hospital-based | FAS-670 A/G | TaqMan | 318 | 318 | 121/91 | 137/161 | 60/66 | 0.736 | (12) |
| Zoodsma | 2005 | Caucasian | Population-based | FAS-670 A/G | TaqMan | 670 | 607 | 143/155 | 363/290 | 164/162 | 0.274 | (13) |
| Chen | 2006 | Asian | Population-based | FAS-670 A/G | MAMA-PCR | 194 | 138 | 68/32 | 94/76 | 32/30 | 0.232 | (22) |
| Ueda | 2006 | Asian | Population-based | FAS-670 A/G | RCR-RFLP | 83 | 95 | 15/23 | 38/54 | 30/18 | 0.172 | (14) |
| Kang | 2007 | Asian | Hospital-based | FAS-670 A/G | RCR-RFLP | 154 | 160 | 48/53 | 73/84 | 33/23 | 0.264 | (15) |
| Zucchi | 2008 | Mixed | Hospital-based | FAS-670 A/G | RCR-RFLP | 91 | 176 | 18/43 | 52/92 | 21/41 | 0.545 | (16) |
| Tamandani | 2008 | Asian | Hospital-based | FAS-670 A/G | RCR-RFLP | 200 | 200 | 26/55 | 167/121 | 7/24 | 0.001 | (17) |
| Chatterjee | 2009 | Black | Hospital-based | FAS-670 A/G | RCR-RFLP | 103 | 99 | 1/2 | 35/43 | 67/54 | 0.047 | (18) |
| Chatterjee | 2009 | Mixed | Hospital-based | FAS-670 A/G | RCR-RFLP | 332 | 321 | 53/39 | 132/146 | 147/136 | 0.985 | (18) |
| GG | AG | AA | ||||||||||
| Sun | 2005 | Asian | Population-based | FAS-1377G/A | RCR-RFLP | 314 | 615 | 144/282 | 144/277 | 26/56 | 0.304 | (11) |
| Lai | 2005 | Asian | Hospital-based | FAS-1377G/A | TaqMan | 318 | 318 | 127/99 | 138/165 | 53/54 | 0.293 | (12) |
| Kang | 2007 | Asian | Hospital-based | FAS-1377G/A | RCR-RFLP | 154 | 158 | 54/56 | 69/82 | 31/20 | 0.233 | (15) |
| Chatterjee | 2009 | black | Hospital-based | FAS-1377G/A | RCR-RFLP | 96 | 90 | 74/75 | 21/14 | 1/1 | 0.707 | (18) |
| Chatterjee | 2009 | Mixed | Hospital-based | FAS-1377G/A | RCR-RFLP | 290 | 295 | 186/196 | 87/86 | 1713 | 0.37 | (18) |
| TT | TC | CC | ||||||||||
| Sun | 2005 | Asian | Population-based | FASL -844T/C | RCR-RFLP | 314 | 615 | 10/40 | 111/291 | 193/284 | 0.002 | (11) |
| Lai | 2005 | Asian | Hospital-based | FASL -844T/C | TaqMan | 303 | 316 | 21/27 | 119/132 | 163/157 | 0.92 | (12) |
| Ivansson | 2007 | Caucasian | Population-based | FASL -844T/C | PCR-RFLP | 1,284 | 280 | 119/24 | 531/112 | 634/144 | 0.738 | (19) |
| Kang | 2007 | Asian | Hospital-based | FASL -844T/C | RCR-RFLP | 154 | 160 | 7/7 | 66/63 | 81/90 | 0.327 | (15) |
| Chatterjee | 2009 | Black | Hospital-based | FASL -844T/C | RCR-RFLP | 103 | 100 | 70/74 | 31/23 | 2/3 | 0.469 | (18) |
| Chatterjee | 2009 | Mixed | Hospital-based | FASL -844T/C | RCR-RFLP | 327 | 315 | 143/139 | 144/136 | 40/40 | 0.457 | (18) |
HWE, Hardy-Weinberg equilibrium; PCR-RFLP, polymerase chain reaction restriction fragment length polymorphism; MAMA-PCR, mismatch amplification mutation assay polymerase chain reaction.
