Abstract
Pseudomonas aeruginosa (P. aeruginosa) is a leading cause of morbidity and mortality in patients with ventilation-associated pneumonia (VAP). It is difficult to treat this infection due to acquired resistance to various antibiotics. In order to understand the potential route of transmission, it is important to have detailed knowledge of the genotypes and antibiotic susceptibility of P. aeruginosa. The aim of this study was to determine antibiotic susceptibility using the broth microdilution minimum inhibitory concentration (MIC) method and to apply the Randomly Amplified Polymorphic DNA (RAPD) typing method for VAP caused by P. aeruginosa in 16 patients (8 men and 8 women; average age at inclusion, 67.8 years; range, 53–76 years). To determine antibiotic susceptibility, imipenem (IPM), cefepime (FEP) and meropenem (MEM) were administered for the treatment of P. aeruginosa, yielding an effectiveness of 75, 62.5, and 62.5%, respectively. According to the National Committee for Clinical Laboratory Standards (NCCLS) breakpoints, 8 (50%) of the 16 mechanical ventilation (MV) isolates were resistant to ceftazidime (CAZ) and pipenacillin (PIP). Amikacin (AK) and aztreonam (AZT) were not as effective against P. aeruginosa (75%). In addition, P. aeruginosa was completely resistant to ciprofloxacin (CIP). The MV isolates were susceptible to polymyxin B (PB). RAPD analysis revealed 12 genotypes for all the isolated P. aeruginosa, separated into 4 patterns. The results demonstrated a high incidence of P. aeruginosa isolated from VAP, with endogenous and cross infections being potential reasons for P. aeruginosa isolated from VAP in the intensive care units.
Keywords: Pseudomonas aeruginosa, antibiotic susceptibility, genotyping, ventilation associated pneumonia
Introduction
Ventilation-associated pneumonia (VAP) remains a frequent health care-associated infection, occurring in 10–20% of Intensive Care Unit (ICU) patients on mechanical ventilation (MV) for ≥48 h. P. aeruginosa is the leading cause of morbidity and mortality in patients with VAP. P. aeruginosa, a lethal bacteria, is resistant to various drugs and capable of producing virulence factors, causing a variety of infections. The pathogenesis of P. aeruginosa infections is involved in the expression of a variety of virulence factors associated with attachment, persistence, evading/destroying host defenses, tissue invasion/penetration and toxin-mediated disease. VAP with P. aeruginosa is an invasive and rapidly progressing infection characterized by acute leukocytosis, fever and increased need for ventilator support (1,2). VAP caused by P. aeruginosa is associated with increased morbidity, mortality and high cost. The purpose of this study was to investigate the antibiotic susceptibility of P. aeruginosa isolates from VAP patients and determine the genotypes of these isolates by the Randomly Amplified Polymorphic DNA (RAPD). We also explored potential routes of transmission between units in the same hospital.
Materials and methods
Patient characteristics
Sixteen MV patients were included in the present study: average age at inclusion, 64.8 years (range, 53–76 years) (Table I). The study was approved by the ethics committee of the Shanghai First People’s Hospital affiliated to Shanghai Jiao Tong University. Written informed patient consent was obtained from the patients prior to their enrollment in the study. A total of 13.3 days (range, 2–30 days) elapsed from initiation of MV to colonization with P. aeruginosa.
Table I.
Clinical characteristics of mechanically ventilated patients at the time P. aeruginosa isolates were obtained.
