Abstract
Enterovirus 71 (EV71) often causes large outbreaks of diseases among children worldwide and its pathogenesis remains unclear. The aim of the present study was to investigate the association between interferon-inducible protein 10 (IP-10) polymorphism in children with EV71 infection. Polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) were performed to analyze the gene polymorphisms of IP-10 (−1596C/T) in 58 EV71-infected and 48 control patients. The results showed that in EV71-infected patients the frequency of carrying CT + TT genotype and T allele is 10.3 and 6.0%, respectively, which is significantly lower than that of the controls (29.2 and 15.6%, respectively). Individuals with T allele had a lower risk of EV71 infection [odds ratio (OR) = 0.35, 95% confidence interval (CI), 0.13–0.89]. The results of this study indicated that −1596T allele for the IP-10 gene may be a beneficial factor for EV71 infection.
Keywords: Enterovirus 71, interferon-inducible protein 10, polymorphism, single nucleotide
Introduction
Enterovirus 71 (EV71) infections may cause hand, foot and mouth disease (HFMD) and even severe neurologic complications and/or pulmonary edema in young children. Although understanding of EV71 infection has recently improved, EV71 pathogenesis has yet to be fully elucidated (1). The outcome of a virus infection is the result of a complex interaction of the virus, environmental factors and host genetics. The outcome of infectious diseases is strongly controlled by the host immune response, while resistance to infection in humans is, to a certain extent, genetically determined (2). Critical host responses to virus infection include the absolute levels of cytokines as well as the control of cytokine secretion such as inducibility and stability of cytokine mRNA (3). Levels and types of cytokines and chemokines released in hosts are mainly determined by gene polymorphisms. Numerous studies have focused on the association between a virus and host response. The aim of the present study was to investigate the association between IP-10 gene polymorphism and EV71 infection in children and improve our knowledge regarding EV71 pathogenesis.
Materials and methods
Study population
The study protocol was approved by the Ethics Committee of the Medical School of Qingdao University (Qingdao, China), and informed consent for the experimental use of specimens was obtained from the participants. A total of 106 patients, aged, 2–10 years, were included in this study. The patients presented to the Department of Pediatrics of the Affiliated Hospital of the Medical School of Qingdao University between May, 2010 and May, 2011. Fifty eight of the 106 participants (male/female, 38/20) were diagnosed with EV71 using reverse transcriptase-polymerase chain reaction (RT-PCR), VPl sequencing and/or virus isolation were included in the EV71 group. The control groups comprised 48 patients (male/female, 30/18) admitted to the Department of Surgery with normal blood test and no infection, allergy or recent use of antibiotics.
Reagents and instruments
Blood Genome DNA Extraction kit, Taq Polymerase, PCR buffer (with Mg2+), dNTP mixture, restriction enzyme XbaI and PCR primers were obtained from Takara Biotechnology Co., Ltd. (Dalian, China); PE2400 PCR system was purchased from Applied Biosystems (Foster City, CA, USA). BioSpectrum 300 Imaging System was obtained from UVP and used to capture gel images (Upland, CA, USA).
DNA extraction, amplification and analysis
Genomic DNA extraction of peripheral blood leukocytes was performed: 3 ml of EDTA-whole blood sample was obtained from each participant, coded and analyzed in a blind manner for genomic DNA extraction using the Blood Genome DNA Extraction kit, according to the manufacturer’s instructions. The quality of the genomic DNA was examined by measuring the optical density (OD) at wavelengths of 260 and 280 nm. The ratio of OD260/OD280 was between 1.70–1.80. The final DNA concentration was 10 ng/μl.
IP-10 genotypes were determined using PCR-restriction fragment length polymorphism (PCR-RFLP) as previously described by Humbert et al(4). The primers for IP-10 (−1595C/T) were: F, 5′GCAGATACTGTCTCAGAACCTG GTA 3′ and R, 5′TGTCACCATCTCTCATTTTGATTGT3′ (5). The PCR reactions were performed in 25 μl containing 10 ng of genomic DNA, 10X PCR buffer (with Mg2+) 2.5 μl, 2.5 mM dNTPs 2.0 μl, 0.625 units TaqDNA polymerase and 0.5 μl of each primer (10 pmol/l). Following initial denaturation at 95°C for 5 min, the PCR reaction was performed for 35 cycles consisting of 94°C for 30 sec, 57°C for 1 min and 72°C for 1 min, followed by a final extension step of 72°C for 10 min. The PCR product was digested using the restriction enzyme XbaI and then resolved on a 2% agarose gel, and visualized by UV following ethidium bromide staining.
