Abstract
Genomic and antigenic characterization of members of the Sandfly fever Naples virus (SFNV) complex reveals the presence of five clades that differ in their geographical distribution. Saint Floris and Gordil viruses, both found in Africa, form one clade; Punique, Granada and Massilia viruses, all isolated in the western Mediterranean, constitute a second; Toscana virus, a third; SFNV isolates from Italy, Cyprus, Egypt and India form a fourth; while Tehran virus and a Serbian isolate Yu 8/76, represent a fifth. Interestingly, this last clade appears not to express the second non-structural protein ORF. Karimabad virus, previously classified as a member of the SFNV complex, and Gabek Forest virus are distinct and form a new species complex (named Karimabad) in the Phlebovirus genus. In contrast with the high reassortment frequency observed in some South American phleboviruses, the only virus of the SFNV complex with evidence of reassortment was Granada virus.
The more than 350 named RNA viruses included in the family Bunyaviridae are divided into five genera: Orthobunyavirus, Nairovirus, Hantavirus, Phlebovirus and Tospovirus (Nichol et al., 2005). Bunyavirus genomes range from 11–19 kilobases in length and comprise three unique molecules of negative or ambisense ssRNA, designated L (large), M (medium) and S (small). Viruses in each genus share similar segment and structural protein sizes and have characteristic terminal sequences at the 3′ and 5′ ends of each segment. Similar to other segmented virus families, genetic reassortment has been demonstrated among related bunyaviruses both in vitro and in vivo (Henderson et al., 1995; Li et al., 1995; Pringle et al., 1984; Rodriguez et al., 1998).
Except for the tospoviruses, which only infect plants, human pathogens are found in each of the other four genera. At present, the genus Phlebovirus comprises approximately 70 named viruses that are classified based on their antigenic, genomic and/or vector relationships into two groups: the Sandfly fever group and the Uukuniemi group (Nichol et al., 2005). Viruses in the Sandfly fever group are transmitted by phlebotomine sandflies and mosquitoes; the Uukuniemi group viruses are tick-borne. Until recently, viruses in the Sandfly fever group were thought to be the only phleboviruses of public health or veterinary importance; however, three new Uukuniemi group viruses, severe fever with thrombocytopenia syndrome virus (Yu et al., 2011), Heartland virus (McMullan et al., 2012) and Bhanja virus (Matsuno et al., 2013) were recently implicated in human disease. Phylogenetic analysis of these and other bunyaviruses has suggested the existence of a third distinct lineage (group) within the genus Phlebovirus, which is composed of Gouleako virus (Marklewitz et al., 2011), and a second mosquito virus (Cumuto) from Trinidad (Auguste et al., in press).
Seven named viruses are currently included within the Sandfly fever Naples species complex, based on their antigenic relationships: Sandfly fever Naples virus (SFNV), Toscana (TOSV), Massilia (MASV), Tehran virus (TEHV), Karimabad (KARV), Granada (GRV) and Punique (PUNV) (Nichol et al., 2005). TOSV infection is a relatively common cause of meningitis and encephalitis in travellers to and residents of the Mediterranean region (Charrel et al., 2005). SFNV is widely distributed in the eastern Mediterranean and Central Asia where it causes an acute febrile illness known as ‘phlebotomus, sandfly or papatacci fever’ that is characterized by severe headache, myalgia, conjunctival injection, malaise, nausea and vomiting, and a marked leucopenia of 3–5 days duration (Tesh, 1988). The other five viruses in the species complex (MASV, TEHV, KARV, GRV and PUNV) have only been isolated from phlebotomine sandflies and have not yet been associated with human disease, although there is serological evidence that some of them also infect people (Collao et al., 2010; Tesh et al., 1976, 1977).
Because of the public health importance of viruses in genus Phlebovirus and in an effort to develop a more precise taxonomic system for the phleboviruses, we have attempted to sequence all of the available named viruses in the genus to clarify their phylogenetic relationships. This is the fifth of a series of publications describing this work (Palacios et al., 2011a, b, 2013a, b), and it covers members of the SFNV species complex.
Viruses used in this study were obtained from the World Reference Center for Emerging Viruses and Arboviruses at the University of Texas Medical Branch. Table 1 provides the names, strain numbers, sources, and dates and locality of isolation, and GenBank accession numbers. In addition to the seven Naples complex viruses noted before, three other tentative members, Saint Floris (SAFV), Gordil (GORV) and Gabek Forest (GFV) viruses, were also included.
Table 1. Names, abbreviations, strain numbers, sources, dates and locality of isolation, and accession numbers of the viruses used in this study.
