Abstract
MicroRNAs (miRNAs) are small, non-coding RNAs of endogenous origin that have been increasingly shown to have altered expressions in various cancer types. The expression levels of miR-375 have not been comprehensively investigated in pancreatic cancer. In this study, total RNA was extracted from 44 pairs of pancreatic cancer tissues and non-tumor adjacent tissues, as well as from four pancreatic cancer cell lines, Panc-1, SW1990, BxpC3 and Patu8988. Following polyadenylation and reverse transcription, the expression levels of miR-375 were determined by real-time PCR and the difference in expression was calculated using the 2−ΔΔCt method. The correlation between the expression levels of miR-375 and clinicopathological characteristics of pancreatic cancer was also assessed. miR-375 expression was frequently downregulated in the pancreatic cancer tissues compared to their non-tumor counterparts (P<0.05; paired t-test). Moreover, a significantly low expression of miR-375 was found in the pancreatic cancer cell lines (Panc-1, P=0.016; SW1990, P=0.016; BxPC3, P=0.018; Patu8988, P=0.017; paired t-test). However, no significant correlations were observed between the low expression of miR-375 and parameters including gender, age, tumor size, tumor location and histological grade (P>0.05). The low expression of miR-375 was correlated with pT stage, lymph node metastases and pTNM stage (P<0.05) (non-parametric test; Mann-Whitney U test between 2 groups and Kruskal-Wallis H test for ≥3 groups). In conclusion, miR-375 is potentially involved in the carcinogenesis of pancreatic cancers and serves as is a potential biomarker for pancreatic cancer.
Keywords: microRNA, miR-375, pancreatic cancer
Introduction
MicroRNAs (miRNAs), which were first identified in 1993 (1), are non-coding RNAs consisting of 21–23 nucleotides. They constitute a recently emerging class of endogenous negative regulators of gene expression which possess a noteworthy evolutionary conservation (2,3). miRNA products are single-stranded RNAs of 19–22 nucleotides cleaved from 70- and 100-nucleotide hairpin pre-miRNA precursors (1,4). miRNAs are thought to modulate gene expression at the post-transcriptional level (5,6). These small molecules exert their regulatory effects by base-pairing to partially complementary mRNAs and function by two mechanisms: degrading target mRNA transcripts or inhibiting mRNA translation (6,7). miRNAs are also associated with the main phenotypes of many cancer cells (including pancreatic cancer), such as proliferation, invasion and apoptosis (8–10). Therefore, study of the functions and mechanisms of miRNAs may lead to new approaches for the classification, diagnosis and treatment of human cancers. The growing interest in these regulatory miRNAs has led to the continued exploration of miRNA expression in cancer samples and the identification of new miRNAs that may act as oncogenes and tumor suppressors (11). Accumulating studies have shown the dysregulation of miRNA expression in various tumor types, including esophageal squamous cell carcinoma (12), lung cancer (13), breast cancer (14), pancreatic adenocarcinoma (15), hepatocellular carcinoma (16), colon cancer (17) and gastric cancer (18). Furthermore, it has been reported that miRNA genes are frequently located at fragile sites and genomic regions involved in cancers, suggesting that aberrant miRNA expression plays an important role in cancer pathogenesis (19).
Pancreatic cancer (PC) is an aggressive malignancy with one of the worst mortality rates. It is the sixth leading cause of death from malignant disease in China and the fourth leading cause of cancer-related death in the United States (20–22). The estimated mortality is almost the same as the estimated incidence, with an overall 5-year survival rate of <5% (22). Therefore, new associated factors and novel therapeutic targets for pancreatic cancer remains to be identified. Although the etiology of pancreatic cancer is attributed to numerous serious factors, the accumulation of genetic and epigenetic changes remains the fundamental mechanism of tumorigenesis.