Analysis for FAS-670 A/G polymorphism
A total of 2,127 cases and 2,408 controls were identified for the analysis on FAS-670 A/G polymorphism and cervical cancer risk. The overall result suggested no statistically significant association of this polymorphism with cervical cancer susceptibility (for G vs. A: OR=1.016; 95% CI, 0.898–1.149; P=0.028 for heterogeneity; for GG vs. AA: OR=0.944; 95% CI, 0.729–1.222; P=0.058 for heterogeneity; for GA vs. AA: OR=1.034; 95% CI, 0.768–1.393; P=0.000 for heterogeneity; for GG+GA vs. AA: OR=1.043; 95% CI, 0.797–1.366; P=0.000 for heterogeneity; for GG vs. GA/AA: OR=0.999; 95% CI, 0.784–1.274; P=0.004 for heterogeneity; Table II). In the subgroup analyses by ethnicity and source of controls, no significant associations were found for the genetic models (Table II). Following the exclusion of studies deviating from HWE in controls (17,18), the non-associations in the above-mentioned genetic models remained.
Table II.
Meta-analysis of FAS/FASL polymorphisms and cervical cancer risk.
| Variables | Na | G vs. A | GG vs. AA | GA vs. AA | GG/GA vs. AA | GG vs. GA/AA | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|||||||
| OR (95% CI) | P-valueb | OR (95% CI) | P-valueb | OR (95% CI) | P-valueb | OR (95% CI) | P-valueb | OR (95% CI) | P-valueb | ||
| FAS-670 A/G | |||||||||||
| Overall | 10 | 1.016 (0.898–1.149)c | 0.028 | 0.944 (0.729–1.222) | 0.058 | 1.034 (0.768–1.393) | 0.000 | 1.043 (0.797–1.366) | 0.000 | 0.999 (0.784–1.274) | 0.004 |
| Studies with HWE | 8 | 0.984 (0.860–1.125) | 0.029 | 0.960 (0.727–1.267) | 0.034 | 0.911 (0.711–1.167) | 0.010 | 0.932 (0.738–1.176) | 0.012 | 1.019 (0.828–1.254) | 0.078 |
| Ethnicity | |||||||||||
| Asian | 6 | 0.996 (0.815–1.217) | 0.009 | 0.909 (0.595–1.386) | 0.020 | 1.011 (0.660–1.547) | 0.000 | 1.032 (0.695–1.531) | 0.000 | 0.931 (0.590–1.469) | 0.001 |
| Mixed | 2 | 0.996 (0.822–1.206) | 0.547 | 0.897 (0.601–1.340) | 0.347 | 0.915 (0.459–1.824) | 0.084 | 0.892 (0.624–1.275) | 0.131 | 1.061 (0.806–1.396) | 0.794 |
| Source of controls | |||||||||||
| PB | 4 | 0.998 (0.795–1.254) | 0.018 | 0.981 (0.610–1.577) | 0.022 | 1.005 (0.718–1.408) | 0.041 | 0.997 (0.712–1.397) | 0.027 | 0.991 (0.671–1.464) | 0.030 |
| HB | 6 | 1.002 (0.892–1.124) | 0.127 | 0.880 (0.680–1.138) | 0.268 | 1.097 (0.646–1.864) | 0.000 | 1.112 (0.699–1.770) | 0.001 | 1.001 (0.703–1.424) | 0.013 |
| FAS-1377 G/A | |||||||||||
| A vs. G | AA vs. GG | AG vs. GG | AA/AG vs. GG | AA vs. AG/GG | |||||||
| Overall | 5 | 0.993 (0.880–1.122) | 0.254 | 1.000 (0.757–1.322) | 0.402 | 0.926 (0.781–1.097) | 0.153 | 0.950 (0.808–1.117) | 0.167 | 1.105 (0.853–1.432) | 0.492 |
| Ethnicity | |||||||||||
| Asian | 3 | 0.952 (0.830–1.092) | 0.195 | 0.949 (0.701–1.284) | 0.201 | 0.854 (0.699–1.044) | 0.149 | 0.879 (0.726–1.063) | 0.165 | 1.076 (0.814–1.420) | 0.215 |
| Source of controls | |||||||||||
| HB | 4 | 1.000 (0.860–1.162) | 0.150 | 1.043 (0.747–1.458) | 0.279 | 0.878 (0.710–1.085) | 0.111 | 0.925 (0.756–1.131) | 0.100 | 1.201 (0.882–1.634) | 0.480 |
| FASL -844 T/C | |||||||||||
| C vs. T | CC vs. TT | TC vs. TT | CC/TC vs. TT | CC vs. TC/TT | |||||||
| Overall | 6 | 1.116 (0.914–1.364) | 0.006 | 1.168 (0.903–1.510) | 0.165 | 1.111 (0.896–1.378) | 0.858 | 1.131 (0.922–1.385) | 0.445 | 1.114 (0.829–1.498) | 0.008 |
| Studies with HWE | 5 | 1.007 (0.895–1.134) | 0.656 | 0.994 (0.749–1.319) | 0.863 | 1.074 (0.856–1.347) | 0.892 | 1.055 (0.851–1.308) | 0.850 | 0.982 (0.829–1.162) | 0.722 |
| Ethnicity | |||||||||||
| Asian | 3 | 1.221 (0.877–1.699) | 0.012 | 1.215 (0.714–2.068) | 0.171 | 1.266 (0.823–1.949) | 0.801 | 1.477 (0.979–2.229) | 0.387 | 1.269 (0.825–1.952) | 0.005 |
| Source of controls | |||||||||||
| PB | 2 | 1.235 (0.713–2.139) | 0.000 | 1.503 (0.501–4.504) | 0.010 | 1.113 (0.751–1.649) | 0.294 | 1.332 (0.590–3.010) | 0.052 | 1.306 (0.656–2.599) | 0.000 |
| HB | 4 | 1.048 (0.904–1.214) | 0.648 | 1.061 (0.744–1.513) | 0.815 | 1.110 (0.859–1.436) | 0.842 | 1.099 (0.861–1.403) | 0.824 | 1.029 (0.823–1.287) | 0.644 |
Number of comparisons.