| Virus name | Abbreviation | Strain | Year of isolation | Source of isolate, location | Area of circulation (isolation, seroprevalence) | Accession numbers* |
| Saint Floris | SAFV | Dak ANBR 512d | 1971 | Tatera sp., Central African Republic | Central African Republic, Somalia, Sudan, Egypt | JF920136 (L) JF920137 (M) JF920138 (S) |
| Gordil virus | GORV | Dak ANBR 496d | 1971 | Lemniscomys striatus, Central African Republic | Central African Republic, Somalia, Sudan | KF297900 (L) KF297901 (M) KF297902 (S) |
| Sandfly Fever Naples | SFNV | R3 | 1985 | Human serum, Cyprus | Cyprus, India, Italy, Turkey, Israel, Egypt, Bangladesh, Iran, Sudan, Ethiopia, Iraq, Pakistan, Turkmenistan, Azerbaijan, Saudi Arabia, Somalia | HM566183 (L) HM566184 (M) HM566182 (S) |
| Sandfly Fever Naples | SFNV | Poona 701795 | 1971 | Human serum, India | HM566176 (L) HM566177 (M) HM566178 (S) | |
| Sandfly Fever Naples | SFNV | Sabin | 1944 | Human serum, Italy | JF939843, HM566169 (L) JF939844, HM566171 (M) JF939845, HM566170 (S) | |
| Sandfly Fever Naples | SFNV | NAMRU 840055 | 1985 | Human serum, Egypt | HM566167 (L) HM566169 (M) HM566168 (S) | |
| Sandfly Fever Naples | SFNV | Yu 8/76 | 1976 | Phlebotomus perfiliewi, Serbia | Serbia | JF920139 (L) JF920140 (M) JF920141 (S) |
| Tehran virus | TEHV | I-47 | 1959 | Phlebotomus papatasi, Iran | Iran | JF939846 (L) JF939847 (M) JF939848 (S) |
| Granada | GRV | GRV25 | 2004 | Phlebotomus perniciosus, man, Spain | Spain | GU135606 (L) GU135607 (M) GU135608 (S) |
| Massilia | MASV | W | 2005 | Phlebotomus spp., France | France | EU725771 (L) EU725772 (M) EU725773 (S) |
| Punique | PUNV | P1/B4 | 2008 | Phlebotomus perniciosus Phlebotomus longicuspis | Tunis | FJ848987 (L) FJ848988 (M) FJ848989 (S) |
| Toscana | TOSV A | ISS.Phl.3 | 1984 | Phlebotomus perniciosus, man, Italy | Italy, France, Spain, Greece, Portugal, Morocco, Tunisia, Bosnia-Herzegovina, Croatia, Kosovo, Turkey | X68414 (L) X89628 (M) X53794 (S) |
| Toscana | TOSV B | H/MTSSA | FJ153281 (L) Fj153283 (M) Fj153282 (S) | |||
| Karimabad | KARV | I-58 | 1959 | Phlebotomus spp., Iran | Iran, Azerbaijan, Uzbekistan, Turkmenistan | KF297912 (L) KF297913 (M) KF297914 (S) |
| Karimabad | KARV | 91019-P | 1975 | Phlebotomus papatasi, Iran | Kyngyzstan, Tajikstan, Russia | KF297906 (L) KF297907 (M) KF297908 (S) |
| Karimabad | KARV | 91045-AG | 1975 | Phlebotomus papatasi, Iran | KF297909 (L) KF297910 (M) KF297911 (S) | |
| Gabek Forest | GFV | Sud AN 754-61 | 1961 | Acomys cahirinus, Sudan | Sudan, Senegal, Cent. African Rep., Nigeria, Benin | KF297903 (L) KF297904 (M) KF297905 (S) |
Bold font highlights sequences obtained in this work.
Antisera for use in serological tests were prepared in adult mice as described previously (Palacios et al., 2011b). The immunization schedule consisted of four intraperitoneal injections of mouse antigen mixed with Freund’s adjuvant, given at weekly intervals. After the final immunization, mice were inoculated with sarcoma 180 cells, and the resulting immune ascitic fluids were collected.
Complement fixation (CF) tests were performed by the microtitre technique (Beaty et al., 1989), using 2 units of guinea pig complement and overnight incubation of the antigens and antibodies at 4 °C. Haemagglutination-inhibition (HI) tests were also performed in microtitre plates, using 4 units of antigen and overnight incubation at 4 °C. Antigens used in the serological tests were prepared from infected newborn mouse brain by the sucrose acetone extraction method (Clarke & Casals, 1958) and were inactivated with 0.05 % β-propriolactone (Sigma) or by gamma irradiation. CF titres were recorded as the highest dilutions giving 3+ or 4+ fixation of complement on a scale of 0 to 4+. In HI tests using 4 units of antigen, antibody titres of 1 : 20 or greater were considered positive.
Results of CF and HI tests are shown in Table 2. By both tests SFNV, TEHV, TOSV and PUNV are closely related antigenically. In fact, except for PUNV, they are indistinguishable by CF test. Likewise, GORV and SAFV are closely related, although they can be differentiated from each other. Both GORV and SAFV also show considerable cross-reactivity with SFNV, TEHV, TOSV and PUNV, indicating that they belong to the Sandfly fever Naples species complex as well. In contrast, KARV and GFV are more distantly related antigenically to the other six viruses. KARV and GFV show some cross-reactivity with most of the Sandfly fever Naples complex viruses in HI tests, but not by CF test. KARV and GFV are more closely related to each other but are also distinct by these serological tests. Given this reactivity pattern, they appear to constitute a novel species complex within the genus Phlebovirus.