Previously, the aberrant expression of several miRNAs was identified in pancreatic carcinoma. miR-375 is one of the most consistently downregulated miRNAs in pancreatic cancer (23,24). The effects of miR-375 may be cell-type specific (23). However, a limited number of studies on pancreatic cancer have focused on the targeting, clinical and prognostic significance of miR-375.
In the present study, we examined miR-375 expression in 44 pancreatic cancer tissues and four pancreatic cancer cell lines and found that miR-375 was frequently down-regulated in pancreatic carcinomas. Additionally, the low expression of miR-375 in 44 pancreatic cancer tissues was relative to the matched adjacent non-tumor tissues by real-time PCR, suggesting that it was significantly associated with pT stage, lymph node metastases and pTNM stage.
Materials and methods
Tissues samples
Pancreatic cancer and matched adjacent non-tumor tissues from 44 patients were obtained post-operatively in 2009 from the Department of General Surgery, The First Affiliated Hospital of Soochow University (Suzhou, China). The patients provided signed, informed consent for their tissues to be used for scientific research. Ethical approval for the study was obtained from the Department of General Surgery, The First Affiliated Hospital of Soochow University (Suzhou, China). Diagnoses were based on pathological and/or cytological evidence. Histological features of the specimens were evaluated by two senior pathologists according to classification criteria from the WHO (World Health Organization) (1990). Cancers were classified using the TNM staging system of the American Joint Committee on Cancer (AJCC; 2010) and the International Union against Cancer (UICC). Tissues were obtained from patients prior to chemotherapy or radiation therapy. Specimens were immediately frozen and stored at −80°C prior to microarray and real-time PCR analyses. One section of each sample was stained with hematoxylin and eosin.
Cell lines and culture conditions
The human pancreatic cancer cell lines (Panc-1, SW1990, BxPC3 and Patu8988) were maintained in DMEM supplemented with 10% fetal bovine serum (FBS). Cells were cultured in a 37°C incubator with 5% CO2. The human Panc-1, SW1990, BxPC3 and Patu8988 pancreatic cancer cell lines were obtained from Shanghai Genechem Co., Ltd. (Genechem, Shanghai, China). Panc-1 cells were cultured in DMEM supplemented with 10% FBS, while SW1990, BxPC3 and Patu8988 were cultured in RMPI-1640 supplemented with 10% FBS. The normal pancreatic tissues (3 matched adjacent non-tumor tissues) were randomly selected from the 44 cases of pancreatic cancer as the controls. All of the cell lines were cultured at 37°C and 5% CO2.
Extraction, polyadenylation and reverse transcriptase reaction
Total RNA was extracted from patients or cell line samples using Trizol (Invitrogen) according to the manufacturer’s protocol. The concentration and purity of RNA were controlled by UV spectrophotometry using a NanoPhotometer UV/Vis spectrophotometer (Implen, Schatzbogen, Munich, Germany).
The reverse transcription was using Taqman assay kits (Applied Biosystems, Foster City, CA, USA) with U6 small nuclear RNA as an internal normalized reference. A 15 μl reverse transcriptase reaction mixture containing 5 μl of the RNA sample, 3 μl RT-primer, 1.5 μl 10X Reverse Transcription Buffer, 1 μl MultiScribe™ Reverse Transcriptase, 0.15 μl 100 mmol/l dNTPs (with dTTP), 0.19 μl RNase inhibitor and 4.16 μl DEPC-treated water was incubated for 5 min at 65°C. Subsequent to the addition of 1 μl RNase H to the mixture, the total reaction mixture was incubated in a 96-well plate of a GeneAmp PCR 9700 Thermocycler (Applied Biosystems, Hayward, CA, USA) for 30 min at 16°C, 30 min at 42°C, 5 min at 85°C, and maintained at 4°C.
Real-time PCR
Real-time PCR was performed using Taqman assay kits (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions, with a PRISM 7900 real-time PCR machine (Applied Biosystems).