P-value of Q-test for heterogeneity test.
Random effects model was used when P value for heterogeneity test was 0.05, otherwise the fixed effects model was used. HB, hospital based case control study; PB, population based case control study; OR, odds ratio; 95% CI, 95% confidence interval.
Analysis for FAS-1377 G/A polymorphism
A total of 1,172 cases and 1,476 controls were identified for the analysis on FAS-1377 G/A polymorphism and cervical cancer risk. The overall result showed that there was no statistically significant association between this polymorphism and cervical cancer susceptibility (Table II). Subsequent subgroup analyses revealed that there was no statistically significant association in each subgroup by ethnicity and source of controls in the genetic models (Table II).
Analysis for FASL-844 T/C polymorphism
A total of 2,485 cases and 1,786 controls were identified for the analysis on FASL-844 T/C polymorphism and cervical cancer risk. The overall result showed that there was no statistically significant association between this polymorphism and cervical cancer susceptibility (Table II). Following exclusion of the study deviating from HWE in controls (11), the results did not alter substantially in any of the genetic models. Subsequent subgroup analyses revealed that there was no statistically significant association in each subgroup by ethnicity and source of controls in the genetic models (Table II).
Publication bias
Both Begg’s funnel plot and Egger test were performed to assess the publication bias of the studies. The shape of the funnel plot did not reveal any evidence of obvious asymmetry (Fig. 1). The Egger test was used to provide statistical evidence of funnel plot symmetry. The results again did not suggest any obvious evidence of publication bias for the genetic models (all P>0.05).
Figure 1.
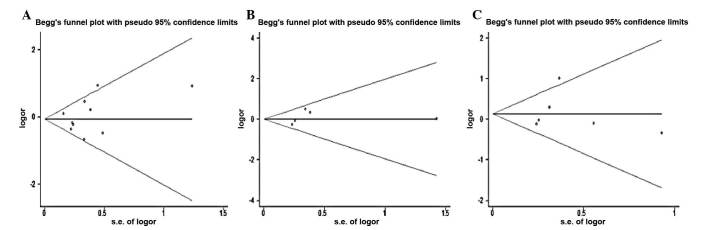
Begg’s funnel plot indicated that no publication bias was observed (A) for GG vs. AA comparison in FAS-670 A/G polymorphism; (B) for AA vs. GG comparison in FAS-1377G/A polymorphism; (C) for CC vs. TT comparison in FASL -844T/C polymorphism. Each point represents a separate study for the indicated association. Log[OR] natural logarithm of odds ratio. Horizontal line indicates the effect size.
Discussion
Genetic variants of the FAS/FASL gene in the etiology of several types of cancer have drawn increasing attention. A number of studies revealed that polymorphic variants of the FAS/FASL polymorphisms were associated with etiology of cervical cancer. However, the results are inconclusive. To gain a better understanding of the association between these polymorphisms and cervical cancer risk, a pooled analysis with a large sample of patients, as well as a subgroup analysis were performed. The aim of our study was to investigate the relationship between FAS/FASL polymorphisms and cervical cancer susceptibility.
In this meta-analysis, we failed to find any association between FAS-670 A/G, FAS-1377 G/A and FASL-844 T/C polymorphisms and cervical cancer risk in the overall analysis. In the subgroup analysis, no association was identified between the three polymorphisms and cervical cancer. These findings indicate that the three polymorphisms may not be risk factors for the development of cervical cancer.