Table 2. Results of complement fixation (CF) and haemagglutination-inhibition (HI) tests.
| Antibody | Complement fixation test* | Haemagglutination-inhibition test† | ||||||||||||||
| Antigen | Antigen 4 units | |||||||||||||||
| SFN | TEH | TOS | PUN | GOR | SAF | KAR | GF | SFN | TEH | TOS | PUN | GOR | SAF | KAR | GF | |
| SFN | 256/32 | 256/32 | 256/32 | 64/32 | 0 | 0 | 0 | 0 | 1 : 320 | 1 : 160 | 1 : 80 | 1 : 80 | 1 : 40 | 1 : 40 | 1 : 40 | 1 : 40 |
| TEH | 64/128 | 256/128 | 128/32 | 32/32 | 0 | 0 | 0 | 0 | 1 : 2560 | 1 : 2560 | 1 : 320 | 1 : 320 | 1 : 320 | 1 : 320 | 1 : 160 | 1 : 40 |
| TOS | 64/128 | 256/128 | 256/128 | 32/32 | 0 | 0 | 0 | 0 | 1 : 640 | 1 : 640 | 1 : 5120 | 1 : 320 | 1 : 320 | 1 : 320 | 1 : 160 | 1 : 160 |
| PUN | 256/≥8 | 256/≥8 | 128/≥8 | 256/≥8 | 0 | 0 | 0 | 0 | 1 : 640 | 1 : 160 | 1 : 40 | 1 : 2560 | 1 : 40 | 1 : 80 | 1 : 40 | 1 : 20 |
| GOR | 0 | 16/8 | 32/8 | 16/≥8 | 512/≥128 | 32/≥128 | 0 | 0 | 1 : 640 | 1 : 640 | 1 : 640 | 1 : 640 | 1 : 5120 | 1 : 640 | 1 : 40 | 1 : 40 |
| SAF | 0 | 8/8 | 16/8 | 16/≥8 | 16/≥128 | 512/≥128 | 0 | 0 | 1 : 640 | 1 : 320 | 1 : 320 | 1 : 320 | 1 : 640 | 1 : 2560 | 0 | 0 |
| KAR | 0 | 0 | 0 | 0 | 0 | 0 | 32/128 | 0 | 1 : 40 | 1 : 20 | 1 : 40 | 1 : 40 | 0 | 1 : 20 | 1 : 320 | 1 : 160 |
| GF | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 256/128 | 1 : 160 | 1 : 40 | 1 : 80 | 1 : 40 | 0 | 1 : 40 | 1 : 640 | 1 : 1280 |
SFN, Sandfly fever Naples (Sabin strain); TEH, Tehran; TOS, Toscana; PUN, Punique; KAR, Karimabad (I-58); GF, Gabek Forest; GOR, Gordil; SAF, Saint Floris.
CF titres are expressed as the highest antibody dilution/highest antigen dilution, 0 = <8/8. Bold numbers represent antibody reactivity with self-antigens.
HI titres are expressed as the highest positive antibody dilution, 0 = <1 : 20.
Genome sequencing was performed as previously described (Cox-Foster et al., 2007; Palacios et al., 2008, 2011b). For the termini of each segment, a primer with the 8 nt conserved sequence was used for a specific reverse transcription with an additional arbitrary nt on the 5′ end (5′-AAGCAGTGGTATCAACGCAGAGTACACACAAAG-3′) where the bold portion highlights the conserved region. This primer is designed to bind to the 3′ end of the genomic RNA and the 3′ of the mRNA. Sequences of the genomes were verified by classical dideoxy sequencing, using primers designed from the draft sequence to create products of 1000 bp with 500 bp overlaps.
Geneious 4.8.3 (Biomatters) was used for sequence assembly and analysis. Topology and targeting predictions were generated by employing SignalP, NetNGlyc, TMHMM (http://www.cbs.dtu.dk/services), the web-based version of TopPred2 (http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html), and integrated predictions in Geneious (Bendtsen et al., 2004; Claros & von Heijne, 1994; Kahsay et al., 2005; Käll et al., 2004; Krogh et al., 2001).
The genome organization of all the viruses in the Sandfly fever Naples antigenic complex is consistent with other members of the genus Phlebovirus. In total, the genomes encode six proteins: encoding an RNA polymerase (L segment), two glycoproteins and a non-structural protein (GN, GC and NSm; M segment), and the nucleocapsid protein (N) and, in an ambisense orientation, a second non-structural protein (NSs; S segment). Several regions of the RNA-dependent RNA polymerase overlap conserved regions found in all available phlebovirus sequences, confirming an association with function (Palacios et al., 2011a, b, 2013a, b). Signal sequences, transmembrane domains, cleavage sites for the cellular signal signallase protease and Golgi retention signals for the GN and GC are conserved in the majority of the viruses in the SFNV virus group. In general, no major differences are observed between SFNV and other previously described phlebovirus (Palacios et al., 2011a, b, 2013a, b). The only observation of note was the absence of a proper ORF for the NSs gene for SFNV strain YU8-76 and TEHV strain I-47. An early stop codon (aa position 215, strain Yu8-76) and a gene truncation (region deleted: 101–270 aa, strain I-47) are observed in the NSs genes of these two isolates.