| Patient | Age (years) | Gender | Admitting diagnosis | Apache II score at ICU admission | Days of ventilation prior to colonisation | ICU days | Died | Cause of death | Antibiotics at time of colonisation |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 60 | M | ARDS | 29 | 2 | 11 | N | IPM | |
| 2 | 59 | F | Pulmonary embolism | 28 | 4 | 12 | N | CAZ | |
| 3 | 53 | M | Traumatic brain injury | 35 | 30 | 30 | Y | Multisystem organ failure | CAZ+AK |
| 4 | 69 | M | AECOPD | 30 | 6 | 6 | N | FEP | |
| 5 | 72 | M | Interstitial lung disease | 32 | 12 | 13 | Y | ARDS | FEP+Azithromycin |
| 6 | 64 | F | Pulmonary-encephalopathy | 24 | 10 | 12 | N | IPM | |
| 7 | 67 | F | Idiopathic pulmonary fibrosis | 24 | 19 | 17 | N | IPM+AK | |
| 8 | 76 | F | ARDS | 28 | 3 | 10 | Y | Multisystem organ failure | MEM |
| 9 | 73 | F | Septicemia | 26 | 6 | 6 | Y | Multisystem organ failure | MEM |
| 10 | 62 | M | ARDS | 29 | 4 | 6 | N | FEP | |
| 11 | 67 | F | IPF | 25 | 18 | 20 | Y | Sepsis | CAZ+AK |
| 12 | 57 | M | Severe asthma | 32 | 4 | 16 | Y | ARDS | IPM |
| 13 | 58 | F | Status asthmaticus | 29 | 6 | 15 | N | CAZ | |
| 14 | 75 | M | AECOPD | 30 | 23 | 25 | N | CAZ+CIP | |
| 15 | 66 | M | Asthma | 31 | 4 | 7 | N | FEP+AK | |
| 16 | 59 | F | ARDS | 32 | 2 | 6 | Y | Sepsis/ARDS | FEP |
Susceptibility was performed as described by the National Committee for Clinical Laboratory Standards (NCCLS, 2012), which was used to classify strains as susceptible (S), intermediate (I) or resistant (R). ICU, Intensive Care Unit; M, male; F, female; N, no; Y, yes; ARDS, acute respiratory distress syndrome; IPM, imipenem; CAZ, ceftazidime; AK, amikacin; AECOPD, acute exacerbation of chronic obstructive pulmonary disease; FEP, cefepime; MEM, meropenem; IPF, idiopathic pulmonary fibrosis; CIP, ciprofloxacin.
Criteria for VAP
VAP was defined as any lower respiratory infection that developed after 2 days of MV. Criteria for VAP were presence or progression of a new infiltrate identified by chest radiography plus two of the following three clinical criteria: i) temperature of >38°C; ii) white blood cell count of >12,000 or <4,000 cells/mm3; and iii) purulent secretions and ≥106 colony-forming units (CFU)/ml P. aeruginosa from the endotracheal aspirate (ETA), and/or ≥104 CFU/ml from a bronchoalveolar lavage (1).
Sample collection
Sixteen clinical samples were collected from MV patients in the PuDong Hospital, Shanghai, China. Daily ETA surveillance cultures were performed on patients ≥18 years of age who were intubated for ≥48 h, irrespective of signs or symptoms of lung infection. Patients with known infection or risk factors for colonization with P. aeruginosa, such as bronchiectasis, chronic tracheostomy and previous intubation during the current hospitalisation or a previous culture positive for P. aeruginosa, were excluded from the present study.
Isolation of the bacterial isolates
The collected samples were inoculated into sterile petri dishes containing blood agarmedia and incubated at 35°C for 18–24 h. The isolated colonies were subcultured and purified for characterization.
Antibiotic sensitivity tests
The antimicrobial susceptibility tests of P. aeruginosa were performed using the broth micro-dilution minimum inhibitory concentration (MIC) method. A sterile plastic tray containing various concentrations of antimicrobial agents was inoculated with a standardized number of test bacteria in Mueller Hinton broth. After overnight incubation at 35°C, the MIC was determined and interpreted as susceptible, intermediate or resistant (3). The ATCC27853 P. aeruginosa strain was used as the quality control strain.