Apoptosis is an important regulatory mechanisms occurring in multicellular organisms that is involved in normal development and tissue homeostasis (28). FAS plays a crucial role in apoptotic signaling and interacts with its natural ligand, FASLG, to initiate the death signal cascade, which results in cell death. Findings of Ueda et al (14) on germ-line polymorphism of Fas promoter -670 demonstrated that the frequency of GG genotype or G allele increased the risk of cervical cancer. Lai et al (12) detected an association between the Fas-670 polymorphism and cervical cancer susceptibility. Zoosdma et al (13) have demonstrated that Fas-670 polymorphism might be involved in the development of adenocarcinoma of the cervix but not in the development of squamous cell carcinoma. Engelmark et al (21), Sun et al (11) and Chatterjee et al (18) did not show a significant association of the FAS/FASL polymorphisms with cervical cancer. Qiu et al (29) indicated that significantly increased risks in FAS-1377 AA carriers were found in the breast cancer subgroup but not in the lung cancer subgroup. Liu et al (30) indicated that the FASL-844C allele was associated with a significantly increased cancer risk overall, but in the stratified analysis by cancer types, significant associations were still not observed in the genetic models. In contrast to these studies, our data failed to reveal significant associations between FAS/FASL polymorphisms and cervical cancer. The cause of these varied results remains unclear, however, discrepancies may exist due to different mechanisms of carcinogenesis in different types of cancer. Cervical cancer is a multi-factorial disease and individual exposures to various environmental factors in combination with genetic susceptibility may have contributed to discrepancies.
However, this study has several limitations. First, significant between-study heterogeneity was detected in some of the comparisons, distorting the findings of the meta-analysis. Second, subgroup analyses concerning other risk factors such as smoking status and HPV infection have not been included in the present study due to insufficient data from the primary literature. Third, further studies estimating the effect of gene-environment interactions may eventually provide an improved, comprehensive understanding of the association between the FAS/FASL polymorphisms and cervical cancer risk. Fourth, relatively small samples of subpopulation may render the interpretation of negative results difficult, requiring more studies with a larger sample size to elucidate those effects. Fifth, the meta-analysis remains a retrospective study that is subject to the methodological deficiencies of the included studies.
In conclusion, our meta-analysis suggests that FAS-670 A/G, FAS-1377 G/A and FASL-844 T/C polymorphisms were not associated with cervical cancer risk. However, large sample studies including different ethnic groups with a careful matching between cases and controls are necessary to confirm our findings.
Acknowledgments
This study was granted by Anhui Provincial Science and Technology Agency Foundation of China (nos. 11070403061 and 09020303042).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Wallin KL, Wiklund F, Luostarinen T, et al. A population-based prospective study of Chlamydia trachomatis infection and cervical carcinoma. Int J Cancer. 2002;101:371–374. doi: 10.1002/ijc.10639. [DOI] [PubMed] [Google Scholar]
- 3.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 4.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 5.Andera L. Signaling activated by the death receptors of the TNFR family. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2009;153:173–180. doi: 10.5507/bp.2009.029. [DOI] [PubMed] [Google Scholar]
- 6.Kim R, Emi M, Tanabe K, Uchida Y, Toge T. The role of Fas ligand and transforming growth factor beta in tumor progression: molecular mechanisms of immune privilege via Fas-mediated apoptosis and potential targets for cancer therapy. Cancer. 2004;100:2281–2291. doi: 10.1002/cncr.20270. [DOI] [PubMed] [Google Scholar]
- 7.Suda T, Takahashi T, Golstein P, Nagata S. Molecular cloning and expression of the Fas ligand, a novel member of the tumor necrosis factor family. Cell. 1993;75:1169–1178. doi: 10.1016/0092-8674(93)90326-l. [DOI] [PubMed] [Google Scholar]
- 8.Huang QR, Morris D, Manolios N. Identification and characterization of polymorphisms in the promoter region of the human Apo-1/Fas (CD95) gene. Mol Immunol. 1997;34:577–582. doi: 10.1016/s0161-5890(97)00081-3. [DOI] [PubMed] [Google Scholar]