For phylogenetic analysis, a set of phlebovirus sequences (131 for the L segment, 169 for the M segment, 191 for the N gene, and 146 for the NS gene) comprising all nt (partial or complete) sequences from GenBank available on May 1, 2013 were aligned, along with our sequences, using the clustal algorithm (as implemented in the mega package Version 5) at the aa level with additional manual editing to ensure the highest possible quality of alignment. Neighbour-joining (NJ) analysis at the aa level was performed due to the observed high variability of the underlying nt sequences. Given the saturation observed in all the alignments, the phylogenetic trees obtained by analysis of all members of the genus were used to define the species complexes, while additional phylogenetic analysis restricted to the SFNV sequences was used to resolve the fine topology of the species complex.
The statistical significance of tree topology was evaluated by bootstrap resampling of the sequences 1000 times. Phylogenetic analyses were performed by using mega software (Tamura et al., 2011).
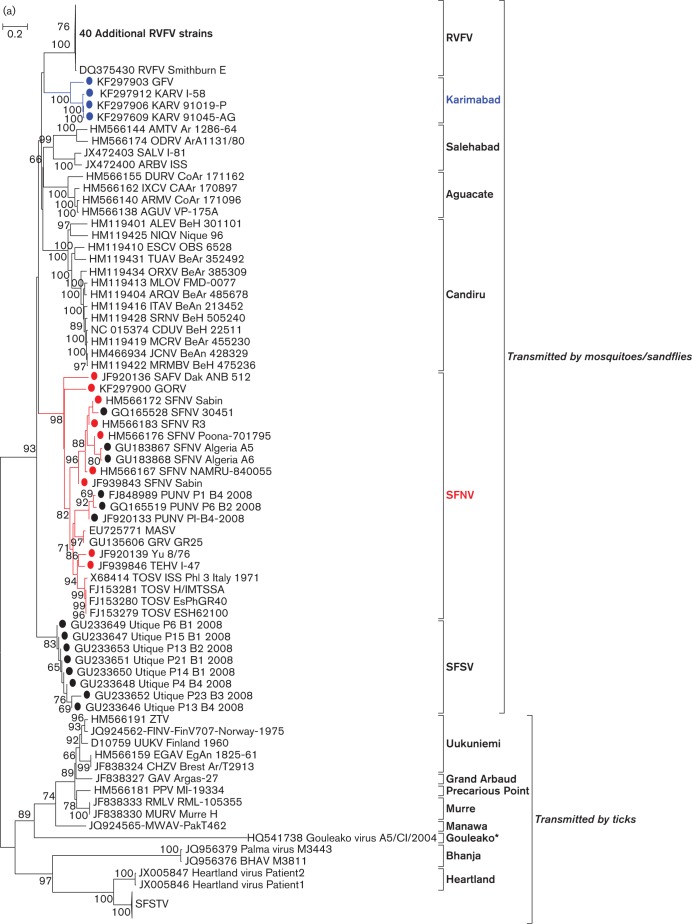
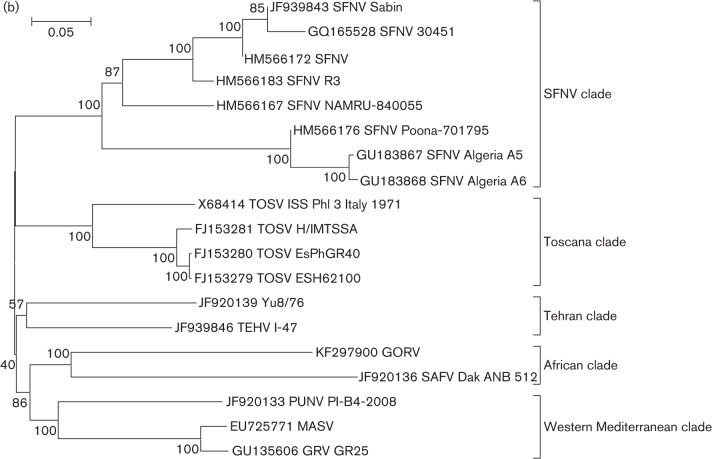
Phylogenetic analyses of the L, M and S gene segment sequences of the 12 new viruses (SAFV, GORV, SFNV strain R3, Poona 701795, Sabin, NAMRU 840055, Yu 8/76, TEHV strain I-47, KARV strain I-58, 91019-P and 91045-AG, and GFV) are consistent with earlier reports, confirming that viruses belonging to the same group cluster together (Charrel et al., 2009; Collao et al., 2010). As anticipated, based on their cross-reactivity in CF tests (Bishop et al., 1980), members of the SFNV species complex generally cluster together with the exception of KARV, which forms its own clade along with the previously uncharacterized GFV (Figs 1 and S1, available in JGV Online). Based on L-, M-, and S-segment sequences, the viruses that form a monophyletic clade with SFNV cluster into five groups: Toscana, Sandfly fever Naples, Yu 8/76/Tehran, Punique/Massilia/Granada and Gordil/Saint Floris. It appears that KARV and GFV show only distant evolutionary relationships with the other members of the SFNV serogroup. Both viruses formed a single clade that has similar levels of diversity compared to other phlebovirus species (data not shown).
Fig. 1.
(a)Phylogenetic analysis of the available sequences of phlebovirus L ORF. Sequences marked with black dots corresponded to partial sequences. Sequences marked with red dots (Sandfly fever Naples species complex) and blue dots (Karimabad species complex) corresponded to sequences obtained during this work. Only partial (when only available for the species) or complete ORF sequences were included in the analysis. Non-coding regions were excluded. *Gouleako virus was actually recovered from mosquitoes. (b) Phylogenetic analysis of all members of the Sandfly fever Naples species complex L segments. Bars indicate XXXX.