RAPD fingerprinting
RAPD was performed according to the method of Libisch et al (4) and Bradbury et al (5), with some modification. The polymerase chain reaction (PCR) mixture was generated using 10X PCR buffer composed of 100 mM Tris-Hcl, 500 mM Kcl, 15 mM MgCl2, 0,01% (W/V) gelatin, pH 8.0, followed by the addition of 32 mM each dGTP, dATP, dCTP and dTTP. Taq DNA polymerase (5 U/μl). Template DNA was then produced using 10–25 ng/μl stock solution containing good quality, protein-free DNA, which was resuspended in high quality sterile, deionized water or Tris-EDTA pH 8.0 RNase (20 ng per 1 ng of DNA). The primer 5′-GTTGGTGGCT-3′ was used in this study at 25 pmol/l (6).
Results
MIC analysis
MICs for isolates from the 16 MV patients are provided in Table II. Imipenem (IPM) was effective against P. aeruginosa (75%). Cefepime (FEP) and meropenem (MEM) were the most commonly used drugs for the treatment of P. aeruginosa with an effectiveness of 62.5%. According to the National Committee for Clinical Laboratory Standards (NCCLS) breakpoints, 8 (50%) of the 16 MV isolates were resistant to ceftazidime (CAZ) and pipenacillin (PIP). Amikacin (AK) and aztreonam (AZT) had a lower effect against P. aeruginosa (75%). In addition, P. aeruginosa was completely resistant to ciprofloxacin (CIP). The MV isolates were susceptible to polymyxin B (PB). According to antibiotic susceptibility, P. aeruginosa isolates were separated into four patterns, the first of which was resistant to AK, CIP and AZT; the second to AK, CIP, AZT and CAZ; the third to PIP, CAZ, FEP, MEM, AK, CIP, and AZT, while the fourth pattern exhibited susceptibility only to PB.
Table II.
Minimum inhibitory concentration (MIC; in μg/ml) of P. aeruginosa isolates from mechanically ventilated patients.
| Patient isolate | PIP | CAZ | FEP | IPM | MEM | AK | CIP | PB | AZT |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 8 (S) | 4 (S) | 4 (S) | 1 (S) | 1 (S) | 18 (M) | 12 (R) | 1 (S) | 20 (M) |
| 2 | 12 (S) | 1 (S) | 4 (S) | 1 (S) | 1 (S) | 18 (M) | 12 (R) | 1 (S) | 28 (M) |
| 3 | 128 (R) | 16 (M) | 32 (R) | 10 (R) | 10 (R) | 132 (R) | 24 (R) | 1 (S) | 32 (R) |
| 4 | 4 (S) | 4 (S) | 1 (S) | 1 (S) | 1 (S) | 18 (M) | 12 (R) | 1 (S) | 32 (R) |
| 5 | 128 (R) | 16 (M) | 32 (R) | 10 (R) | 10 (R) | 132 (R) | 24 (R) | 1 (S) | 32 (R) |
| 6 | 12 (S) | 1 (S) | 4 (S) | 1 (S) | 1 (S) | 18 (M) | 16 (R) | 1 (S) | 28 (M) |
| 7 | 10 (S) | 1 (S) | 2 (S) | 1 (S) | 1 (S) | 12 (S) | 16 (R) | 1 (S) | 8 (S) |
| 8 | 128 (R) | 16 (M) | 32 (R) | 10 (R) | 10 (R) | 132 (R) | 24 (R) | 1 (S) | 32 (R) |
| 9 | 128 (R) | 16 (M) | 32 (R) | 10 (R) | 12 (R) | 132 (R) | 32 (R) | 1 (S) | 64 (R) |
| 10 | 64 (M) | 32 (R) | 2 (S) | 1 (S) | 1 (S) | 12 (S) | 16 (R) | 1 (S) | 8 (S) |
| 11 | 128 (R) | 16 (M) | 32 (R) | 1 (S) | 4 (M) | 132 (R) | 24 (R) | 1 (S) | 32 (R) |
| 12 | 64 (M) | 32 (R) | 2 (S) | 1 (S) | 1 (S) | 12 (S) | 16 (R) | 1 (S) | 8 (S) |
| 13 | 8 (S) | 4 (S) | 4 (S) | 1 (S) | S(1) | 18 (M) | 12 (R) | 1 (S) | 20 (M) |
| 14 | 10 (S) | 2 (S) | 2 (S) | 1 (S) | 1 (S) | 12 (S) | 12 (R) | 1 (S) | 16 (M) |
| 15 | 12 (S) | 4 (S) | 1 (S) | 1 (S) | 1 (S) | 64 (R) | 12 (R) | 1 (S) | 8 (S) |
| 16 | 128 (R) | 16 (M) | 32 (R) | 1 (S) | 4 (M) | 132 (R) | 24 (R) | 1 (S) | 32 (R) |
PIP, pipenacillin; CAZ, ceftazidime; FEP, cefepime; IPM, imipenem; MEM, meropenem; AK, amikacin; CIP, ciprofloxacin; PB, polymyxin B; AZT, aztreonam.