The 20-μl mixture of PCR consisted of 10 μl Taqman 2X Universal PCR Master Mix, 1 μl Taqman MicroRNA assay (20X), 5 μl reverse-transcribed product, and 4 μl DEPC. Threshold cycle data were determined by setting a default threshold. The reactive condition was 40 amplification cycles of 95°C for 10 min, 90°C for 15 sec and 60°C for 1 min in a 96-well optical plate using a 7500 Fast Real-Time PCR System (Applied Biosystems). The U6 RNA was adopted as an endogenous reference compared to the expression levels of miR-375, and the 2−ΔΔCt method was used to calculate the relative expression levels of miR-375 in cancerous samples compared to their non-tumor counterparts. Samples were performed in triplicate. The products of real-time PCR were confirmed by TA cloning and a sequencing assay.
Statistical analysis
The expression levels in pancreatic cancer tissues relative to the non-tumor controls were analyzed using the 2−ΔΔCt method. Briefly, the threshold cycle (Ct) of fluorescence for each sample was determined. ΔCt indicated the difference in expression levels with the Ct value between miR-375 and U6 (ΔCt = CtmiR-375 − CtU6), and ΔΔCt indicated the difference in the ΔCt value between cancer tissue and the matched control (ΔΔCt = ΔCtcancer − ΔCtcontrol). The 2−ΔΔCt value (fold value) was also calculated. When the fold value was <1, there was a low expression of miR-375 in the cancer tissues and cancer cell lines compared to their non-tumorous counterparts.
The statistical differences in miR-375 expression in cancer tissues and cell lines relative to the matched adjacent non-tumor tissues were analyzed by a paired t-test. Moreover, the association between miR-375 expression and clinicopathological parameters was analyzed by a non-parametric test (Mann-Whitney U test between 2 groups and Kruskal-Wallis H test for ≥3 groups). P<0.05 was considered to indicate a statistically significant difference. Statistical analysis was performed using the Statistical Program for Social Sciences (SPSS) software 17.0 (SPSS Inc., Chicago, IL, USA).
Results
Expression of miR-375 is frequently downregulated in pancreatic cancer tissues
Quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR) analysis of miR-375 was performed in 44 pairs of pancreatic cancer tumor tissues and matched adjacent non-tumor tissues. The results showed that miR-375 was significantly down-regulated in pancreatic cancer tumor tissues. The value of ΔCt (mean ± SD) was −0.208±1.529 in pancreatic cancer tissues and −1.992±3.447 in their matching adjacent non-tumor tissues (P=0.021, paired t-test; Fig. 1). miR-375 was 0.413±0.026 in 44 cases of pancreatic cancer tissues and 1.538±0.061 in their matching adjacent non-tumor tissues. miR-375 was significantly downregulated in pancreatic cancer tissues with a median 3.4-fold reduction relative to their matched adjacent non-tumor tissues (Fig. 2). Among 44 pancreatic cancer patients, 32 (72.7%) cases revealed a >50% reduction in the expression levels of miR-375 compared to their matched adjacent non-tumor tissues.
Figure 1.
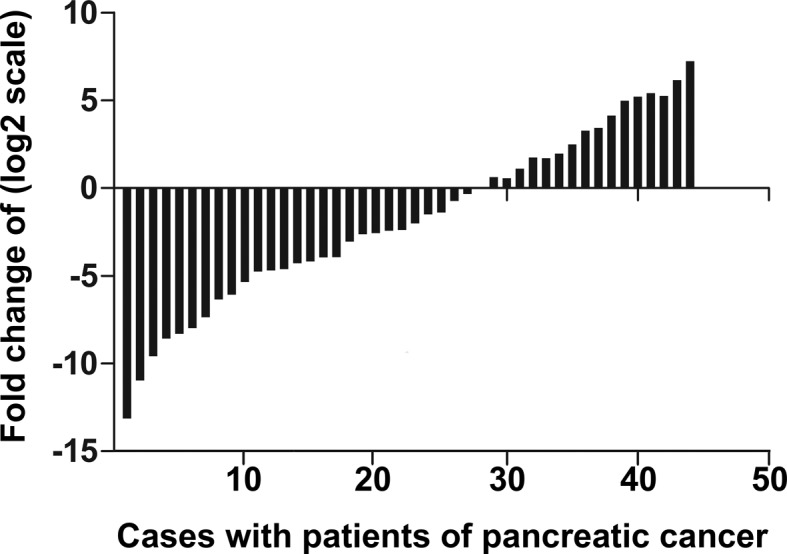
Quantification of miR-375 was measured by quantitative real-time reverse transcription-polymerase chain reaction. Each sample was analyzed in triplicate and repeated three times. Data are presented as the log2 of fold-change of pancreatic cancer tissues relative to matched adjacent non-tumor tissues.