Systematic screening for the presence of recombination patterns was pursued by using the nt alignments and the Recombination Detection Program (RDP, (Martin & Rybicki, 2000) and Bootscan (Salminen et al., 1995), MaxChi (Smith, 1992), Chimaera (Posada & Crandall, 2001), LARD (Holmes, 1998) and phylip Plot (Felsenstein, 1989).
With the known exception of GRV (Collao et al., 2010), no evidence of SFNV reassortment was found in topological analysis of phylogenetic trees (Figs 2 and S2) or by RDP, Bootscan, MaxChi, LARD and phylip Plot analysis (data not shown).
Phylogenetic analysis of the complete genomes of the isolates of the Sandfly fever Naples species complex allowed us to identify geographical and genetic correlations. The African viruses SAFV and GORV, previously reported to be related based on serological criteria (Tesh et al., 1976, 1982), form one clade. GRV, MASV and PUNV, all recently discovered viruses detected in the western Mediterranean region, form a second phylogenetic cluster, suggesting a common ancestor. TOSV, a human pathogen detected throughout the Mediterranean region forms a third clade. The widely distributed SFNV isolates from Cyprus, Italy, Egypt and India form a fourth clade. TEHV and the Serbian isolate Yu 8/76 form a fifth clade. In this new classification scheme, strains R3, Poona 701795, Sabin and NAMRU 840055 would be representatives of the SFNV clade, while I-47 (TEHV) and Yu 8/76 would be representatives of a new TEHV clade. Interestingly, this last clade apparently does not require expression of the NSs ORF, since its replication is not impaired by the presence of either an early stop codon or a large truncation. NSs proteins have been reported to be an important determinant in the pathogenesis of phleboviruses (Billecocq et al., 2004; Bouloy et al., 2001; Ikegami et al., 2006; Sall et al., 1997; Vialat et al., 2000). The main supporting argument for this statement is the finding and description of a naturally attenuated RVFV strain (clone 13) that has a large in-frame deletion in the NSs coding region of the S segment (Muller et al., 1995). The truncated NSs protein of clone 13 is expressed and remains in the cytoplasm, where it is degraded rapidly by the proteasome.
KARV and GFV are distinct and form a new species complex within the Sandfly fever group. KARV was originally isolated in Iran in 1959 (Berge, 1975); subsequent KARV isolations and serological evidence of human infection have been reported from Iran, Azerbeijan, Uzbekistan, Turkmenistan, Kyrgyzstan, Tajikistan and Russia (Gaidamovich et al., 1991; Tesh et al., 1976, 1977). GFV was initially isolated in Sudan in 1961 but many subsequent isolates have been obtained in Nigeria, Benin, Senegal and Central African Republic (Kemp et al., 1974). Serological evidence of human infection with GFV has been reported in Sudan, Egypt and Nigeria (Tesh et al., 1976). Since KARV was the first of the two viruses to be isolated, Karimabad should be the species designation of this new complex.
Segment reassortment in bunyaviruses has been reported with increasing frequency, especially in the genus Orthobunyavirus (Bowen et al., 2001; Briese et al., 2006, 2007; Burt et al., 2009; Collao et al., 2010; Iroegbu & Pringle, 1981; Kondiah et al., 2010; Nunes et al., 2005; Saeed et al., 2001; Yanase et al., 2006, 2010). Previously, we reported that the frequency of reassortment in the Candiru species complex of the genus Phlebovirus (five of thirteen named viruses) was unprecedented (Palacios et al., 2011a). In contrast, our analysis of members of the Uukuniemi group did not indicate any reassortment events (Palacios et al., 2013b). With the exception of the aforementioned reassortment of GRV, no additional reassortment was detected among members of the SFNV species group.
The antigenic relatedness between KARV and some members of the SFNV species complex illustrates one of the problems of bunyavirus classification, using serological data from a single type of test (i.e. HI or CF). There is sometimes a poor correlation between results of CF and HI tests. In the case of the phleboviruses, the HI tends to be broadly reactive, whereas the CF test is more species-specific. The results of CF tests generally correlate more closely with the genetic data. The relationship of KARV with SFNV is an example. Other examples of this discordance within the genus Phlebovirus are the relationships between the Uukuniemi, Bhanja and Severe fever with thrombocytopenia species groups (Matsuno et al., 2013; Palacios et al., 2013b), and the relationships observed within the Salehabad species group (Palacios et al., 2013a). For this reason, we previously suggested that a classification system for the family Bunyaviridae should be based on genetic as well as serological (CF) data (Palacios et al., 2013b).
Acknowledgements
This work was supported by the Defense Threat Reduction Agency Project no. 1881290, and the United States Department of Defense, Google.org, National Institutes of Health award AI57158 (Northeast Biodefense Center - Lipkin), and USAID PREDICT funding source code 07-301-7119-52258 (Center for Infection and Immunity). R. T., A. T. R. and H. G. were supported by NIH contract HHSN272201000040I/HHSN27200004/DO4.