RAPD analysis of strains
RAPD analysis revealed different genotypes for the isolated P. aeruginosa (Fig. 1). There was a difference in the intensity of bands within the same pattern. However, many isolates were identical in genotype-exhibited variability in the antibiotic susceptibility pattern. Lanes 2 and 14 are RAPD fingerprints of long-term colonization of the strains at different time periods for the different patients. Patients with severe exacerbation of chronic obstructive pulmonary disease (COPD) usually requires MV at an increased risk of P. aeruginosa infection and colonization. Lanes 5 and 16 are the genotyping of seven types of antibiotic-resistant strains in the nine types of antibiotics. In 3 of the 16 patients included in the present study, identical clone strains were identified by RAPD genotyping (lanes 1, 3 and 11). From the isolated P. aeruginosa, the proportion of clone strains was 18.75%.
Figure 1.
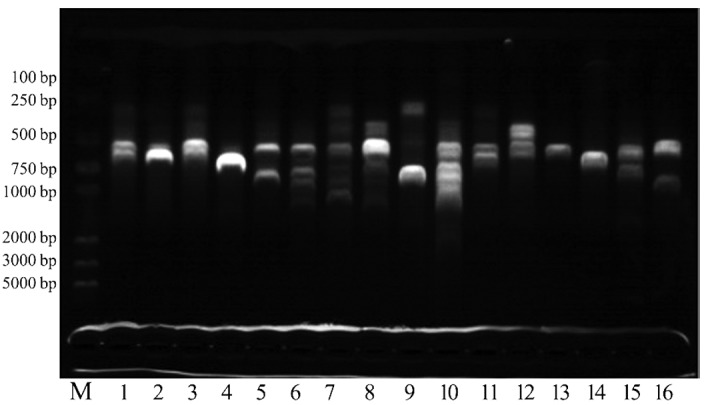
Randomly Amplified Polymorphic DNA (RAPD) analysis of P. aeruginosa strains from newly colonized mechanically ventilated patients. Lane M, DNA molecular weight marker and lanes 1–16, P. aeruginosa DNAs.
Discussion
P. aeruginosa is one of the most important clinical pathogens in the nosocomial setting and a common causative agent of bacteremia, pneumonia, and urinary tract infections (4). VAP is the most frequently occurring and severe infection acquired in the ICU, leading to prolonged MV and excess mortality. VAP is preceded by lower respiratory tract colonization by pathogenic microorganisms that derive from endogenous or exogenous sources. Most VAP are the results of exogenous nosocomial colonization, particularly pneumonias caused by resistant bacteria, such as P. aeruginosa (7). This study found that the proportion of P. aeruginosa clone strains is very high (18.75%), and that exogenous cross infection is a potentially significant reason for VAP caused by P. aeruginosa. Gil-Perotin et al (8) reported that biofilm formation is a type of colonization. A potential association of airway colonization, biofilm and VAP development presents cause for concern. Patient information revealed that P. aeruginosa infection was 41.18% in a variety of processes including tracheal incision, gastric canal, catheter, drainage tube and deep vein puncture, and 52.94% relative to MV. However, Farah and Makhoul (10) reported that, although the stomach colonized by enteric Gram-negative bacilli is a potential reservoir, it is not the primary source for lower airway colonization, thus this gastropulmonary route is not currently considered a significant route for the development of VAP (9). Several factors favor flora modification, including the severity of underlying disease, previous hospital stay, poor nutritional status, and antibiotic treatment. Patients with severe exacerbation of COPD and asthma usually require MV (10), while patients with COPD are at an increased risk of contracting P. aeruginosa infection. The P. aeruginosa responsible for VAP also vary depending on patient characteristics and clinical circumstances; however, these differences appear to be due primarily to the duration of MV and/or degree of previous antibiotic exposure of these patients.