Figure 2.
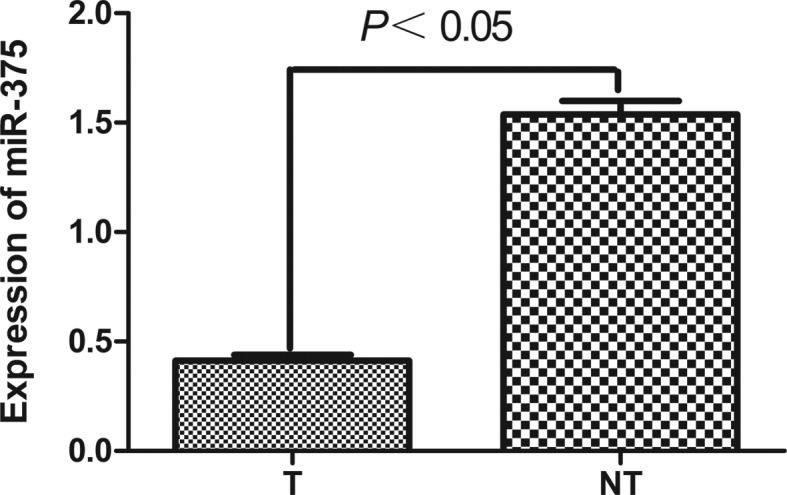
miR-375 was differentially expressed between pancreatic cancer tissues (T) and matched non-tumor adjacent tissues (NT). miR-375 was significantly downregulated in pancreatic cancer tissues compared to the matching adjacent non-tumor tissues (P<0.05; t-test).
Expression of miR-375 is significantly downregulated in pancreatic cancer cell lines
To confirm the association between miR-375 expression and pancreatic cancer, we measured miR-375 expression in three pancreatic cancer cell lines using qRT-PCR. We found a significantly low expression of miR-375 in Panc-1 (P=0.016), SW1990 (P=0.016), BxPC3 (P=0.018) and Patu8988 cells (P=0.017) relative to three matching adjacent non-tumor tissues randomly selected from the patients (Fig. 3).
Figure 3.
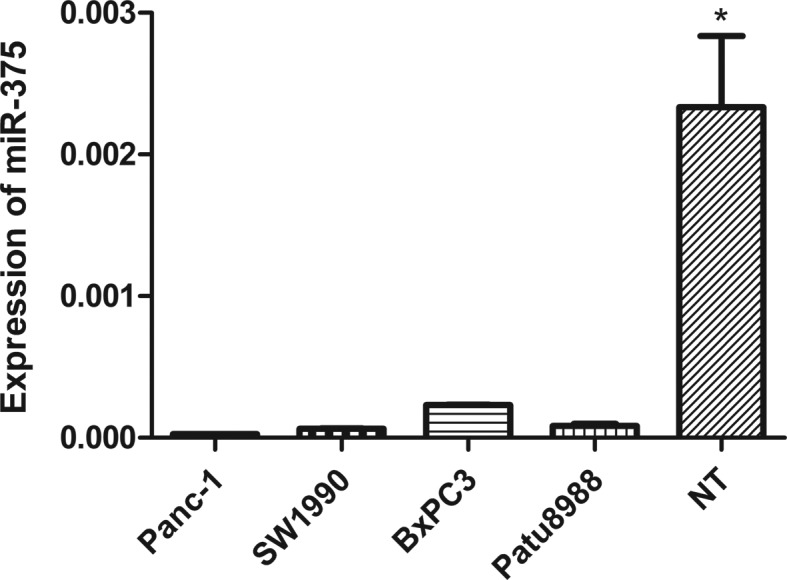
Expression levels of miR-375 in 3 pancreatic cancer cell lines (Panc-1, SW1990, BxPC3 and Patu8988). Quantification of miRNAs was measured by quantitative real-time reverse transcription-polymerase chain reaction. Data are presented in the pancreatic cancer cell lines relative to the normal pancreatic tissues. miR-375 was significantly downregulated in four pancreatic cancer cell lines compared to non-tumor adjacent tissues (NT) (*P<0.05; t-test).