Footnotes
Two supplementary figures are available with the online version of this paper.
References
- Auguste A. J., Carrington C. V. F., Forrester N. L., Popov V. L., Guzman H., Widen S. G., Wood T. G., Weaver S. C., Tesh R. B. Characterization of a novel Negevirus and a novel Bunyavirus isolated from Culex declaratory mosquitoes in Trinidad. J Gen Virol (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty B., Calisher C. H., Shope R. E. (1989). Arboviruses. In Diagnostic Procedures for Viral, Rickettsial and Chlamydial Infections, pp. 979–1005 Edited by Schmidt N. J., Emmons R. W. Washington, DC: American Public Health Association [Google Scholar]
- Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004). Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340, 783–795 10.1016/j.jmb.2004.05.028 [DOI] [PubMed] [Google Scholar]
- Berge T. (1975). International Catalogue of Arboviruses Including Certain Other Viruses of Vertebrates. Atlanta: CDC; [DOI] [PubMed] [Google Scholar]
- Billecocq A., Spiegel M., Vialat P., Kohl A., Weber F., Bouloy M., Haller O. (2004). NSs protein of Rift Valley Fever Virus blocks interferon production by inhibiting host gene transcription. J Virol 78, 9798–9806 10.1128/JVI.78.18.9798-9806.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D. H., Calisher C. H., Casals J., Chumakov M. P., Gaidamovich S. Y., Hannoun C., Lvov D. K., Marshall I. D., Oker-Blom N. & other authors (1980). Bunyaviridae. Intervirology 14, 125–143 10.1159/000149174 [DOI] [PubMed] [Google Scholar]
- Bouloy M., Janzen C., Vialat P., Khun H., Pavlovic J., Huerre M., Haller O. (2001). Genetic evidence for an interferon-antagonistic function of Rift Valley Fever Virus nonstructural protein NSs. J Virol 75, 1371–1377 10.1128/JVI.75.3.1371-1377.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen M. D., Trappier S. G., Sanchez A. J., Meyer R. F., Goldsmith C. S., Zaki S. R., Dunster L. M., Peters C. J., Ksiazek T. G., Nichol S. T., RVF Task Force (2001). A reassortant bunyavirus isolated from acute hemorrhagic fever cases in Kenya and Somalia. Virology 291, 185–190 10.1006/viro.2001.1201 [DOI] [PubMed] [Google Scholar]
- Briese T., Bird B., Kapoor V., Nichol S. T., Lipkin W. I. (2006). Batai and Ngari viruses: M segment reassortment and association with severe febrile disease outbreaks in East Africa. J Virol 80, 5627–5630 10.1128/JVI.02448-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese T., Kapoor V., Lipkin W. I. (2007). Natural M-segment reassortment in Potosi and Main Drain viruses: implications for the evolution of orthobunyaviruses. Arch Virol 152, 2237–2247 10.1007/s00705-007-1069-z [DOI] [PubMed] [Google Scholar]
- Burt F. J., Paweska J. T., Ashkettle B., Swanepoel R. (2009). Genetic relationship in southern African Crimean-Congo haemorrhagic fever virus isolates: evidence for occurrence of reassortment. Epidemiol Infect 137, 1302–1308 10.1017/S0950268808001878 [DOI] [PubMed] [Google Scholar]
- Charrel R. N., Gallian P., Navarro-Mari J. M., Nicoletti L., Papa A., Sánchez-Seco M. P., Tenorio A., de Lamballerie X. (2005). Emergence of Toscana virus in Europe. Emerg Infect Dis 11, 1657–1663 10.3201/eid1111.050869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrel R. N., Moureau G., Temmam S., Izri A., Marty P., Parola P., da Rosa A. T., Tesh R. B., de Lamballerie X. (2009). Massilia virus, a novel Phlebovirus (Bunyaviridae) isolated from sandflies in the Mediterranean. Vector Borne Zoonotic Dis 9, 519–530 10.1089/vbz.2008.0131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke D. H., Casals J. (1958). Techniques for hemagglutination and hemagglutination-inhibition with arthropod-borne viruses. Am J Trop Med Hyg 7, 561–573 [DOI] [PubMed] [Google Scholar]
- Claros M. G., von Heijne G. (1994). TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci 10, 685–686 [DOI] [PubMed] [Google Scholar]
- Collao X., Palacios G., de Ory F., Sanbonmatsu S., Pérez-Ruiz M., Navarro J. M., Molina R., Hutchison S. K., Lipkin W. I. & other authors (2010). Granada virus: a natural phlebovirus reassortant of the sandfly fever Naples serocomplex with low seroprevalence in humans. Am J Trop Med Hyg 83, 760–765 10.4269/ajtmh.2010.09-0697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox-Foster D. L., Conlan S., Holmes E. C., Palacios G., Evans J. D., Moran N. A., Quan P. L., Briese T., Hornig M. & other authors (2007). A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318, 283–287 10.1126/science.1146498 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1989). phylip - Phylogeny Inference Package (Version 3.2). Cladistics 5, 164–166 [Google Scholar]