At present, the most reliable treatment for P. aeruginosa VAP is a carbapenem and fourth-generation cephalosporins, usually combined with AK, which could achieve synergy in the treatment of P. aeruginosa. In our treatment programs, FEP, MEM and IPM were the most commonly administered drugs, combined with AK or azithromycin for the treatment of P. aeruginosa, with IPM being the most reliable treatment. Patients with P. aeruginosa VAP may experience favorable survival when treated with adjunctive aerosolized antibiotics, despite greater severity of illness and a greater incidence of multidrug-resistant (MDR) infection (11). However, some strains of P. aeruginosa responsible for VAP are resistant to the common active antibiotics. Thus, the only antimicrobial therapy that remains potentially active is colistin. Experimental results of this study have demonstrated that the MV isolates were susceptible to PB. Findings of Bassetti et al (12) demonstrated that only polymyxins are capable of providing an overall coverage of ≥90%. Therefore, polymyxin inhalation therapy can be applied to P. aeruginosa colonization and infection.
Despite the use of combination therapy, studies have reported on the emergence of acquired MDR clinical isolates of P. aeruginosa. This pathogen builds resistance against antibiotics through various mechanisms including blockade of entry, the use of efflux pumps, enzymatic degradation, and target alteration (13). Previous exposure to antibiotic therapy appears to be one of the major causes leading to infection with MDR pathogens. In this experimental study, seven types of antibiotic resistant strains were identified in the nine types of antibiotics studied that may be associated with previous use of antibiotics.
However, the development of lung tissue infection occurs in only some patients on MV and is the result of a complex interaction between the host defenses of the patient and the quantity and virulence of the P. aeruginosa colonizing the lower respiratory tract. Results of the study by Ruiz et al (14) have shown that the origin of hospital P. aeruginosa infections is due mainly to the growth of bacterial strains acquired by patients prior to hospital admission or from patient to patient through healthcare workers. Standard preventative measures, such as hand hygiene using hydro alcoholic solutions, are integrated in general programs to combat against nosocomial infections and, more generally, against all infections associated with health care. Therefore, it is imperative that knowledge regarding health care personnel hand hygiene be promulgated, and that there is firm administration of the use of antibiotics in order to reduce rates of pathogens, which may significantly improve department epidemic status of P. aeruginosa in ICU. Knowledge of the nature and the relative importance of each risk factor is essential for the development of effective measures to prevent VAP. These data suggest that cross infections were potentially significant reasons of P. aeruginosa in the hospital. Limiting the route of transmission among patient-to-patient and patient-to-health care workers could decrease the infection rate of P. aeruginosa and become one of the most critical factors involved in the decrease of the infection rate of P. aeruginosa.
In conclusion, antibiotic susceptibility and genotyping of 16 P. aeruginosa clinical samples from VAP in ICU were carried out in the present study. The results obtained can be useful in the selection of antibiotics and in understanding the potential route of transmission in order to decrease the infection rate of P. aeruginosa in MV patients in ICU.
Acknowledgments
This study is supported by National Natural Science Foundation (81072448), key projects of Shanghai Science and Technology Foundation Office (11JC1410300), Shanghai JiaoTong University training fund major projects.