Correlation between miR-375 expression levels and clinicopathological characteristics in pancreatic cancer patients
The non-parametric test between the relative expression levels of miR-375 in pancreatic cancer cases and its clinicopathological characteristics showed that there was no significant correlation between the low expression of miR-375 and parameters such as gender, age, tumor size, tumor location and histological grade (P>0.05). However, the low expression of miR-375 was correlated with pT stage, lymph node metastases and pTNM stage (P<0.05) (Table I).
Table I.
Association between the expression of miR-375 with clinicopathological characteristics in patients with pancreatic cancer.
| Parameters | No. | miR-375a | P-value |
|---|---|---|---|
| Total | 44 | ||
| Age (years) | 0.659 | ||
| ≤60 | 20 | 0.00175 (0.000075–0.011896) | |
| >60 | 24 | 0.00180 (0.000071–0.011022) | |
| Gender | 0.124 | ||
| Male | 30 | 0.0022 (0.000058–0.003811) | |
| Female | 14 | 0.0015 (0.000076–0.013231) | |
| Tumor size (cm) | 0.090 | ||
| ≤2 | 1 | 0.1826 | |
| >2 | 43 | 0.0017 (0.000074–0.011266) | |
| pT stage | 0.002b | ||
| T1 | 1 | 0.1826 | |
| T2 | 22 | 0.0090 (0.00078–0.013505) | |
| T3 | 21 | 0.0012 (0.000049–0.001853) | |
| T4 | 0 | - | |
| pN stage | 0.018b | ||
| N0 | 28 | 0.0066 (0.001811–0.013365) | |
| N1 | 16 | 0.000067 (0.000028–0.000077) | |
| pTNM stage | 0.001b | ||
| I | 16 | 0.0644 | |
| IA | 1 | 0.3533 | |
| IB | 15 | 0.0664 (0.041764–0.071184) | |
| II | 28 | 0.0018 | |
| IIA | 12 | 0.0178 (0.011330–0.026199) | |
| IIB | 16 | 0.0014 (0.000670–0.001585) | |
| III | 0 | - | |
| IV | 0 | - | |
| Histological grade | 0.097 | ||
| Poorly differentiated | 9 | 0.0009 (0.00025–0.006525) | |
| Moderately differentiated | 34 | 0.0018 (0.00007–0.012628) | |
| Well differentiated | 1 | 0.0259 |
Data are the median relative expression (25–75th percentile);
P<0.05.
Discussion
Due to the asymptomatic onset of pancreatic cancer, most patients are in advanced or metastatic condition at the time of diagnosis, resulting in a poor prognosis. The majority of patients diagnosed with have pancreatic cancer succumb to the disease within 12 months, and few survive 5 years after diagnosis. The poor prognosis of these patients is due to its late clinical presentation with symptoms, early and aggressive local invasion, and high metastatic potential (25).
miRNA alterations have been shown to play an important role in different steps of tumor formation and progression (26). miRNA expression has been studied by cloning and sequencing, northern blotting, in situ hybridization, microarrays, real-time PCR and other techniques. A number of studies have analyzed the global expression pattern of miRNAs in pancreatic carcinoma (23,24,27,28). However, real-time PCR has an advantage in that it is a more quantitative and more sensitive method compared to high-throughput assays.
