Abstract
The Middle East respiratory syndrome coronavirus (MERS-CoV) is a newly emerging highly pathogenic virus causing almost 50 % lethality in infected individuals. The development of a small-animal model is critical for the understanding of this virus and to aid in development of countermeasures against MERS-CoV. We found that BALB/c, 129/SvEv and 129/SvEv STAT1 knockout mice are not permissive to MERS-CoV infection. The lack of infection may be due to the low level of mRNA and protein for the MERS-CoV receptor, dipeptidyl peptidase 4 (DPP4), in the lungs of mice. The low level of DPP4 in the lungs likely contributes to the lack of viral replication in these mouse models and suggests that a transgenic mouse model expressing DPP4 to higher levels is necessary to create a mouse model for MERS-CoV.
The newly emerged Middle East respiratory syndrome coronavirus (MERS-CoV) has infected 106 people in eight countries, of whom 49 have died, as of August 2013, with increasing numbers of cases added weekly (www.cdc.gov). MERS-CoV has spread person-to-person, with several clusters of infection in Saudi Arabia, the UK, Italy and France (Mailles et al., 2013; Memish et al., 2013), with the largest cluster of 23 infected people identified in a hospital in Al Hassa, Saudi Arabia (Memish et al., 2013). The symptoms of MERS-CoV infection are similar to those of the severe acute respiratory syndrome (SARS), with fever, cough and pneumonia leading to respiratory failure. However, patients with MERS have also developed kidney failure, which was not reported for SARS. A recent study on a patient from the United Arab Emirates showed that MERS-CoV RNA could be detected, albeit at low levels, in urine and stool samples (Drosten et al., 2013), suggesting that MERS-CoV may not be restricted to the lung. Recent serology studies have also demonstrated that camels in Oman and Egypt are seropositive for MERS-CoV and have antibodies that can neutralize MERS-CoV infection in cell culture; however, MERS-CoV viral RNA has yet to be detected in these camels (Perera et al., 2013; Reusken et al., 2013).
MERS-CoV is hypothesized to be of bat origin due to its similarity to other bat CoVs, but the reservoir species is currently unknown. Cell-culture experiments suggest a broad host range for MERS-CoV, with infections seen in human, bat and monkey cells (Muller et al., 2012 ). The receptor for MERS-CoV was recently identified as dipeptidyl peptidase 4 (DPP4); antibodies to DPP4 efficiently block infection of permissive cell lines and transfection of a human DPP4 expression plasmid into non-permissive cells allows for MERS-CoV infection (Raj et al., 2013).
The study of SARS-CoV pathogenesis progressed rapidly due to the development of a mouse-adapted virus of SARS-CoV, called MA15, that produced lethal lung disease in BALB/c mice (Roberts et al., 2007) and weight loss and inflammation in 129S6/SvEv and C57BL/6 mice (Sheahan et al., 2008). Importantly, both WT and MA15 SARS-CoV produce enhanced disease in an innate immune-deficient mouse strain where STAT1 is deleted (129/STAT1−/−) (Frieman et al., 2010). An equally robust small-animal model is critical for the study of MERS-CoV pathogenesis and disease.
While MERS-CoV appears to replicate in various human and other mammalian cell types in vitro (Fuk-Woo Chan et al., 2013; Kindler et al., 2013; Muller et al., 2012; Zielecki et al., 2013), the only reported animal model for MERS-CoV is the rhesus macaque (Macaca mulatta), in which it replicates and causes pneumonia and pulmonary infiltration (Munster et al., 2013). Given the practical, ethical and financial issues associated with primate research, there is a need for a small-animal model of MERS-CoV. A recent study has shown that MERS-CoV does not replicate or cause disease in Syrian hamsters (de Wit et al., 2013). MERS-CoV infection of mice has not been reported; therefore, we tested MERS-CoV replication in three different mouse species (BALB/c, 129S6/SvEv and 129/STAT1−/−) with the goal of creating a small-animal model of disease to begin to understand the pathogenesis of MERS-CoV and/or act as a vehicle for mouse adaptation of MERS-CoV.