Statistical analysis
Statistical analysis was performed using the SPSS software (version 17.0; SPSS, Inc., Chicago, IL, USA). The χ2 test was used to evaluate differences in frequency distributions of genotype and allele of the IP-10 polymorphisms between the EV71 and control groups.
The verification of the Hardy-Weinberg equilibrium of genotypes was performed using the χ2 test. The odds ratios (ORs) for the risk of EV71 infection and their 95% confidence intervals (CIs) were calculated.
Results
Characteristics of the study subjects
The genotype frequencies of the two polymorphisms among the controls were in agreement with the Hardy-Weinberg equilibrium (P>0.05) (Table I), indicating that study subjects included in the present study were representative of the target population.
Table I.
Hardy-Weinberg test of IP-10 genotype distribution.
| IP-10 genotype, n
|
||||||
|---|---|---|---|---|---|---|
| Group | N | CC | CT | TT | χ2 | P-value |
| EV71 | 58 | 52 | 5 | 1 | 3.34 | 0.07 |
| Control | 48 | 34 | 13 | 1 | 0.04 | 0.85 |
IP-10, interferon-inducible protein 10; EV71, Enterovirus 71.
Genotype distributions of IP-10 polymorphisms
The PCR product of IP-10 (−1596C/T) was 499 bp. Following XbaI digestion, the product included one single band at 499 bp (CC type), two bands at 325 and 174 bp (TT type) and three bands at 499, 325 and 174 bp (CT type) (Fig. 1). The genotype and allele frequencies of the EV71 and control groups are shown in Table II. The percentage of CT + TT genotype and the frequency of T allele in the EV71 group (10.3 and 6%, respectively) was significantly lower compared to the control group (29.2 and 15.6%, respectively). Individuals with T allele had a lower risk of EV71 infection (OR=0.35, 95% CI, 0.13–0.89).
Figure 1.
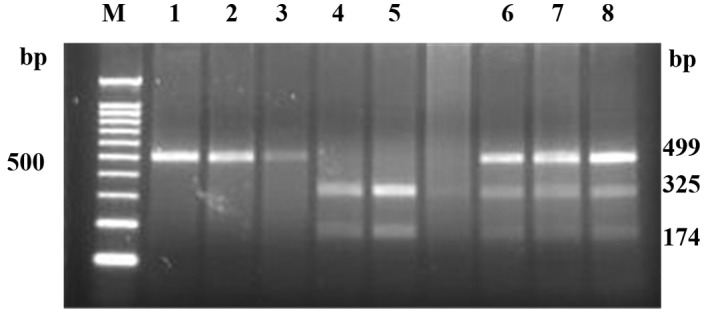
IP-10 (−1596 C/T) PCR product digestion results. M, Marker (DNA standards); lanes 1–4, EV71 groups. Lane 1, 2 and 3 are CC type; lane 4 is TT type. Lanes 5–8, control groups. Lane 5 is TT type; lanes 6, 7 and 8 are CT type.
Table II.
Genotypes and allele frequencies of EV71 and control groups.
| Genotype, n (%)
|
Allele, n (%)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Group | N | CC | CT + TT | OR | 95% CI | C | T | OR | 95% CI |
| EV71 | 58 | 52 (89.7) | 6 (10.3)a | 0.28 | 0.10–0.79 | 109 (94.0) | 7 (6.0)b | 0.35 | 0.13–0.89 |
| Control | 48 | 34 (70.8) | 14 (29.2) | 81 (84.4) | 15 (15.6) | ||||
P=0.014,
P=0.023. EV71, Enterovirus 71; OR, odds ratio; CI, confidence interval.