Szafranska et al(23) have performed the first comprehensive miRNA expression profile study in tissues from normal pancreas (n=7), chronic pancreatitis (n=7), pancreatic cancer (n=10) and 33 human tissues of different non-pancreatic origin, to identify miRNA candidates with a potential for future clinical application from a pool of 377 known and novel miRNAs. Their findings demonstrated that miR-375 may be used to classify normal, chronic pancreatitis and cancerous tissues, and discriminate between neoplastic and non-neoplastic processes in pancreatic cancer. miR-375 expression was high in normal pancreas but was significantly lower in both diseased tissues and absent in cell lines. miR-375 has been previously described to be expressed in mouse pancreatic islet cells suppressing glucose-induced insulin secretion and miR-375 was recently identified as a pancreatic islet-specific miRNA regulating insulin secretion (29). Thus, it is likely, that the lower content of miR-375 in chronic pancreatitis and pancreatic cancer tissues reflects the reduced number of islet cells present in these tissues.
Therefore, we used real-time PCR to assess the expression levels of miR-375 in a large number of cases and clarified the correlation between miR-375 and clinicopathological characteristics in pancreatic carcinoma.
The non-parametric test between the relative expression levels of miR-375 in pancreatic cancer cases and its clinicopathological characteristics showed that there was no significant correlation between the low expression of miR-375 and parameters such as gender, age, tumor size, tumor location and histological grade (P>0.05). However, the low expression of miR-375 was correlated with pT stage, lymph node metastases and pTNM stage (P<0.05).
To the best of our knowledge, we performed the largest study to date that assesses the expression levels of miR-375 in pancreatic cancers by real-time PCR. The results showed that miR-375 was frequently downregulated in 44 cancer and matching adjacent non-tumor tissue pairs. The significantly reduced expression of miR-375 was found in four pancreatic cancer cell lines. Our results were consistent with previous studies on the global expression pattern of miRNAs in pancreatic carcinoma (25,28). Therefore, as the low expression of miR-375 is frequently observed in pancreatic cancers, it may be crucial in the process of carcinogenesis.
This study demonstrated that miR-375 was significantly downregulated in pancreatic cancer. Due to the resected tissues samples being obtained, according to the TNM staging system of the American Joint Committee on Cancer (AJCC; 2010) and the International Union against Cancer (UICC), there were no T4 stage patients, and only one case of T1 stage. miR-375 was mainly associated with stages T2 and T3, thus miR-375 was associated with whether the pancreatic cancer invaded the adjacent organs or vessels. miR-375 expression was correlated with pT stage, suggesting that miR-375 is involved in the carcinogenesis, development and metastasis of pancreatic carcinoma. miR-375 was also correlated with pN or lymph node metastases, thus miR-375 is crucial in the metastasis of pancreatic cancer. There was a decrease in the expression level of miR-375, but an increase in pTNM stage suggesting that miR-375 is associated with the progression and metastasis of pancreantic cancer. Findings of this study therefore suggest that miR-375 is involved in the carcinogenesis, development and metastasis of pancreatic carcinoma.
miRNA expression can be reduced by a number of factors, including transcriptional factors, mutations, deletions and methylation. However, miR-375 plays an anti-oncogene role.