References
- 1.Fricks-Lima J, Hendrickson CM, Allgaier M, et al. Differences in biofilm formation and antimicrobial resistance of Pseudomonas aeruginosa isolated from airways of mechanically ventilated patients and cystic fibrosis patients. Int J Antimicrob Agents. 2011;37:309–315. doi: 10.1016/j.ijantimicag.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garnacho-Montero J, Sa-Borges M, Sole-Violan J, et al. Optimal management therapy for Pseudomonas aeruginosa ventilator-associated pneumonia: an observational, multicenter study comparing monotherapy with combination antibiotic therapy. Critical Care Med. 2007;35:1888–1895. doi: 10.1097/01.CCM.0000275389.31974.22. [DOI] [PubMed] [Google Scholar]
- 3.Sevillano E, Valderrey C, Canduela MJ, Umaran A, Calvo F, Gallego L. Resistance to antibiotics in clinical isolates of Pseudomonas aeruginosa. Pathol Biol (Paris) 2006;54:493–497. doi: 10.1016/j.patbio.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 4.Libisch B, Watine J, Balogh B, et al. Molecular typing indicates an important role for two international clonal complexes in dissemination of VIM-producing Pseudomonas aeruginosa clinical isolates in Hungary. Res Microbiol. 2008;159:162–168. doi: 10.1016/j.resmic.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Bradbury R, Champion A, Reid DW. Poor clinical outcomes associated with a multi-drug resistant clonal strain of Pseudomonas aeruginosa in the Tasmanian cystic fibrosis population. Respirology. 2008;13:886–892. doi: 10.1111/j.1440-1843.2008.01383.x. [DOI] [PubMed] [Google Scholar]
- 6.Mahenthiralingam E, Campbell ME, Foster J, Lam JS, Speert DP. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J Clin Microbiol. 1996;34:1129–1135. doi: 10.1128/jcm.34.5.1129-1135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crnich J, Safdar N, Maki DG. The role of the intensive care unit environment in the pathogenesis and prevention of ventilator-associated pneumonia. Respir Care. 2005;50:813–838. [PubMed] [Google Scholar]
- 8.Gil-Perotin S, Ramirez P, Marti V, et al. Implications of endotracheal tube biofilm in ventilator-associated pneumonia response: a state of concept. Crit Care. 2012;16:R93. doi: 10.1186/cc11357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trouillet JL. Ventilator-associated pneumonia: a comprehensive review. Hosp Pract (Minneap) 2012;40:165–175. doi: 10.3810/hp.2012.04.982. [DOI] [PubMed] [Google Scholar]
- 10.Farah R, Makhoul N. Optimal time needed for withdrawal of mechanical ventilation in patients with chronic obstructive pulmonary disease. Isr Med Assoc J. 2011;13:609–612. [PubMed] [Google Scholar]
- 11.Arnold HM, Sawyer AM, Kollef MH. Use of adjunctive aerosolized antimicrobial therapy in the treatment of Pseudomonas aeruginosa and Acinetobacter baumannii ventilator-associated pneumonia. Respir Care. 2012;57:1226–1233. doi: 10.4187/respcare.01556. [DOI] [PubMed] [Google Scholar]
- 12.Bassetti M, Taramasso L, Giacobbe DR, Pelosi P. Management of ventilator-associated pneumonia: epidemiology, diagnosis and antimicrobial therapy. Expert Rev Anti Infect Ther. 2012;10:585–596. doi: 10.1586/eri.12.36. [DOI] [PubMed] [Google Scholar]
- 13.Mahida K, Kwon DH. Co-existence of multidrug-resistant and -susceptible strains of Pseudomonas aeruginosa from a single clinical isolate. Curr Microbiol. 2010;61:19–24. doi: 10.1007/s00284-009-9570-0. [DOI] [PubMed] [Google Scholar]
- 14.Ruiz L, Dominguez MA, Ruiz N, Vinas M. Relationship between clinical and environmental isolates of Pseudomonas aeruginosa in a hospital setting. Arch Med Res. 2004;35:251–257. doi: 10.1016/j.arcmed.2004.02.005. [DOI] [PubMed] [Google Scholar]