- Gaidamovich S., Khutoretskaya N., Asyamov Y. V., Tsyupa V. I., Melnikova E. (1991). Sandfly fever in Central Asia and Afghanistan. In Hemorrhagic Fever with Renal Syndrome, Tick- and Mosquito-Borne Viruses, pp. 287–293 Edited by Calisher C. H. Springer: Vienna [Google Scholar]
- Henderson W. W., Monroe M. C., St Jeor S. C., Thayer W. P., Rowe J. E., Peters C. J., Nichol S. T. (1995). Naturally occurring Sin Nombre virus genetic reassortants. Virology 214, 602–610 10.1006/viro.1995.0071 [DOI] [PubMed] [Google Scholar]
- Holmes E. C. (1998). Molecular epidemiology of dengue virus–the time for big science. Trop Med Int Health 3, 855–856 10.1046/j.1365-3156.1998.00332.x [DOI] [PubMed] [Google Scholar]
- Ikegami T., Won S., Peters C. J., Makino S. (2006). Rescue of infectious Rift Valley fever virus entirely from cDNA, analysis of virus lacking the NSs gene, and expression of a foreign gene. J Virol 80, 2933–2940 10.1128/JVI.80.6.2933-2940.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iroegbu C. U., Pringle C. R. (1981). Genetic interactions among viruses of the Bunyamwera complex. J Virol 37, 383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahsay R. Y., Gao G., Liao L. (2005). An improved hidden Markov model for transmembrane protein detection and topology prediction and its applications to complete genomes. Bioinformatics 21, 1853–1858 10.1093/bioinformatics/bti303 [DOI] [PubMed] [Google Scholar]
- Käll L., Krogh A., Sonnhammer E. L. (2004). A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338, 1027–1036 10.1016/j.jmb.2004.03.016 [DOI] [PubMed] [Google Scholar]
- Kemp G. E., Causey O. R., Setzer H. W., Moore D. L. (1974). Isolation of viruses from wild mammals in West Africa, 1966–1970. J Wildl Dis 10, 279–293 [DOI] [PubMed] [Google Scholar]
- Kondiah K., Swanepoel R., Paweska J. T., Burt F. J. (2010). A Simple-probe real-time PCR assay for genotyping reassorted and non-reassorted isolates of Crimean-Congo hemorrhagic fever virus in southern Africa. J Virol Methods 169, 34–38 10.1016/j.jviromet.2010.06.010 [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305, 567–580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Li D., Schmaljohn A. L., Anderson K., Schmaljohn C. S. (1995). Complete nucleotide sequences of the M and S segments of two hantavirus isolates from California: evidence for reassortment in nature among viruses related to hantavirus pulmonary syndrome. Virology 206, 973–983 10.1006/viro.1995.1020 [DOI] [PubMed] [Google Scholar]
- Marklewitz M., Handrick S., Grasse W., Kurth A., Lukashev A., Drosten C., Ellerbrok H., Leendertz F. H., Pauli G., Junglen S. (2011). Gouleako virus isolated from West African mosquitoes constitutes a proposed novel genus in the family Bunyaviridae. J Virol 85, 9227–9234 10.1128/JVI.00230-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D., Rybicki E. (2000). RDP: detection of recombination amongst aligned sequences. Bioinformatics 16, 562–563 10.1093/bioinformatics/16.6.562 [DOI] [PubMed] [Google Scholar]
- Matsuno K., Weisend C., Travassos da Rosa A. P., Anzick S. L., Dahlstrom E., Porcella S. F., Dorward D. W., Yu X. J., Tesh R. B., Ebihara H. (2013). Characterization of the Bhanja serogroup viruses (Bunyaviridae): a novel species of the genus Phlebovirus and its relationship with other emerging tick-borne phleboviruses. J Virol 87, 3719–3728 10.1128/JVI.02845-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullan L. K., Folk S. M., Kelly A. J., MacNeil A., Goldsmith C. S., Metcalfe M. G., Batten B. C., Albariño C. G., Zaki S. R. & other authors (2012). A new phlebovirus associated with severe febrile illness in Missouri. N Engl J Med 367, 834–841 10.1056/NEJMoa1203378 [DOI] [PubMed] [Google Scholar]
- Muller R., Saluzzo J. F., Lopez N., Dreier T., Turell M., Smith J., Bouloy M. (1995). Characterization of clone 13, a naturally attenuated avirulent isolate of Rift Valley fever virus, which is altered in the small segment. Am J Trop Med Hyg 53, 405–411 [DOI] [PubMed] [Google Scholar]
- Nichol S. T., Beaty B. J., Elliott R. M., Goldbach R., Plyusin A., Schmaljohn C., Tesh R. (2005). Family Bunyaviridae. London: Elsevier Academic Press [Google Scholar]
- Nunes M. R., Travassos da Rosa A. P., Weaver S. C., Tesh R. B., Vasconcelos P. F. (2005). Molecular epidemiology of group C viruses (Bunyaviridae, Orthobunyavirus) isolated in the Americas. J Virol 79, 10561–10570 10.1128/JVI.79.16.10561-10570.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios G., Druce J., Du L., Tran T., Birch C., Briese T., Conlan S., Quan P. L., Hui J. & other authors (2008). A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med 358, 991–998 10.1056/NEJMoa073785 [DOI] [PubMed] [Google Scholar]