Results of bioinformatic algorithms, such as Target Scan 4.2, demonstrated that 3-phosphoinositide-dependent protein kinase-1 (PDK1) is a potential target gene of miR-375. Previous studies have confirmed this role in pancreatic (30), gastric (31) and esophageal cancer (32). Various growth factors activate the phosphatidylinositol 3-kinase (PI3K) pathway, which in turn phosphorylates phosphatidylinositol-4,5-biphosphate (PIP2) to generate phosphatidylinositol-3,4,5-triphosphate (PIP3). One of the most studied signaling events controlled by PIP3 is the activation of a group of AGC family protein kinases, including isoforms of protein kinase B (Pkb/Akt) and the ribosomal S6 kinase (S6K), which play crucial roles in regulating physiological processes relevant to metabolism, cell growth, proliferation and survival (33,34). PDK1 is a PH domain-containing protein that is activated following PI3K activity, which in turn phosphorylates Akt1 at threonine 308 (or cognate locations on other isoforms) together with a large variety of other AGC kinase substrates. Although this kinase is important in PI3K-Akt-mTOR signaling, activating mutations of the gene encoding PDK1 have not been described. Findings of a recent study (35) suggest that PDK1 expression levels control the growth, proliferation and survival of developing cells in pancreatic cancer, however, this has yet to be adequately investigated. PDK1 activation is dependent primarily on cytoplasmic membrane localization, and is considered to be constitutively active. Thus, while it is unlikely that activating mutations in the kinase domain occur, it is possible that membrane-targeting PDK1 mutations may result in pathway activation (36).
PDK1 is a gene that has been identified as a direct target of miR-375 (30). PDK-1 is a key component in Akt signaling, a well-documented pathway that regulates cancer cell survival and proliferation.
The reduced expression of miR-375 has been reported in pancreatic cancer (27), hepatocellular carcinoma (37) as well as head or neck squamous cell carcinoma (38). Apart from its role in cancer, miR-375 is an important regulator in mammalian pancreatic islet-cell development and regulation of insulin secretion (39), indicating its diverse role in normal physiology. The target genes of miR-375 may function cooperatively through different cell mechanisms. PDK1 as a target of miR-375 provides new insights into the molecular networks of miR-375. However, further studies are required to investigate the targets of miR-375 that may favor the process of tumorigenesis. The introduction of a single miRNA that modulates complex downstream signals and, in turn, retards the process of tumorigenesis would be useful.
At present, the yes-associated protein (YAP) (40), JAK2 (18) and 14-3-3 ζ (30) are other genes that have been identified as a direct target of miR-375.
In conclusion, the results of this study suggest that miR-375 interferes with the PI3K-Akt-mTOR signaling. Therefore, miR-375 is a potential therapeutic target against the PI3K-Akt-mTOR signaling axis for the prevention of pancreatic cancer development and progression.
Acknowledgments
This study was supported by the Young Scientist Fund of the National Natural Science Foundation of China (no. 81201905)
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegansheterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 3.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng Y. Principles of micro-RNA production and maturation. Oncogene. 2006;25:6156–6162. doi: 10.1038/sj.onc.1209908. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 8.Zaman MS, Chen Y, Deng G, et al. The functional significance of microRNA-145 in prostate cancer. Br J Cancer. 2010;103:256–264. doi: 10.1038/sj.bjc.6605742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu S, Lu Z, Liu C, et al. miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Cancer Res. 2010;70:6015–6025. doi: 10.1158/0008-5472.CAN-09-4531. [DOI] [PubMed] [Google Scholar]
- 10.Schickel R, Park SM, Murmann AE, Peter ME. miR-200c regulates induction of apoptosis through CD95 by targeting FAP-1. Mol Cell. 2010;38:908–915. doi: 10.1016/j.molcel.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 12.Mori Y, Ishiguro H, Kuwabara Y, et al. MicroRNA-21 induces cell proliferation and invasion in esophageal squamous cell carcinoma. Mol Med Rep. 2009;2:235–239. doi: 10.3892/mmr_00000089. [DOI] [PubMed] [Google Scholar]