Eight-week-old BALB/c (Charles River, strain BALB/cAnNCrl), 129S6/SvEv (Taconic #129SVE) and 129/STAT1−/− (Taconic #2045-F) mice were infected with the MERS-CoV (strain hCoV-EMC/2012) obtained from Erasmus Medical College (van Boheemen et al., 2012). Prior to intranasal inoculation, mice were anaesthetized by intraperitoneal injection using a mix of xylazine (0.38 mg/mouse) and ketamine (1.3 mg/mouse), diluted in PBS to make a total volume of 50 µl per mouse. Once anaesthetized, five mice per group were intranasally inoculated with PBS, 120 or 1200 TCID50 of MERS-CoV diluted into PBS for a total inoculum of 50 µl. During the experiment, mice were weighed prior to infection and every day of the experiment to assess MERS-CoV-induced weight loss. BALB/c mice were euthanized at days 2 and 4 post-infection (p.i.) and 129S6/SvEv and 129/STAT1−/− mice were euthanized on day 2 or 9 p.i. using isofluorane (Butler Animal Health Supply). Lungs and kidneys were harvested for further analysis of MERS-CoV replication and pathology.
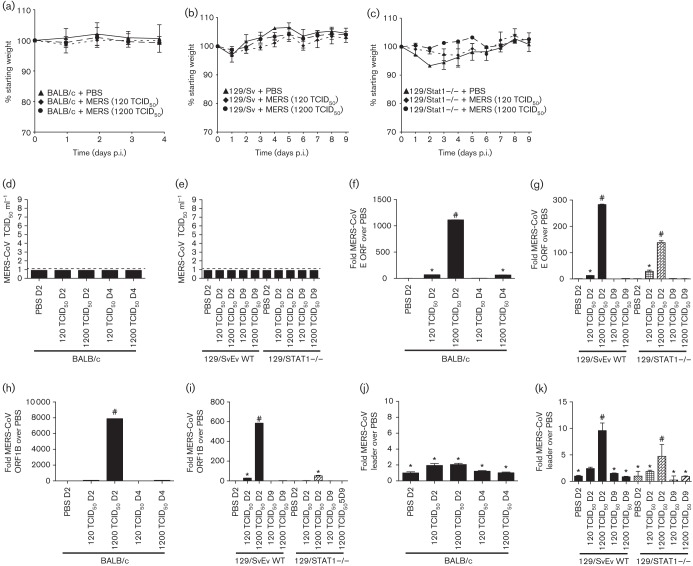
A key characteristic of SARS-CoV infection of mice is the loss of weight during infection. Daily weight-loss curves for MERS-CoV infection in BALB/c, 129S6/SvEv and 129/STAT1−/− mice are shown in Fig. 1(a–c). There was no significant weight loss, nor significant difference from PBS-infected controls observed for WT BALB/c (Fig. 1a) or WT 129S6/SvEv (Fig. 1b) mice. Furthermore, the innate immune-deficient 129/STAT1−/− mice display no weight loss during infection with MERS-CoV (Fig. 1c).
Fig. 1.
MERS-CoV pathogenesis in mice. Weight-loss curves of BALB/c (a), 129Sv/Ev (b) or 129/STAT1−/− mice (c) after either mock infection with PBS or MERS-CoV at two inoculum doses. (d, e) Lung homogenate from infected mice in (a–c) was assayed for the presence of virus at multiple time points after infection. Dotted line notes the level of detection in our TCID50 assays. RNA was extracted from mouse lungs during infection and assayed for amount of viral RNA present during infection by real-time PCR analysis of the envelope ORF (f, g) or 1B(h, i). RNA from BALB/c mice (f, h) and RNA from 129Sv/Ev and 129/STAT1−/− mice (g, i) were assayed. Three mice at each time point and condition were used for each averaged value. Primers specific to the leader primer of MERS-CoV were used for real-time PCR analysis to identify subgenomic RNA as a reporter of viral replication (j, k). *P-value >0.5, P-value <0.5.