- Palacios G., da Rosa A. T., Savji N., Sze W., Wick I., Guzman H., Hutchison S., Tesh R., Lipkin W. I. (2011a). Aguacate virus, a new antigenic complex of the genus Phlebovirus (family Bunyaviridae). J Gen Virol 92, 1445–1453 10.1099/vir.0.029389-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios G., Tesh R., Travassos da Rosa A., Savji N., Sze W., Jain K., Serge R., Guzman H., Guevara C. & other authors (2011b). Characterization of the Candiru antigenic complex (Bunyaviridae: Phlebovirus), a highly diverse and reassorting group of viruses affecting humans in tropical America. J Virol 85, 3811–3820 10.1128/JVI.02275-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios G., Savji N., Travassos da Rosa A., Desai A., Sanchez-Seco M. P., Guzman H., Lipkin W. I., Tesh R. (2013a). Characterization of the Salehabad virus species complex of the genus Phlebovirus (Bunyaviridae). J Gen Virol 94, 837–842 10.1099/vir.0.048850-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios G., Savji N., Travassos da Rosa A., Guzman H., Yu X., Desai A., Rosen G. E., Hutchison S., Lipkin W. I., Tesh R. (2013b). Characterization of the Uukuniemi virus group (Phlebovirus: Bunyaviridae): evidence for seven distinct species. J Virol 87, 3187–3195 10.1128/JVI.02719-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D., Crandall K. A. (2001). Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci U S A 98, 13757–13762 10.1073/pnas.241370698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle C. R., Lees J. F., Clark W., Elliott R. M. (1984). Genome subunit reassortment among Bunyaviruses analysed by dot hybridization using molecularly cloned complementary DNA probes. Virology 135, 244–256 10.1016/0042-6822(84)90134-X [DOI] [PubMed] [Google Scholar]
- Rodriguez L. L., Owens J. H., Peters C. J., Nichol S. T. (1998). Genetic reassortment among viruses causing hantavirus pulmonary syndrome. Virology 242, 99–106 10.1006/viro.1997.8990 [DOI] [PubMed] [Google Scholar]
- Saeed M. F., Wang H., Suderman M., Beasley D. W., Travassos da Rosa A., Li L., Shope R. E., Tesh R. B., Barrett A. D. (2001). Jatobal virus is a reassortant containing the small RNA of Oropouche virus. Virus Res 77, 25–30 10.1016/S0168-1702(01)00262-3 [DOI] [PubMed] [Google Scholar]
- Sall A. A., de A Zanotto P. M., Zeller H. G., Digoutte J. P., Thiongane Y., Bouloy M. (1997). Variability of the NS(S) protein among Rift Valley fever virus isolates. J Gen Virol 78, 2853–2858 [DOI] [PubMed] [Google Scholar]
- Salminen M. O., Carr J. K., Burke D. S., McCutchan F. E. (1995). Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res Hum Retroviruses 11, 1423–1425 10.1089/aid.1995.11.1423 [DOI] [PubMed] [Google Scholar]
- Smith J. M. (1992). Analyzing the mosaic structure of genes. J Mol Evol 34, 126–129 10.1007/BF00182389 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). mega5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesh R. B. (1988). Phlebotomus Fevers. Boca Raton: CRC Press [Google Scholar]
- Tesh R. B., Saidi S., Gajdamovic S. J., Rodhain F., Vesenjak-Hirjan J. (1976). Serological studies on the epidemiology of sandfly fever in the Old World. Bull World Health Organ 54, 663–674 [PMC free article] [PubMed] [Google Scholar]
- Tesh R., Saidi S., Javadian E., Nadim A. (1977). Studies on the epidemiology of sandfly fever in Iran. I. Virus isolates obtained from Phlebotomus. Am J Trop Med Hyg 26, 282–287 [DOI] [PubMed] [Google Scholar]
- Tesh R. B., Peters C. J., Meegan J. M. (1982). Studies on the antigenic relationship among phleboviruses. Am J Trop Med Hyg 31, 149–155 [DOI] [PubMed] [Google Scholar]
- Vialat P., Billecocq A., Kohl A., Bouloy M. (2000). The S segment of Rift Valley Fever Phlebovirus (Bunyaviridae) carries determinants for attenuation and virulence in mice. J Virol 74, 1538–1543 10.1128/JVI.74.3.1538-1543.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanase T., Kato T., Yamakawa M., Takayoshi K., Nakamura K., Kokuba T., Tsuda T. (2006). Genetic characterization of Batai virus indicates a genomic reassortment between orthobunyaviruses in nature. Arch Virol 151, 2253–2260 10.1007/s00705-006-0808-x [DOI] [PubMed] [Google Scholar]
- Yanase T., Aizawa M., Kato T., Yamakawa M., Shirafuji H., Tsuda T. (2010). Genetic characterization of Aino and Peaton virus field isolates reveals a genetic reassortment between these viruses in nature. Virus Res 153, 1–7 10.1016/j.virusres.2010.06.020 [DOI] [PubMed] [Google Scholar]
- Yu X. J., Liang M. F., Zhang S. Y., Liu Y., Li J. D., Sun Y. L., Zhang L., Zhang Q. F., Popov V. L. & other authors (2011). Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med 364, 1523–1532 10.1056/NEJMoa1010095 [DOI] [PMC free article] [PubMed] [Google Scholar]