Discussion
Interferon-γ-inducible protein 10 (IP-10, also termed CXCL10) is one of the CXC chemokines (α subunit). Luster et al(6) first reported that IP-10, which has a relative molecular mass of 12,378, could be secreted by interferon (IFN)-γ-treated U937 cells. IP-10 has a significant amino-acid homology to platelet factor-4 (PF-4) (6). It is known to play an important role in autoimmune disease, transplantation, infection, allergic inflammatory disease and cardiovascular disease (7). Findings of previous studies have shown that the two major functions of IP-10 involve the recruiting of activated T cells into sites of tissue inflammation (8) and inhibition of angiogenesis (9). Th1 reactions depend on IP-10, which acts as a chemoattractor for monocytes/macrophages, T and NK cells, but not neutrophils. Numerous studies have shown that the chemokine CXCL10/IP-10 along with CXCL9/Mig and CXCL11/I-TAC demonstrate the ability to recruit various leukocyte subsets, the capacity to induce the proliferation of vascular pericytes as well as powerful antitumor effects. Thus, these chemokines have been suggested to be potential therapeutic targets in cancer, allograft rejection, diabetes, multiple sclerosis and autoimmune disorders of the thyroid (10). IP-10 has also been found to be associated with hepatic inflammation and to be a potential marker of treatment outcome (11). Previous studies have also shown that the CXCL10/IP-10 level is elevated in the sera and livers of progressed HBV carriers, and that the −1596T-201A haplotype is associated with higher CXCL10/IP-10 transcription in IFN-γ-stimulated peripheral blood mononuclear cells (5).
In the present study, the association between IP-10 gene polymorphism and EV71 infection in children was investigated. The results have shown that individuals with T allele have a lower risk of EV71 infection indicating that −1596T allele for IP-10 gene may be a beneficial factor for EV71 infection. Genetic susceptibility may vary due to ethnicity, complexity between virus and host as well as interaction between multiple genes and haploid mutation. Future studies may focus on screening more candidate polymorphic genes for EV71 susceptibility to help understand the genetic susceptibility and pathophysiology of EV71.
References
- 1.Khong WX, Foo DG, Trasti SL, Tan EL, Alonso S. Sustained high levels of interleukin-6 contribute to the pathogenesis of enterovirus 71 in a neonate mouse model. J Virol. 2011;85:3067–3076. doi: 10.1128/JVI.01779-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Deventer SJ. Cytokine and cytokine receptor polymorphisms in infectious disease. Intensive Care Med. 2000;26(Suppl 1):S98–S102. doi: 10.1007/s001340051125. [DOI] [PubMed] [Google Scholar]
- 3.Booy R, Nadel S, Hibberd M, Levin M, Newport MJ. Genetic influence on cytokine production in meningococcal disease. Lancet. 1997;349:1176. doi: 10.1016/s0140-6736(05)63055-3. [DOI] [PubMed] [Google Scholar]
- 4.Humbert R, Adler DA, Disteche CM, Hassett C, Omiecinski CJ, Furlong CE. The molecular basis of the human serum paraoxonase activity polymorphism. Nat Genet. 1993;3:73–76. doi: 10.1038/ng0193-73. [DOI] [PubMed] [Google Scholar]
- 5.Deng G, Zhou G, Zhang R, et al. Regulatory polymorphisms in the promoter of CXCL10 gene and disease progression in male hepatitis B virus carriers. Gastroenterology. 2008;134:716–726. doi: 10.1053/j.gastro.2007.12.044. [DOI] [PubMed] [Google Scholar]
- 6.Luster AD, Unkeless JC, Ravetch JV. Gamma-interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature. 1985;315:672–676. doi: 10.1038/315672a0. [DOI] [PubMed] [Google Scholar]
- 7.Gerard C, Rollins BJ. Chemokines and disease. Nat Immunol. 2001;2:108–115. doi: 10.1038/84209. [DOI] [PubMed] [Google Scholar]
- 8.Dufour JH, Dziejman M, Liu MT, Leung JH, Lane TE, Luster AD. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J Immunol. 2002;168:3195–3204. doi: 10.4049/jimmunol.168.7.3195. [DOI] [PubMed] [Google Scholar]
- 9.Angiolillo AL, Sgadari C, Taub DD, et al. Human interferon-inducible protein 10 is a potent inhibitor of angiogenesis in vivo. J Exp Med. 1995;182:155–162. doi: 10.1084/jem.182.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lazzeri E, Romagnani P. CXCR3-binding chemokines: novel multifunctional therapeutic targets. Curr Drug Targets Immune Endocr Metabol Disord. 2005;5:109–118. doi: 10.2174/1568008053174723. [DOI] [PubMed] [Google Scholar]
- 11.Zeremski M, Petrovic LM, Talal AH. The role of chemokines as inflammatory mediators in chronic hepatitis C virus infection. J Viral Hepat. 2007;14:675–687. doi: 10.1111/j.1365-2893.2006.00838.x. [DOI] [PubMed] [Google Scholar]