We next tested MERS-CoV-infected mouse lungs for the presence of virus by TCID50 assay. Briefly, VeroE6 cells were seeded into 96-well plates (USA Scientific) at 1×104 cells per well and were cultured overnight. Cells were infected with a fivefold dilution series of virus-containing media in triplicate and cultured for a further 48 h. Plates were fixed in 4 % paraformaldehyde for 5 min at room temperature and then stained with 0.05 % crystal violet in 20 % methanol for 30 min at room temperature. Plates were then washed twice in water and cell death was assessed by the presence (live cells) or absence (dead cells) of crystal violet stain. Viral load was calculated based on the dilution at which the media killed 50 % of the cells using the TCID50 formula (Reed & Muench, 1938).
Lungs from MERS-CoV-infected BALB/c, 129S6/SvEv and 129/STAT1−/− mice were assayed by TCID50 virus growth assay to assess whether virus was replicating and persisting during infection. We found that there was no detectable virus above the level of detection in the lungs of BALB/c mice at 2 and 4 days p.i. nor 129S6/SvEv and 129/STAT1−/− mice at 2 and 9 days p.i. (Fig. 1d, e).
We also tested for both genomic and actively transcribing MERS-CoV RNA in lungs during infection by real-time PCR with primers specific for genomic (Corman et al., 2012b) and replicating MERS-CoV RNA by homogenizing lung tissue in Trizol (Sigma Aldrich) for RNA extraction. For this purpose, we used two different primer sets for detection of MERS-CoV genomic RNA, corresponding to ORF1B and MERS-E (MERS-CoV-E forward: GCAACGCGCGATTCAGTT; MERS-CoV-E reverse: GCCTCTACACGGGACCCATA; MERS-CoV-ORF1B forward: TTCGATGTTGAGGGTGCTCAT; and MERS-CoV-ORF1B reverse: TCACACCAGTTGAAAATCCTAATT) (Corman et al., 2012a). We also assayed for actively transcribing MERS-CoV RNA by assaying with primers to the leader sequence that will only recognize MERS-CoV subgenomic mRNA due to the presence of the specific leader sequence found only in an actively transcribed MERS-CoV leader sequence containing mRNA (forward: CTATCTCACTTCCCCTCGTTCTC; reverse: GAGGGGGTTTACTATCCTGG). Mouse actin primers (forward: ATGGAGGGGAATACAGCCC; reverse: TTCTTTGCAGCTCCTTCGTT) were used as a normalization control.
RNA was extracted from lungs of WT BALB/c, 129S6/SvEv and 129/STAT1−/− mice at the time points noted above and converted into randomly primed cDNA using Revertaid Reverse Transcriptase (Thermo Scientific) according to the manufacturer’s instructions. When cDNA samples were probed for MERS-E (Fig. 1f, g) or MERS-ORF1B (Fig. 1h, i) transcripts, which probe for only viral genomic RNA levels, there were significantly higher levels compared to mock infected controls. This indicates that MERS-CoV genome is present in the lungs of infected mice early in infection. However, when the same samples were probed for MERS-CoV leader sequence containing mRNA, which assays for replicating virus, there were no statistically significant sustained levels of MERS-mRNA detected above background mRNA of PBS inoculated mice (Fig. 1j, k), suggesting that there is minimal active MERS-CoV transcription occurring in the lungs of WT or innate immune-deficient mice and it does not progress to a continuous production of infectious virus.
We analysed lung histology of each mouse strain to assess whether the mice displayed any histological signs of responding and, potentially, clearing of the infection. Paraformaldehyde-fixed lungs were embedded in paraffin and sectioned before staining with haematoxylin and eosin (H&E) (Fig. 2). Analysis of the lungs shows that in 129S6/SvEv and the innate immune-deficient 129/STAT1−/− mice there are only minor signs of pathological lesions or inflammatory response to the infection (Fig. 2b). Lungs of 129/STAT1−/− mice infected with the high dose of MERS-CoV displayed a few lesions of focal interstitial pneumonitis composed of neutrophils and macrophages at 9 days p.i., but not consistently in all mice. In BALB/c mice infected with the high dose of MERS-CoV, we noted perivascular cuffing with scattered neutrophils at 2 and 4 days p.i. with foci of pneumonia around proximal airways more predominant at 4 days p.i. (Fig. 2a). The histological features observed are consistent with immune response to antigen present in the inoculum; however, no cytopathic effect nor signs of MERS-CoV infection (apoptotic cells, syncytia formation) were noted.