- 9.Sibley K, Rollinson S, Allan JM, et al. Functional FAS promoter polymorphisms are associated with increased risk of acute myeloid leukemia. Cancer Res. 2003;63:4327–4330. [PubMed] [Google Scholar]
- 10.Wu J, Metz C, Xu X, et al. A novel polymorphic CAAT/enhancer- binding protein beta element in the FasL gene promoter alters Fas ligand expression: a candidate background gene in African American systemic lupus erythematosus patients. J Immunol. 2003;170:132–138. doi: 10.4049/jimmunol.170.1.132. [DOI] [PubMed] [Google Scholar]
- 11.Sun T, Zhou Y, Li H, et al. FASL -844C polymorphism is associated with increased activation-induced T cell death and risk of cervical cancer. J Exp Med. 2005;202:967–974. doi: 10.1084/jem.20050707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai HC, Lin WY, Lin YW, et al. Genetic polymorphisms of FAS and FASL (CD95/CD95L) genes in cervical carcinogenesis: an analysis of haplotype and gene-gene interaction. Gynecol Oncol. 2005;99:113–118. doi: 10.1016/j.ygyno.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 13.Zoodsma M, Nolte IM, Schipper M, et al. Interleukin-10 and Fas polymorphisms and susceptibility for (pre)neoplastic cervical disease. Int J Gynecol Cancer. 2005;15(Suppl 3):282–290. doi: 10.1111/j.1525-1438.2005.00433.x. [DOI] [PubMed] [Google Scholar]
- 14.Ueda M, Terai Y, Kanda K, et al. Fas gene promoter -670 polymorphism in gynecological cancer. Int J Gynecol Cancer. 2006;16(Suppl 1):179–182. doi: 10.1111/j.1525-1438.2006.00505.x. [DOI] [PubMed] [Google Scholar]
- 15.Kang S, Dong SM, Seo SS, Kim JW, Park SY. FAS-1377 G/A polymorphism and the risk of lymph node metastasis in cervical cancer. Cancer Genet Cytogenet. 2008;180:1–5. doi: 10.1016/j.cancergencyto.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Zucchi F, da Silva ID, Ribalta JC, et al. Fas/CD95 promoter polymorphism gene and its relationship with cervical carcinoma. Eur J Gynaecol Oncol. 2009;30:142–144. [PubMed] [Google Scholar]
- 17.Kordi Tamandani DM, Sobti RC, Shekari M. Association of Fas-670 gene polymorphism with risk of cervical cancer in North Indian population. Clin Exp Obstet Gynecol. 2008;35:183–186. [PubMed] [Google Scholar]
- 18.Chatterjee K, Engelmark M, Gyllensten U, et al. Fas and FasL gene polymorphisms are not associated with cervical cancer but differ among Black and Mixed-ancestry South Africans. BMC Research Notes. 2009;2:238. doi: 10.1186/1756-0500-2-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivansson EL, Gustavsson IM, Magnusson JJ, et al. Variants of chemokine receptor 2 and interleukin 4 receptor, but not inter-leukin 10 or Fas ligand, increase risk of cervical cancer. Int J Cancer. 2007;121:2451–2457. doi: 10.1002/ijc.22989. [DOI] [PubMed] [Google Scholar]
- 20.Ueda M, Hung YC, Terai Y, et al. Fas gene promoter -670 polymorphism (A/G) is associated with cervical carcinogenesis. Gynecol Oncol. 2005;98:129–133. doi: 10.1016/j.ygyno.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Engelmark MT, Renkema KY, Gyllensten UB. No evidence of the involvement of the Fas-670 promoter polymorphism in cervical cancer in situ. Int J Cancer. 2004;112:1084–1085. doi: 10.1002/ijc.20515. [DOI] [PubMed] [Google Scholar]
- 22.Chen YQ, Lu LG, Tian QF, et al. The research of Fas-670 polymorphism and cervical cancer susceptibility. Natl Med J China. 2006;86:2792–2794. [Google Scholar]
- 23.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 24.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 25.Haber M. Exact significance levels of goodness-of-fit tests for the Hardy-Weinberg equilibrium. Hum Hered. 1981;31:161–166. doi: 10.1159/000153199. [DOI] [PubMed] [Google Scholar]
- 26.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 27.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reed JC. Mechanisms of apoptosis. Am J Pathol. 2000;157:1415–1430. doi: 10.1016/S0002-9440(10)64779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu LX, Shi J, Yuan H, et al. FAS -1,377 G/A polymorphism is associated with cancer susceptibility: evidence from 10,564 cases and 12,075 controls. Hum Gen. 2009;125:431–435. doi: 10.1007/s00439-009-0639-4. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Wen Q-J, Yin Y, et al. FASLG polymorphism is associated with cancer risk. Eur J Cancer. 2009;45:2574–2578. doi: 10.1016/j.ejca.2009.04.001. [DOI] [PubMed] [Google Scholar]