- 13.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 14.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 15.Ikenaga N, Ohuchida K, Mizumoto K, et al. MicroRNA-203 expression as a new prognostic marker of pancreatic adenocarcinoma. Ann Surg Oncol. 2010;17:3120–3128. doi: 10.1245/s10434-010-1188-8. [DOI] [PubMed] [Google Scholar]
- 16.Murakami Y, Yasuda T, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 17.Dai X, Chiang Y, Wang Z, et al. Expression levels of microRNA-375 in colorectal carcinoma. Mol Med Rep. 2012;5:1299–1304. doi: 10.3892/mmr.2012.815. [DOI] [PubMed] [Google Scholar]
- 18.Ding L, Xu Y, Zhang W, et al. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010;20:784–793. doi: 10.1038/cr.2010.79. [DOI] [PubMed] [Google Scholar]
- 19.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. 2004;363:1049–1057. doi: 10.1016/S0140-6736(04)15841-8. [DOI] [PubMed] [Google Scholar]
- 21.Guo X, Cui Z. Current diagnosis and treatment of pancreatic cancer in China. Pancreas. 2005;31:13–22. doi: 10.1097/01.mpa.0000168220.97967.d1. [DOI] [PubMed] [Google Scholar]
- 22.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 23.Szafranska AE, Davison TS, John J, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 24.Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 25.Park JY, Helm J, Coppola D, Kim D, Malafa M, Kim SJ. MicroRNAs in pancreatic ductal adenocarcinoma. World J Gastroenterol. 2011;17:817–827. doi: 10.3748/wjg.v17.i7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 27.Lee EJ, Gusev Y, Jiang J, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Li M, Wang H, et al. Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis. World J Surg. 2009;33:698–709. doi: 10.1007/s00268-008-9833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poy MN, Hausser J, Trajkovski M, et al. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci USA. 2009;106:5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El Ouaamari A, Baroukh N, Martens GA, Lebrun P, Pipeleers D, van Obberghen E. miR-375 targets 3′-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes. 2008;57:2708–2717. doi: 10.2337/db07-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsukamoto Y, Nakada C, Noguchi T, et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010;70:2339–2349. doi: 10.1158/0008-5472.CAN-09-2777. [DOI] [PubMed] [Google Scholar]
- 32.Li X, Lin R, Li J. Epigenetic silencing of microRNA-375 regulates PDK1 expression in esophageal cancer. Dig Dis Sci. 2011;56:2849–2856. doi: 10.1007/s10620-011-1711-1. [DOI] [PubMed] [Google Scholar]
- 33.Kozma SC, Thomas G. Regulation of cell size in growth, development and human disease: PI3K, PKB and S6K. Bioessays. 2002;24:65–71. doi: 10.1002/bies.10031. [DOI] [PubMed] [Google Scholar]
- 34.Mora A, Komander D, van Aalten DM, Alessi DR. PDK1, the master regulator of AGC kinase signal transduction. Semin Cell Dev Biol. 2004;15:161–170. doi: 10.1016/j.semcdb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 35.Westmoreland JJ, Wang Q, Bouzaffour M, Baker SJ, Sosa-Pineda B. Pdk1 activity controls proliferation, survival, and growth of developing pancreatic cells. Dev Biol. 2009;334:285–298. doi: 10.1016/j.ydbio.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storz P, Toker A. 3′-phosphoinositide-dependent kinase-1 (PDK-1) in PI 3-kinase signaling. Front Biosci. 2002;7:d886–d902. doi: 10.2741/storz. [DOI] [PubMed] [Google Scholar]
- 37.Ladeiro Y, Couchy G, Balabaud C, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 38.Avissar M, Christensen BC, Kelsey KT, Marsit CJ. MicroRNA expression ratio is predictive of head and neck squamous cell carcinoma. Clin Cancer Res. 2009;15:2850–2855. doi: 10.1158/1078-0432.CCR-08-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 40.Liu AM, Poon RT, Luk JM. MicroRNA-375 targets Hippo-signaling effector YAP in liver cancer and inhibits tumor properties. Biochem Biophys Res Commun. 2010;394:623–627. doi: 10.1016/j.bbrc.2010.03.036. [DOI] [PubMed] [Google Scholar]