Fig. 2.
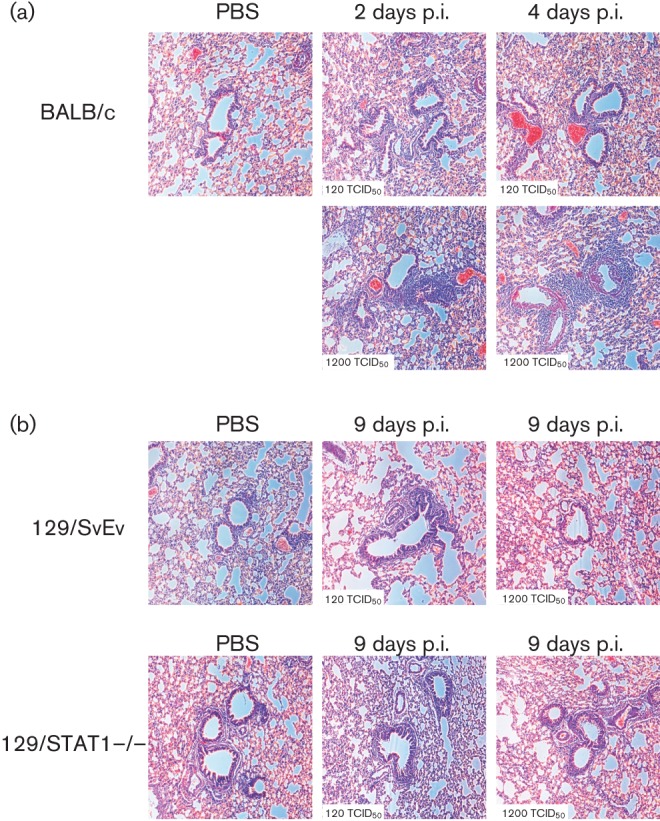
Histological analysis of MERS-CoV infected mice. (a) Histology of H&E stained lungs of BALB/c mice infected with MERS-CoV at 2 and 4 days p.i. (b) Histology of H&E stained lungs of 128/SvEv and 129/STAT1−/− mice infected with MERS-CoV at 9 days p.i.
The lack of MERS-CoV replication in mice could be explained by a mouse host factor that inhibits MERS-CoV from replicating in cells. MERS-CoV spike may not be able to bind to mouse DPP4 and allow for infection or DPP4 protein could not be expressed in the lungs of mice. We assayed for the expression of DPP4 mRNA and protein in mice using antibodies specific for mouse DPP4 (Biolegend, Cat. 137801) and real-time PCR to assess the expression level of DPP4 in mouse lung. As a positive control, we labelled 4 % paraformaldehyde-fixed and paraffin-embedded BALB/c mouse intestine with the anti-DPP4 antibody. In control sections with no primary antibody we see no labelling; however, in sections labelled with anti-DPP4 we see strong labelling in intestinal brush border cells (Fig. 3a). When we used the same antibody on BALB/c lungs, we find very little positive staining in Clara cells or type II alveolar cells, which is where DPP4 is expressed in human lung (Fig. 3b). Additionally, we assayed for DPP4 mRNA levels in the intestines and lungs of BALB/c mice (Fig. 3c). We find low levels of DPP4 in lung and significantly more DPP4 in the intestines. While the differential levels are not to the extent seen by immunohistochemical (IHC) staining in Fig. 3a, we hypothesize that the stability of DPP4 mRNA may diminish the signal in both tissues compared to protein levels. This suggests that if there are no other mouse factors inhibiting MERS-CoV infection and replication, then the development of a transgenic mouse model expressing the human DPP4 receptor in the homologous cell type as that expressed in humans would be able to create a small-animal model for MERS-CoV.
Fig. 3.
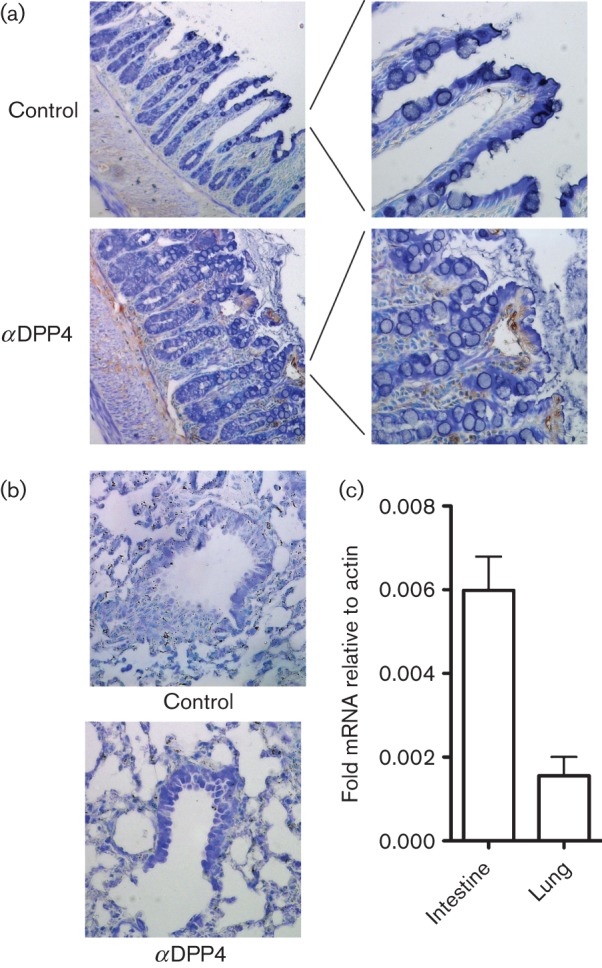
DPP4 expression levels in mice. IHC staining of BALB/c mouse intestines (a) and lung (b) was performed with anti-DPP4 antibody at 1/100 dilution using antigen retrieval. We find significant levels of DPP4 protein expression in mouse intestines compared to secondary only control tissue; however, in mouse lungs we observe minimal DPP4 staining in airways or alveolar cells. (c) Real-time PCR analysis of DPP4 mRNA from intestines and lungs of mice compared to actin mRNA levels as a control.
In this study we aimed to identify whether MERS-CoV replicated in WT or innate immune-deficient mice. We were unable to detect any significant MERS-CoV replication in any of the mouse strains used. Using STAT1 deletion 129 mice demonstrates that the lack of replication and MERS-CoV in 129S6/SvEv mice is not due to STAT1-dependent innate immune responses. The doses used for inoculations were lower than the maximum to assay for relevant levels of inoculation; however, they were still higher than a hypothetical physiological dose. Furthermore, while there was some evidence of a focal inflammatory response in mouse lungs, there was no recapitulation of the human disease in any of these mouse strains. Further studies are needed to identify whether transgenic mice expressing the human DPP4 protein in the correct cell type will allow the development of a small-animal model for this novel CoV.
Acknowledgements
We thank the members of the Frieman laboratory for their helpful advice and assistance. This work was supported by National Institutes of Health grant NIH RO1 AI 095569-01 to M. B. F.
References
- Chan J. F., Chan K. H., Choi G. K., To K. K., Tse H., Cai J. P., Yeung M. L., Cheng V. C., Chen H. & other authors (2013). Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: implications for disease pathogenesis and clinical manifestation. J Infect Dis 207, 1743–1752 10.1093/infdis/jit123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V. M., Eckerle I., Bleicker T., Zaki A., Landt O., Eschbach-Bludau M., van Boheemen S., Gopal R., Ballhause M. & other authors (2012a). Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Eurosurveillance 17, 39. [DOI] [PubMed] [Google Scholar]
- Corman V. M., Muller M. A., Costabel U., Timm J., Binger T., Meyer B., Kreher P., Lattwein E., Eschbach-Bludau M. & other authors (2012b). Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Eurosurveillance 17, 49. [DOI] [PubMed] [Google Scholar]
- de Wit E., Prescott J., Baseler L., Bushmaker T., Thomas T., Lackemeyer M. G., Martellaro C., Milne-Price S., Haddock E. & other authors (2013). The Middle East respiratory syndrome coronavirus (MERS-CoV) does not replicate in Syrian hamsters. PLoS ONE 8, e69127 10.1371/journal.pone.0069127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Seilmaier M., Corman V. M., Hartmann W., Scheible G., Sack S., Guggemos W., Kallies R., Muth D. & other authors (2013). Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect Dis 13, 745–751 10.1016/S1473-3099(13)70154-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M. B., Chen J., Morrison T. E., Whitmore A., Funkhouser W., Ward J. M., Lamirande E. W., Roberts A., Heise M. & other authors (2010). SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism. PLoS Pathog 6, e1000849 10.1371/journal.ppat.1000849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler E., Jónsdóttir H. R., Muth D., Hamming O. J., Hartmann R., Rodriguez R., Geffers R., Fouchier R. A., Drosten C. & other authors (2013). Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoo-notic potential. mBio 4, e00611–e00612 10.1128/mBio.00611-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailles A., Blanckaert K., Chaud P., van der Werf S., Lina B., Caro V., Campese C., Guéry B., Prouvost H. & other authors (2013). First cases of Middle East Respiratory Syndrome Coronavirus (MERS-CoV) infections in France, investigations and implications for the prevention of human-to-human transmission, France, May 2013. Eurosurveillance 18, 24. [PubMed] [Google Scholar]
- Memish Z. A., Zumla A. I., Al-Hakeem R. F., Al-Rabeeah A. A., Stephens G. M. (2013). Family cluster of Middle East respiratory syndrome coronavirus infections. N Engl J Med 368, 2487–2494 10.1056/NEJMoa1303729 [DOI] [PubMed] [Google Scholar]
- Müller M. A., Raj V. S., Muth D., Meyer B., Kallies S., Smits S. L., Wollny R., Bestebroer T. M., Specht S. & other authors (2012). Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines. MBio 3, e00515-12 10.1128/mBio.00515-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V. J., de Wit E., Feldmann H. (2013). Pneumonia from human coronavirus in a macaque model. N Engl J Med 368, 1560–1562 10.1056/NEJMc1215691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R., Wang P., Gomaa M., El-Shesheny R., Kandeil A., Bagato O., Siu L., Shehata M., Kayed A. & other authors (2013). Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Eurosurveillance 18, 18 10.2807/1560-7917.ES2013.18.36.20574 [DOI] [PubMed] [Google Scholar]
- Raj V. S., Mou H., Smits S. L., Dekkers D. H., Müller M. A., Dijkman R., Muth D., Demmers J. A., Zaki A. & other authors (2013). Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495, 251–254 10.1038/nature12005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L. J., Muench H. (1938). A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27, 493–497 [Google Scholar]
- Reusken C. B., Haagmans B. L., Müller M. A., Gutierrez C., Godeke G. J., Meyer B., Muth D., Raj V. S., Vries L. S. & other authors (2013). Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis 13, 859–866 10.1016/S1473-3099(13)70164-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Deming D., Paddock C. D., Cheng A., Yount B., Vogel L., Herman B. D., Sheahan T., Heise M. & other authors (2007). A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog 3, e5 10.1371/journal.ppat.0030005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T., Morrison T. E., Funkhouser W., Uematsu S., Akira S., Baric R. S., Heise M. T. (2008). MyD88 is required for protection from lethal infection with a mouse-adapted SARS-CoV. PLoS Pathog 4, e1000240 10.1371/journal.ppat.1000240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boheemen S., de Graaf M., Lauber C., Bestebroer T. M., Raj V. S., Zaki A. M., Osterhaus A. D., Haagmans B. L., Gorbalenya A. E. & other authors (2012). Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio 3, e00473-12 10.1128/mBio.00473-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielecki F., Weber M., Eickmann M., Spiegelberg L., Zaki A. M., Matrosovich M., Becker S., Weber F. (2013). Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J Virol 87, 5300–5304 10.1128/JVI.03496-12 [DOI] [PMC free article] [PubMed] [Google Scholar]